Abstract
Resistance to anticancer therapeutics is a major global concern. Thus, new anticancer agents should be aimed against novel protein targets to effectively mitigate the increased resistance. This study evaluated the potential of secondary metabolites from a bacterial endophyte, as new anticancer agents, against a novel protein target, fibroblast growth factor. In silico genomic characterization of the Bacillus sp. strain MHSD_37 was used to identify potential genes involved in encoding secondary metabolites with biological activity. The strain was also exposed to stress and liquid chromatography–mass spectrometry used for the identification and annotation of secondary metabolites of oligopeptide class with anticancer activity. Selected metabolites were evaluated for their anticancer activity through molecular docking and Absorption, Distribution, Metabolism, Excretion and Toxicity (ADMET) properties analysis. Phylogenetic analysis revealed that strain MHSD_37 shared close evolutionary relationships with Bacillus at the species level, with no identified relationships at the sub-species level. Both in silico genomic characterization and spectrometry analysis identified secondary metabolites with potential anticancer activity. Molecular docking analysis illustrated that the metabolites formed complexes with the target protein, fibroblast growth factor, which were stabilized by hydrogen bonds. Moreover, the ADMET analysis showed that the metabolites passed the toxicity test for use as a potential drug. Thereby, Bacillus sp. strain MHSD_37 is a potential novel strain with oligopeptide metabolites that can be used as new anticancer agents against novel protein targets.
1. Introduction
The significant global prevalence of cancer can be attributed to an increase in risk factors which expose humans to carcinogens. The main risk factors include smoking, obesity, and excessive alcohol consumption [1]. The risk factors subject normal cells to biological and chemical stress that introduce genetic changes and, consequently, proliferation and disruption of normal cell growth [2]. The number of new cancer cases was estimated at 18.1 million in 2020 [3]. Lung, female breast, and prostate cancer contributed to at least 40% of the total global cases [4]. Furthermore, approximately 2 million new cases and 600,000 cancer deaths were reported in the United States. Interestingly, lung cancer contributed to 21% of cancer deaths [5].
Cancer prevalence and deaths remain significantly high despite the availability of a range of therapies for managing the disease [3,4,5]. Cancer management interventions can be pharmacological or non-pharmacological [6]. Pain is a common symptom of cancer, and, thus, non-pharmacological strategies such as physical therapy, diet, and cold/hot therapy are used for pain management [7,8]. Chemotherapy, radiation therapy, and surgery are the principal strategies in pharmacological intervention [9,10,11]. Chemotherapy drugs, however, cause severe organ toxicity, thereby limiting their administration at higher doses [12]. Moreover, the occurrence of drug resistance severely impacts the effectiveness of cancer drugs [13,14]. Thus, knowledge about the mechanisms of anticancer therapy resistance is essential for the development of novel cancer therapeutic agents.
The major mechanisms of anticancer therapy resistance are drug efflux, mutation of drug targets, interference with apoptosis and DNA replication, and drug inactivation [15,16,17]. The modification of drug targets and interference with DNA replication are the two key strategies of drug resistance employed by cancer cells at the molecular level [15,18]; the former involves the overexpression or mutation of drug targets. The overexpression and gene amplification of the HER2-specific peptide have been identified as a resistance mechanism for the anti-HER2 agent, Trastuzumab [19]. Interference in DNA replication is a result of drug-topoisomerase-II-complex-induced DNA damage, during anticancer therapy [20].
Anti-apoptosis mediated anticancer therapy resistance is a result of the suppression of apoptosis in cancer cells [21]. The B-cell lymphoma/leukemia 2 (BCL-2) family of proteins has been linked with anti-apoptotic characteristics in tumor cells [22]. Drug efflux, detoxification, and inactivation are the main cancer therapy resistance strategies that develop at the initiation of or during the treatment stage [23,24]. Drug efflux is mediated through the expression of P-glycoprotein efflux pumps [25]. P-glycoprotein is an ATP-binding transporter, involved in the generation and maintenance of colorectal cancer drug resistance [26]. The transporter is capable of the rapid removal of anticancer drugs from target tumor cells [27]. Glutathione S-transferase has been reported to be important for the development of anticancer drug resistance through detoxification [28,29].
Furthermore, the development of new cancer therapies can be achieved by targeting factors involved in the pathophysiology of cancer [30]. New drug therapies can be targeted at inhibiting the development, growth, and the spread of cancer [31]. The drugs can be targeted towards the inhibition of angiogenesis, which is crucial for the development of blood vessel to supply cancer cells and support growth thereof [32]. Cancer cells release angiogenic factors, including transforming growth factor-β (TGF-β), angiogenin, vascular endothelial growth factor (VEGF), and fibroblast growth factor (FGF) [32,33,34]. These factors are responsible for initiating the proliferation, migration, and invasion of endothelial cells within new vascular structures [35]. Therefore, agents that target these factors will prevent the proliferation and migration of cancer cells and subsequent invasion of healthy cells.
DNA transcription is important for the growth, survival, invasion, metastasis, angiogenesis, and apoptosis of tumor cells [36,37,38]. Therefore, replication and transcriptional factors are potential targets of new cancer therapeutics [39]. The therapies can be targeted towards cyclic-dependent kinase and RNA polymerases [40]. Enzymes are another important group of potential targets for the development of new cancer therapeutics [41,42]. Aromatase enzyme is a potential target for breast cancer therapy. Aromatase is involved in the synthesis of estrogens which is responsible for the growth of breast cancer cells [43]. In addition, Protein kinase C is another key enzyme in tumor cells, with roles in the cell cycle, cell division, differentiation, and proliferation [44,45].
Therefore, this study will explore the potential of secondary metabolites, of bacterial endophyte origin, as anticancer agents. Bacterial endophytes are a rich source of secondary metabolites of biological importance due their existence in harsh environments [46]. Secondary metabolites were identified with liquid chromatography–mass spectrometry (LC–MS) following the exposure of the bacterial endophyte, Bacillus sp. strain MHSD_37, to stress. The metabolites identified were screened against FGF, using computational analysis, to gain insight into their therapeutic potential as new anticancer agents.
2. Materials and Methods
2.1. Plant Material Collection and Bacterial Endophyte Isolate
Leaves from Solanum nigrum, a medicinal plant, were collected from a sandy location in Botlokwa, Ga-Ramatšowe, Limpopo Province, South Africa (−23.491054, 29.746048), in March 2017. The leaves were stored in a sterile polyethylene bag and transported to the laboratory at a temperature of 4 °C. The identification of the plant material was performed at the University of Johannesburg Herbarium (JRAU) and a sample of the plant material specimen was subsequently deposited in JRAU with voucher specimen number Serepa-Dlamini 209 and species name Solanum nigrum. The remainder of the leaves were immediately processed in the laboratory and the bacterial endophytes were isolated sequentially by washing the leaves in water for 1 min, 70% ethanol for 1.5 min, 1% sodium hypochlorite (NaOCl, Merck, Saint Louis, MO, USA) for 3 min, and a final wash in sterile distilled water three times. The water from the final wash was plated as a negative control. The surface sterilized leaves were ground in 2 mL of saline using a sterile pestle and mortar and the resultant homogenate was streaked onto nutrient agar (NA, Becton Dickson, Franklin Lakes, NJ, USA) plates under sterile conditions. Bacterial growth was monitored daily after incubating the plates for 24–48 h at 28 °C. The grown colonies were re-cultured three times in NA and under the conditions mentioned above to get pure colonies with uniform morphology. A 30% glycerol (glycerol diluted in sterile distilled water (v/v), Merck, Saint Louis, MO, USA) stock culture was prepared for each bacterial endophyte and stored at −80 °C for future use.
2.2. Bacterial Strain Maintenance
A 30% glycerol (Merck, Saint Louis, MO, USA) stock of the bacterial cultures was plated on NA (Becton Dickson, Franklin Lakes, NJ, USA) plates and incubated for 24 h at 28 °C for routine culture maintenance. The bacteria were grown on nutrient broth (NB, Becton Dickson, Franklin Lakes, NJ, USA) at 28 °C, agitating at 150 rpm for 24 h.
2.3. Genome Extraction, Library Preparation, and Sequencing
The NucleoSpin microbial DNA extraction kit (Macherey-Nagel, Düren, Germany) was used to extract the genomic DNA from the solid colonies as per the manufacturer’s protocol. The DNA was sequenced at the Biotechnology Platform, Agricultural Research Council, Onderstepoort, South Africa, a commercial service provider. Paired-end libraries (2 × 150 bp) were generated using the MGIEasy Universal DNA Library preparation kit (MGI Tech Co., Guangdong, China), and the sequencing was performed on the MGIEasy® platform.
2.4. Genome Assembly and Annotation
Genome quality control, trimming, and assembly were performed on GALAXY, accessible from https://usegalaxy.org/ (accessed on 15 July 2023) [47]. FastQC (v 0.72.0) was used for the quality control of raw sequence reads which were then trimmed with the Trimmomatic program (version 0.38.0) [48]. The reads were de novo assembled using Unicycler (v 0.4.8.0) [49], and the quality was assessed with Quast (Galaxy v 5.0.2) [50]. The draft genome was annotated using the National Center for Biotechnology Information—Prokaryotic Genome Annotation Pipeline (PGAP) [51] and Rapid Annotations using Subsystems Technology (RAST accessed on 21 July 2023) [52].
2.5. Phylogenetic Analysis
A whole genome-based taxonomic analysis was done using a free bioinformatics platform, Type Strain Genome Server (TYGS), accessible from https://tygs.dsmz.de (accessed on 21 July 2023) [53]. A pairwise comparison among a set of genomes was performed with the Genome Blast Distance Phylogeny, and accurate intergenomic distances were inferred under the algorithm trimming and distance formula d2 [54]. The average nucleotide identity (ANI) values between the strain and closely related species were calculated with Orthologous Average Nucleotide Identity Tool (OAT) software (Version 0.90) [55]. The Genome-to-Genome Distance Calculator was accessed from https://ggdc.dsmz.de/ (accessed on 21 July 2023) [53].
2.6. Liquid Chromatography–Mass Spectrometry Analysis (LC–MS)
The bacterial excretome, following exposure to lead (Pb), was analyzed with a liquid chromatography-quadrupole time-of-flight tandem mass spectrometer (LC–MS-9030 q-TOF, Shimadzu Corporation, Kyoto, Japan) fitted with a Shim-pack Velox C18 column (100 mm × 2.1 mm with particle size of 2.7 m). The column oven temperature was maintained at 50 °C. The injection volume was 5 µL, and the samples were analytically separated over a 30 min binary gradient. A constant flow rate of 0.04 mL/min was applied using a binary solvent mixture of water with 0.1% formic acid (Eluent AMerck, Saint Louis, MO, USA) and 0.1% formic acid in acetonitrile (Eluent B, Merck, Saint Louis, MO, USA). The gradient technique was gradually increased from 3 to 30 min to facilitate the separation of the compounds within the samples. Eluent B was kept at 5% from 0 to 3 min, gradually increased from 5 to 40% between 3 and 5 min, and finally increased to 40–95% between 5- and 23-min. Eluent B was subsequently kept isocratic at 95% between 23 and 25 min. The gradient was returned to original conditions of 5% at 25–27 min, and re-equilibration at 5% occurred at 27–30 min. The liquid chromatographic eluents were subsequently subjected to a Quadruple Time-of-Flight high-definition mass spectrometer for analysis in positive electrospray ionization (ESI) mode with the following conditions: 400 °C heat block temperature, 250 °C Desolvation Line (DL) temperature, 42 °C flight tube temperature, and 3 L/min nebulization and dry gas flow. The data was acquired using the Data-dependent acquisition (DDA) mode, which simultaneously generated MS1 and MS2 data for all ions within a mass-to-charge ratio (m/z) range of 100–1500 (precursor m/z isolation window) and an intensity threshold above 5000. The MS2 Experiments were conducted utilizing argon gas as the collision gas and a collision energy of 35 eV with a spread of 5 and sodium iodide (Merck, Saint Louis, MO, USA) as a calibration solution to monitor high mass precision. Metabolite annotation was completed at Metabolomics Standards Initiative (MSI) levels 2 and 3. The former is based on the retention time, mass-to-charge ratio (m/z), and fragmentation patterns matching data available from the databases in Sirius [56,57,58]. The fragments with no matches to anything on the databases were classified according to their compound class according to the molecular networking from Canopus on Sirius [57,58]. Literature data was subsequently used to identify the biological activities of the annotated metabolites.
2.7. Secondary Metabolites and Proteins Selection
Four secondary metabolites belonging to the oligopeptides class were selected based on their potential identified anticancer activity, from literature data, and lack of an associated patent. The simple data forma (SDF) files for the 3-D structures of the selected metabolites of the oligopeptide class were retrieved from PubChem (https://pubchem.ncbi.nlm.nih.gov/, accessed on 18 December 2023) and named metabolite 1–4, and their PubChem IDs were also used to differentiate among the 4 metabolites. The collected structures of the metabolites were further optimized using Avogadro. The crystallography structure of the FGF protein target (PDB: 1CVS) was retrieved from the protein data bank (PDB) database (www.rcsb.org, accessed on 18 December 2023).
2.8. Molecular Docking
Molecular docking studies were carried out to estimate the binding energies of the metabolites towards the therapeutic protein targets, FGF, by using the computational program AutoDock Vina 1.1.2 [59]. The protein’s 3-D structure was modelled using ChimeraX _(Version 1.7) to generate a fine structure. The non-standard residues from the target protein were removed. The SDF and PDB files of the metabolites and the target protein were converted into a PDBQT format with the AutoDock (Version 1.5.7) tools. A grid box was created for the random docking of the protein. The grid box dimensions, spacing, and the grid map coordinate center for the random docking of the target protein were 60 Å × 60 Å × 60 Å, 0.375 Å, and x = 67.596, y = 23.207, and z = 118.874, respectively. Molecular docking analysis was performed with the Lamarckian genetic algorithm (LGA), and the docked structures were analyzed by using ChimeraX.
2.9. Absorption, Distribution, Metabolism, Elimination, and Toxicity (ADMET) Analysis
The four metabolites used in the dock studies were screened for their absorption, distribution, metabolism, elimination, and toxicity (ADMET) using the online tool http://biosig.unimelb.edu.au/pkcsm/prediction (accessed on 3 January 2024) to predict their important pharmacokinetic properties. The ADMET properties included absorption, human intestinal absorption, water solubility, Caco-2 permeability, P-glycoprotein substrate, P-glycoprotein I and II inhibitors, skin permeability, and distribution: steady state volume of distribution (VDss), fraction unbound, blood-brain barrier (BBB) permeability, and minnow toxicity [60].
3. Results
3.1. Phylogeny Characterisation of Strain MHSD_37 and Secondary Metabolite Biosynthesis Gene Clusters Analysis
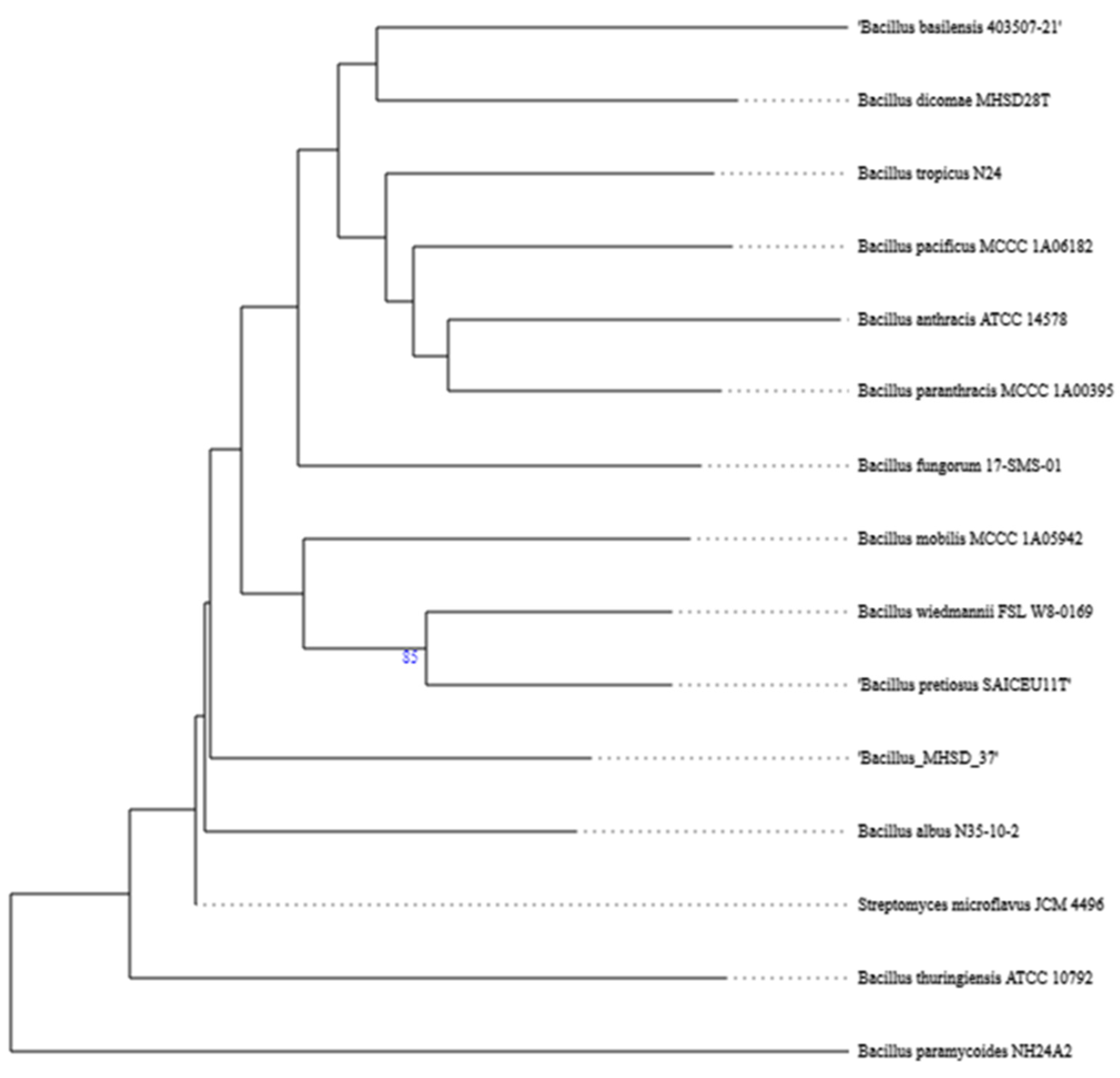
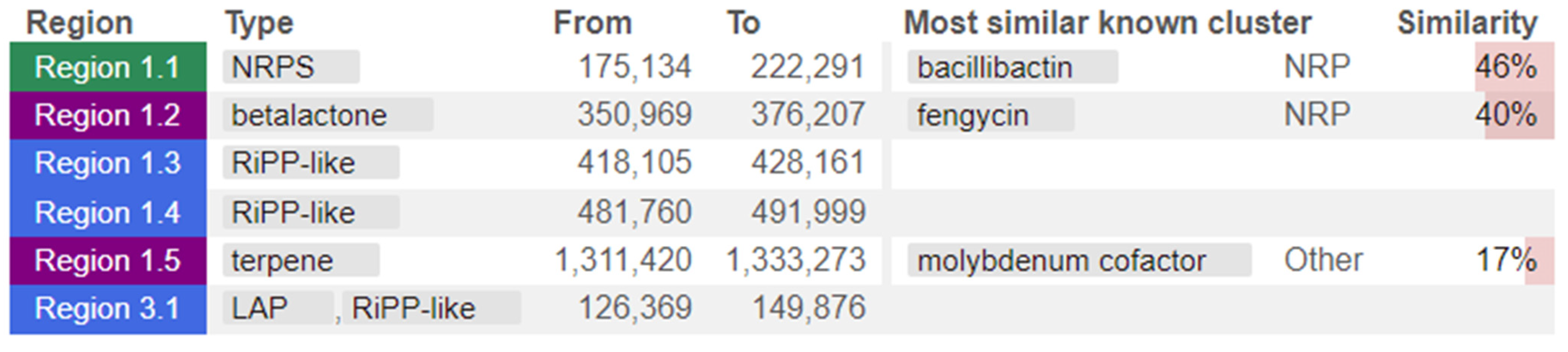
The whole genome of strain MHSD_37 was subjected to taxonomic analysis to determine the strain’s evolutionary descendancy. The evolutionary relationships of strain MHSD_37 are illustrated in the evolutionary tree in Figure 1. MHSD_37 shares the most recent common ancestor with a range of Bacillus sp. and is closely related to Bacillus albus N35-10-2 at the species level, thus it had the highest digital DNA–DNA hybridization (dDDH) with the strain. The strain was, however, not closely related to any of the Bacilli at the sub-species level, according to the evolutionary tree, which implies that strain MHSD_37 observed ANI values were below the species boundary value (ANI > 95–96%). Thus, strain MHSD_37 is a potential novel Bacillus sp. The antiSMASH analysis identified three regions which encode for the synthesis of secondary metabolites (Figure 2). Two of the regions encoded for metabolites with at least 40% similarity to bacillibactin and fengycin.
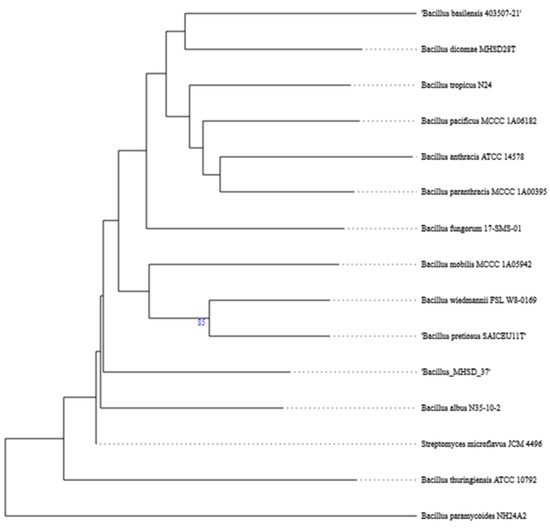
Figure 1.
Phylogenetic tree illustrating the evolutionary relationships of strain MHSD_37.
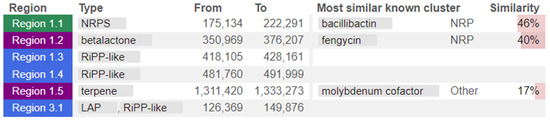
Figure 2.
Strain MHSD_37 genome annotated regions encoding for secondary metabolites. The colors illustrate regions from the different class of secondary metabolites.
3.2. Identification and Annotation of Secondary Metabolites Using LC–MS
The untargeted metabolomics approach was applied to identifying the range of secondary metabolites synthesized and secreted by strain MHSD_37 when exposed to stress in the form of the toxic heavy metal Pb. Table 1 is a summary of biological active secondary metabolites selected from the global view profiled through untargeted metabolomics. Strain MHSD_37 synthesized secondary metabolites with antimalarial, antiviral, antibacterial, and anticancer activity (Table 1). The annotated metabolites belong to the classes: diterpenoids, terpene glycosides, alpha amino acids, and oligopeptides. Interestingly, the strain synthesized a notable number of oligopeptides with anticancer activity (Table 1).

Table 1.
Annotated secondary metabolites identified from the excretome of strain MHSD_37.
3.3. In Silico Analysis of the Anticancer Potential of Secondary Metabolites from MHSD_37
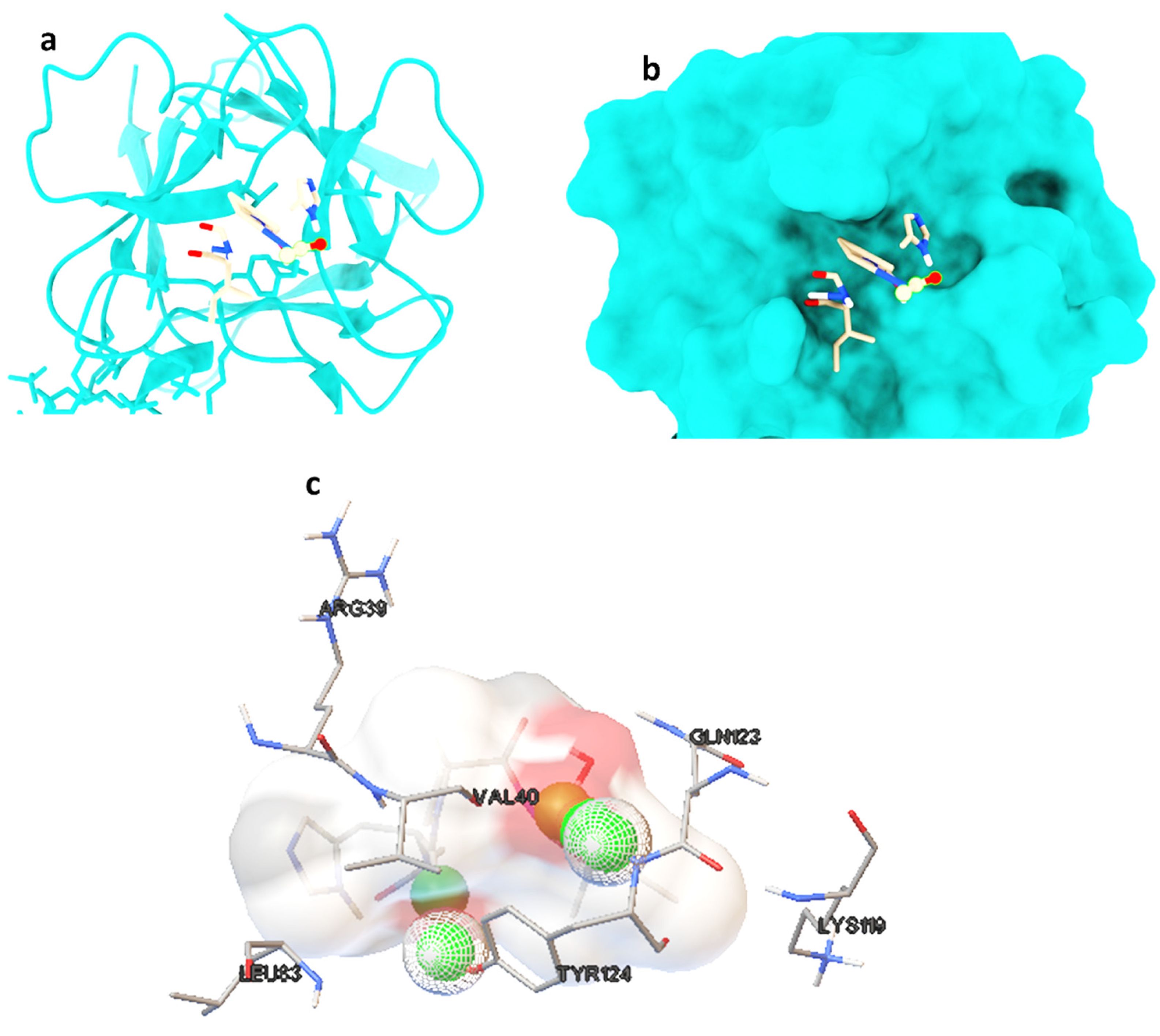
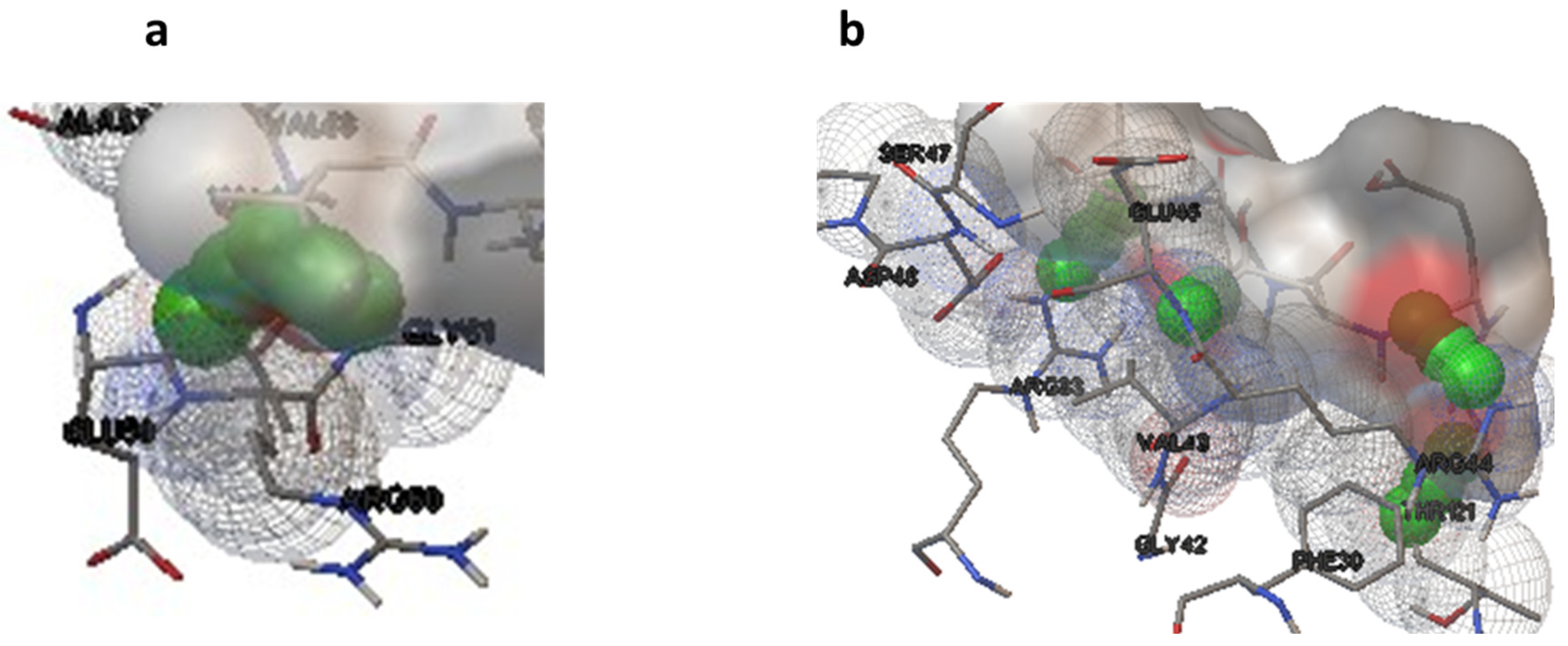
Molecular docking studies were performed on selected oligopeptides identified to have potential anticancer but with no known annotated identity or patent. A total of four oligopeptides were identified to have potential anticancer activity (Table 1). Their 3-D structures were obtained from PubChem and subsequently used for screening against the protein target, FGF. The screening was achieved through random molecular docking using the computational program AutoDock. Table 2 is a summary of the docking scores of the metabolites and their important molecular interactions with FGF. The metabolites formed molecular interactions dominated by hydrogen bonds to stabilize the interaction complex with the protein target. Metabolite 1 and 3 had the highest binding energy of −3.13 and −3.46 kcal/mol (Table 2), respectively. Figure 3 is an illustration of a 3-D best complex between the ligand and FGF (Figure 3a), docked pose of the ligand with FGF (Figure 3b), and hydrogen bond between the ligand and FGF in the best conformation (Figure 3c). Metabolite 1 formed a hydrogen bond with TYR 124 (TYR 124:HN and TYR 124:HH) as illustrated in Figure 3c. Metabolite 3 was also stabilized by a single hydrogen bond, LEU 98:HN, at its best conformation (Figure 3c). Metabolite 4 had the lowest binding energy of −1.06 kcal/mol (Table 2). The metabolite had two hydrogen bonds, THR 1:H1 and GLY 61:HN, with the protein target FGF (Figure 4a). Metabolite 2, which had the second lowest binding energy (−1.34 kcal/mol), had three hydrogen bonds, ARG 33:H, ARG33:HH1, and GLU 45:HN, with the protein target (Figure 4b).

Table 2.
Binding energies and important interactions for the best complex of metabolites.
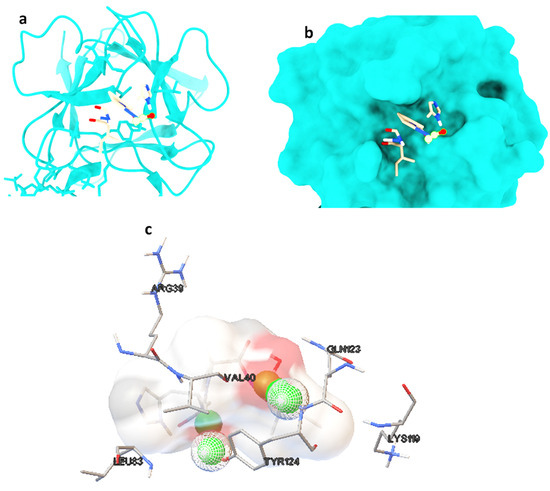
Figure 3.
The best ligand-target protein complex (a), ligand docked pose of metabolite 1 with FGF (b), and hydrogen bond between the ligand and FGF in the best conformation (c). The hydrogen bonds are represented by the green balls.
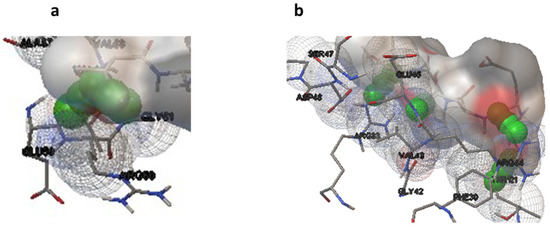
Figure 4.
Hydrogen bonds for the best conformation of metabolite 4 (a) and 2 (b). The hydrogen bonds are represented by the green balls.
3.4. ADMET Screening of the Secondary Metabolites
The metabolites were further screened for their therapeutic suitability, based on their pharmacokinetic profiles and toxicity properties, using in silico ADMET analysis. Table 3 is a summary of the ADMET properties for the four secondary metabolites. All four metabolites had an intestinal absorption of less than 30%, with metabolite 1 having the highest absorption of 27% (Table 3). The metabolites also had low Caco 2 permeability, ranging between −2.811 and −3.491. The predicted T. pyriformis toxicity was 0.285 log mM for all four metabolites. The minnow toxicity ranged between 3.444 and 7.633 log mM for the metabolites (Table 3).

Table 3.
ADMET properties for the metabolites from strain MHSD_37 with potential anticancer activity against FGF.
4. Discussion
Cancer remains a prevalent disease and predominant cause of death globally, despite the availability of a wide array of therapies [3,4,5]. Cancer cells have developed resistant mechanisms which have had a significant impact on the effectiveness of cancer therapeutics [14,15,16,17]. New cancer therapeutics should, thus, be focused on novel protein targets. A case in point is FGF, because of the protein’s involvement in tumor cell growth and spread [35]. On the other hand, bacterial endophytes are a rich source of biologically active secondary metabolites with potential anticancer activity [58,59]. Bacterial endophytes thus are an important source in the continued search of new cancer therapeutics targeting novel target proteins. Furthermore, bioinformatics and computational biology offer cost effective and efficient approaches for the screening of novel anticancer agents.
This study applied whole genome sequencing and assembly to identify and characterize a bacterial endophyte isolated from the leaves of Solanum nigrum. The plant is widely found in wastelands due to its resilience and is used in traditional medicine for the treatment of tumors, asthma, inflammation, bacterial, and viral infections [90]. De novo sequencing and assembly provide researchers with tools to identify and characterize bacteria at the sub-species level [91]. The isolated strain was a Bacillus species with endophytic characteristics. The strain was, however, not closely related to any known Bacillus at the sub-species level, thereby illustrating that it is a potentially novel subspecies of Bacillus [92].
The genome annotation from antiSmash identified three regions encoding for secondary metabolites. Two of the regions encoded for secondary metabolites with a significant similarity to bacillibactin and fengycin, which have been reportedly found to be predominant in Bacillus [86,87,93]. The former is a siderophore with antimicrobial activity and is also widely applied as a biocontrol agent [93]. Fengycin is an antimicrobial lipopeptide, has been shown to have fungicidal activity, and has potential in agricultural applications [54]. Bacterial endophytes have been shown to produce bacillibactin and fengycin as defense mechanisms against competing microbes, thereby enabling them to maintain their dominance and establish a symbiotic relationship with the host [86,93].
The LC–MS analysis further established the existence of a wide range of secondary metabolites. The metabolites of oligopeptide class were identified as having potential anticancer activity. Oligopeptides have been successfully evaluated for their anticancer activity [94,95]. An oligopeptide isolated from Anthopleura anjunae was reported to induce apoptosis in prostate cancer cells, with the apoptotic cells showing an increase in Pro-apoptotic proteins, such as Bax, cytochrome-C, caspase-3, and caspase-9 [94]. A cationic peptide was reported to have dual potential as a drug delivery system for targeted therapy and an anticancer therapeutic agent [95].
The protein ligand interactions illustrated that the metabolites and the target protein, FGF, formed hydrogen bonds to stabilize the protein-ligand complexes. The binding energies of the complexes ranged between −1.06 and −3.46 kcal/mol. Binding affinities of between −3.51 and −8.42 Kcal/mol have been reported for metabolites from Ficus carica, with a range of anticancer target proteins [96]. In addition to efficacy, safety is also an important factor for successful drug development; therefore, ADMET properties analysis is crucial during drug discovery and development [97]. Although the secondary metabolites from MHSD_37 had poor intestinal solubility and Caco 2 permeability, they showed low toxicity. The poor solubilities are, however, not a major hindrance because the most important and challenging issue is the delivery to the tumor site and the maintenance of low toxicity. Furthermore, more emphasis should be placed on enhancing the production cost, selectivity, and proteolytic stability [98].
5. Conclusions
The bacterial endophyte, Bacillus sp. strain MHSD_37 is a rich source of secondary metabolites with a range of biological activities including anticancer, antimicrobial, and antiviral activities. The strain synthesizes metabolites of the oligopeptide class with potential anticancer activity. Molecular docking studies, based on oligopeptides with no patents, showed that the strain synthesized oligopeptides which could target the FGF protein, an important growth factor involved in anticancer drug resistance. Moreover, the ADMET analysis illustrated that the oligopeptides had low toxicity. Therefore, strain MHSD-37 is a promising source of secondary metabolites for the development of novel anticancer agents.
Author Contributions
Conceptualization, P.M. and M.H.S.-D.; methodology, P.M. and M.H.S.-D.; formal analysis, P.M. and M.H.S.-D.; resources, M.H.S.-D. and P.M.; data curation, P.M.; writing—original draft preparation, P.M.; writing—review and editing, P.M. and M.H.S.-D.; visualization, P.M.; project administration, M.H.S.-D.; funding acquisition, M.H.S.-D. All authors have read and agreed to the published version of the manuscript.
Funding
The authors would like to acknowledge the South African National Research foundation (NRF), Thuthuka grant no. TTK210216586709 and Postdoctoral grant no. PSTD2204082662, for the financial support for this study.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data from the Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession JAVBIS000000000, BioSample accession number SAMN36845528, and BioProject accession number PRJNA1002565. The version described in this paper is JAVBIS000000000.
Acknowledgments
The work was supported by the National Research Foundation South Africa and the University of Johannesburg Global Excellence and Stature Fellowship.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Zhang, Z.; Zhou, L.; Xie, N.; Nice, E.C.; Zhang, T.; Cui, Y.; Huang, C. Overcoming cancer therapeutic bottleneck by drug repurposing. Signal Transduct. Target. Ther. 2020, 5, 113. [Google Scholar] [CrossRef]
- Gusev, E.Y.; Zotova, N.V. Cellular stress and general pathological processes. Curr. Pharm. Des. 2019, 25, 251–297. [Google Scholar] [CrossRef]
- Available online: https://www.wcrf.org/cancer-trends/worldwide-cancer-data/ (accessed on 25 January 2024).
- Available online: https://www.cancerresearchuk.org/health-professional/cancer-statistics/worldwide-cancer/incidence#heading-Zero (accessed on 25 January 2024).
- Available online: https://seer.cancer.gov/statfacts/html/common.html (accessed on 25 January 2024).
- Chan, C.W.; Tai, D.; Kwong, S.; Chow, K.M.; Chan, D.N.; Law, B.M. The effects of pharmacological and non-pharmacological interventions on symptom management and quality of life among breast cancer survivors undergoing adjuvant endocrine therapy: A systematic review. Int. J. Environ. Res. 2020, 17, 2950. [Google Scholar] [CrossRef]
- Singh, P.; Chaturvedi, A. Complementary and alternative medicine in cancer pain management: A systematic review. Indian J. Palliat. Care. 2015, 21, 105. [Google Scholar] [CrossRef]
- Gowan, J.; Roller, L. Disease state management: Anticholinergic medicines-practice considerations and alternatives. AJP Aust. J. Pharm. 2022, 103, 76–84. [Google Scholar]
- Wang, X.; Zhang, H.; Chen, X. Drug resistance and combating drug resistance in cancer. Cancer Drug Resist. 2019, 2, 141. [Google Scholar] [CrossRef]
- Song, G.; Cheng, L.; Chao, Y.; Yang, K.; Liu, Z. Emerging nanotechnology and advanced materials for cancer radiation therapy. Adv. Mater. 2017, 29, 1700996. [Google Scholar] [CrossRef]
- Sharma, P.; Mehta, M.; Dhanjal, D.S.; Kaur, S.; Gupta, G.; Singh, H.; Thangavelu, L.; Rajeshkumar, S.; Tambuwala, M.; Bakshi, H.A.; et al. Emerging trends in the novel drug delivery approaches for the treatment of lung cancer. Chem. Biol. Interact. 2019, 309, 108720. [Google Scholar] [CrossRef]
- Oun, R.; Moussa, Y.E.; Wheate, N.J. The side effects of platinum-based chemotherapy drugs: A review for chemists. Dalton. Trans. 2018, 47, 6645–6653. [Google Scholar] [CrossRef] [PubMed]
- Cai, M.; Song, X.; Li, X.; Chen, M.; Guo, J.; Yang, D.; Chen, Z.; Zhao, S. Current therapy and drug resistance in metastatic castration-resistant prostate cancer. Drug Resist. Updates 2023, 68, 100962. [Google Scholar] [CrossRef] [PubMed]
- Haider, T.; Pandey, V.; Banjare, N.; Gupta, P.N.; Soni, V. Drug resistance in cancer: Mechanisms and tackling strategies. Pharmacol. Rep. 2023, 72, 1125–1151. [Google Scholar] [CrossRef]
- Mansoori, B.; Mohammadi, A.; Davudian, S.; Shirjang, S.; Baradaran, B. The different mechanisms of cancer drug resistance: A brief review. Adv. Pharm. Bull. 2017, 7, 339. [Google Scholar] [CrossRef]
- Nikolaou, M.; Pavlopoulou, A.; Georgakilas, A.G.; Kyrodimos, E. The challenge of drug resistance in cancer treatment: A current overview. Clin. Exp. Metastasis 2018, 35, 309–318. [Google Scholar] [CrossRef]
- Nussinov, R.; Tsai, C.J.; Jang, H. Anticancer drug resistance: An update and perspective. Drug Resist. Updates 2021, 59, 00796. [Google Scholar] [CrossRef]
- Chen, Y.; Zhang, Y. Application of the CRISPR/Cas9 system to drug resistance in breast cancer. Adv. Sci. 2018, 5, 1700964. [Google Scholar] [CrossRef]
- Morales-Cruz, M.; Delgado, Y.; Castillo, B.; Figueroa, C.M.; Molina, A.M.; Torres, A.; Milián, M.; Griebenow, K. Smart targeting to improve cancer therapeutics. Drug Des. Dev. Ther. 2019, 13, 3753–3772. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, J.; Weng, Z.; Li, Q.; Zhao, L.; Yu, N.; Deng, L.; Xu, W.; Yang, Y.; Zhu, Z.; et al. A new anti-HER2 antibody that enhances the anti-tumor efficacy of trastuzumab and pertuzumab with a distinct mechanism of action. Mol. Immunol. 2020, 119, 48–58. [Google Scholar] [CrossRef]
- Dogan, E.; Kara, H.G.; Kosova, B.; Cetintas, V.B. Targeting Apoptosis to Overcome Chemotherapy Resistance; Exon Publications: Brisbane City, QLD, Australia, 2022; pp. 163–180. [Google Scholar]
- Davids, M.S.; Letai, A. Targeting the B-cell lymphoma/leukemia 2 family in cancer. J. Clin. Oncol. 2012, 30, 3127. [Google Scholar] [CrossRef] [PubMed]
- Pan, S.T.; Li, Z.L.; He, Z.X.; Qiu, J.X.; Zhou, S.F. Molecular mechanisms for tumour resistance to chemotherapy. Clin. Exp. Pharmacol. Physiol. 2016, 43, 723–737. [Google Scholar] [CrossRef] [PubMed]
- Vaidya, F.U.; Sufiyan Chhipa, A.; Mishra, V.; Gupta, V.K.; Rawat, S.G.; Kumar, A.; Pathak, C. Molecular and cellular paradigms of multidrug resistance in cancer. Cancer Rep. 2022, 5, 1291. [Google Scholar] [CrossRef] [PubMed]
- Sajid, A.; Lusvarghi, S.; Murakami, M.; Chufan, E.E.; Abel, B.; Gottesman, M.M.; Durell, S.R.; Ambudkar, S.V. Reversing the direction of drug transport mediated by the human multidrug transporter P-glycoprotein. Proc. Natl. Acad. Sci. USA 2020, 117, 29609–29617. [Google Scholar] [CrossRef]
- Goebel, J.; Chmielewski, J.; Hrycyna, C.A. The roles of the human ATP-binding cassette transporters P-glycoprotein and ABCG2 in multidrug resistance in cancer and at endogenous sites: Future opportunities for structure-based drug design of inhibitors. Cancer Drug Resist. 2021, 4, 784. [Google Scholar] [CrossRef]
- Robinson, K.; Tiriveedhi, V. Perplexing role of P-glycoprotein in tumor microenvironment. Front. Oncol. 2020, 10, 265. [Google Scholar] [CrossRef]
- Pal, D.; Rai, A.; Checker, R.; Patwardhan, R.S.; Singh, B.; Sharma, D.; Sandur, S.K. Role of protein S-Glutathionylation in cancer progression and development of resistance to anti-cancer drugs. Arch. Biochem. Biophys. 2021, 704, 108890. [Google Scholar] [CrossRef]
- Lv, N.; Huang, C.; Huang, H.; Dong, Z.; Chen, X.; Lu, C.; Zhang, Y. Overexpression of Glutathione S-Transferases in Human Diseases: Drug Targets and Therapeutic Implications. Antioxidants 2023, 12, 1970. [Google Scholar] [CrossRef] [PubMed]
- Zhong, L.; Li, Y.; Xiong, L.; Wang, W.; Wu, M.; Yuan, T.; Yang, W.; Tian, C.; Miao, Z.; Wang, T.; et al. Small molecules in targeted cancer therapy: Advances, challenges, and future perspectives. Signal Transduct. Target. Ther. 2021, 6, 201. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Huang, Q.; Xiao, W.; Zhao, Y.; Pi, J.; Xu, H.; Zhao, H.; Xu, J.; Evans, C.E.; Jin, H. Advances in anti-tumor treatments targeting the CD47/SIRPα axis. Front. Immunol. 2020, 11, 18. [Google Scholar] [CrossRef] [PubMed]
- Badodekar, N.; Sharma, A.; Patil, V.; Telang, G.; Sharma, R.; Patil, S.; Vyas, N.; Somasundaram, I. Angiogenesis induction in breast cancer: A paracrine paradigm. Cell Biochem. Funct. 2021, 39, 860–873. [Google Scholar] [CrossRef] [PubMed]
- Elebiyo, T.C.; Rotimi, D.; Evbuomwan, I.O.; Maimako, R.F.; Iyobhebhe, M.; Ojo, O.A.; Oluba, O.M.; Adeyemi, O.S. Reassessing vascular endothelial growth factor (VEGF) in anti-angiogenic cancer therapy. Cancer Treat. Res. Commun. 2022, 32, 100620. [Google Scholar] [CrossRef]
- Zhang, T.; Wang, X.F.; Wang, Z.C.; Lou, D.; Fang, Q.Q.; Hu, Y.Y.; Zhao, W.Y.; Zhang, L.Y.; Wu, L.H.; Tan, W.Q. Current potential therapeutic strategies targeting the TGF-β/Smad signaling pathway to attenuate keloid and hypertrophic scar formation. Biomed. Pharmacother. 2020, 129, 110287. [Google Scholar] [CrossRef] [PubMed]
- Stephen, N.M.; Deepika, U.R.; Maradagi, T.; Sugawara, T.; Hirata, T.; Ganesan, P. Insight on the cellular and molecular basis of blood vessel formation: A specific focus on tumor targets and therapy. MedComm–Oncol. 2023, 2, 22. [Google Scholar] [CrossRef]
- Usman, S.; Waseem, N.H.; Nguyen TK, N.; Mohsin, S.; Jamal, A.; Teh, M.T.; Waseem, A. Vimentin is at the heart of epithelial mesenchymal transition (EMT) mediated metastasis. Cancers 2021, 13, 4985. [Google Scholar] [CrossRef] [PubMed]
- Stabrauskiene, J.; Kopustinskiene, D.M.; Lazauskas, R.; Bernatoniene, J. Naringin and naringenin: Their mechanisms of action and the potential anticancer activities. Biomedicines 2022, 10, 1686. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.H.; Li, M.Y.; Wang, Z.; Zuo, H.X.; Wang, J.Y.; Xing, Y.; Jin, C.; Xu, G.; Piao, L.; Piao, H.; et al. Convallatoxin promotes apoptosis and inhibits proliferation and angiogenesis through crosstalk between JAK2/STAT3 (T705) and mTOR/STAT3 (S727) signaling pathways in colorectal cancer. Phytomedicine 2020, 68, 153172. [Google Scholar] [CrossRef] [PubMed]
- Labrie, M.; Brugge, J.S.; Mills, G.B.; Zervantonakis, I.K. Therapy resistance: Opportunities created by adaptive responses to targeted therapies in cancer. Nat. Rev. Cancer 2022, 22, 323–339. [Google Scholar] [CrossRef]
- Kifle, Z.D.; Tadele, M.; Alemu, E.; Gedamu, T.; Ayele, A.G. A recent development of new therapeutic agents and novel drug targets for cancer treatment. SAGE Open Med. 2021, 9, 20503121211067083. [Google Scholar] [CrossRef]
- Marak, B.N.; Dowarah, J.; Khiangte, L.; Singh, V.P. A comprehensive insight on the recent development of cyclic dependent kinase inhibitors as anticancer agents. Eur. J. Med. Chem. 2020, 203, 112571. [Google Scholar] [CrossRef]
- Maity, T.K.; Kim, E.Y.; Cultraro, C.M.; Venugopalan, A.; Khare, L.; Poddutoori, R.; Marappan, S.; Syed, S.D.; Telford, W.G.; Samajdar, S.; et al. Novel CDK12/13 Inhibitors AU-15506 and AU-16770 Are Potent Anti-Cancer Agents in EGFR Mutant Lung Adenocarcinoma with and without Osimertinib Resistance. Cancers 2023, 15, 2263. [Google Scholar] [CrossRef]
- Molehin, D.; Filleur, S.; Pruitt, K. Regulation of aromatase expression: Potential therapeutic insight into breast cancer treatment. Mol. Cell Endocrinol. 2021, 531, 111321. [Google Scholar] [CrossRef]
- Yang, Y.; Ye, W.L.; Zhang, R.N.; He, X.S.; Wang, J.R.; Liu, Y.X.; Wang, Y.; Yang, X.M.; Zhang, Y.J.; Gan, W.J. The role of TGF-β signaling pathways in cancer and its potential as a therapeutic target. Evid. Based. Complement. Altern. Med. 2021, 2021, 15–31. [Google Scholar] [CrossRef]
- Erkisa, M.; Sariman, M.; Geyik, O.G.; Geyik, C.; Stanojkovic, T.; Ulukaya, E. Natural products as a promising therapeutic strategy to target cancer stem cells. Curr. Med. Chem. 2022, 29, 741–783. [Google Scholar] [CrossRef]
- Basit, A.; Shah, S.T.; Ullah, I.; Ullah, I.; Mohamed, H.I. Microbial Bioactive Compounds Produced by Endophytes (Bacteria and Fungi) and Their Uses in Plant Health. In Plant Growth-Promoting Microbes for Sustainable Biotic and Abiotic Stress Management; Springer: Berlin/Heidelberg, Germany, 2021; pp. 285–318. [Google Scholar]
- Afgan, E.; Baker, D.; Batut, B.; van den Beek, M.; Bouvier, D.; Čech, M. The Galaxy Platform for Accessible, Reproducible and Collaborative Biomedical Analyses: 2018 Update. Nucleic Acids Res. 2018, 46, W537–W544. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Mikheenko, A.; Prjibelski, A.; Saveliev, V.; Antipov, D.; Gurevich, A. Versatile genome assembly evaluation with QUAST-LG. Bioinformatics 2018, 34, i142–i150. [Google Scholar] [CrossRef]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef] [PubMed]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75–90. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Carbasse, J.S.; Peinado-Olarte, R.L.; Göker, M. TYGS and LPSN: A database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acids Res. 2022, 50, D801–D807. [Google Scholar] [CrossRef]
- Lee, I.; OukKim, Y.; Park, S.-C.; Chun, J. OrthoANI: An Improved Algorithm and Software for Calculating Average Nucleotide Identity. Int. J. Syst. Evol. Microbiol. 2016, 66, 1100–1103. [Google Scholar] [CrossRef]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef]
- Djoumbou Feunang, Y.; Eisner, R.; Knox, C.; Chepelev, L.; Hastings, J.; Owen, G.; Fahy, E.; Steinbeck, C.; Subramanian, S.; Bolton, E.; et al. ClassyFire: Automated chemical classification with a comprehensive, computable taxonomy. J. Cheminform. 2016, 8, 61–81. [Google Scholar] [CrossRef]
- Kim, H.W.; Wang, M.; Leber, C.A.; Nothias, L.-F.; Reher, R.; Kang, K.B.; van der Hooft, J.J.J.; Dorrestein, P.C.; Gerwick, W.H.; Cottrell, G.W. NPClassifier: A Deep Neural Network-Based Structural Classification Tool for Natural Products. J. Nat. Prod. 2021, 84, 2795–2807. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Pires, D.E.V.; Blundell, T.L.; Ascher, D.B. pkCSM: Predicting small-molecule pharmacokinetic properties using graph-based signatures. J. Med. Chem. 2015, 58, 4066–4072. [Google Scholar] [CrossRef]
- Walker, K.; Long, R.; Croteau, R. The final acylation step in taxol biosynthesis: Cloning of the taxoid C13-side-chain N-benzoyltransferase from Taxus. Proc. Natl. Acad. Sci. USA 2002, 14, 166–171. [Google Scholar] [CrossRef] [PubMed]
- Nishiyama, Y.; Murakami, T.; Kurita, K.; Yamamoto, N. Low-molecular-weight anti-HIV-1 peptides from the amino-terminal sequence of RANTES: Possible lead compounds for coreceptor-directed anti-HIV-1 agents. Bioorganic Med. Chem. Lett. 1999, 9, 1357–1360. [Google Scholar] [CrossRef] [PubMed]
- Paudel, B.; Maharjan, R.; Rajbhandari, P.; Aryal, N.; Aziz, S.; Bhattarai, K.; Baral, B.; Malla, R.; Bhattarai, H.D. Maculosin, a non-toxic antioxidant compound isolated from Streptomyces sp. KTM18. Pharm. Biol. 2021, 59, 933–936. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.Y.; Zhao, D.G.; Gao, K. Structural investigation and biological activity of sesquiterpene lactones from the traditional Chinese herb Inula racemosa. J. Nat. Prod. 2013, 76, 564–570. [Google Scholar] [CrossRef]
- El-Gendy, B.e.l.-D.; Rateb, M.E. Antibacterial activity of diketopiperazines isolated from a marine fungus using t-butoxycarbonyl group as a simple tool for purification. Bioorganic Med. Chem. Lett. 2015, 25, 3125–3128. [Google Scholar] [CrossRef]
- Kher, S.S.; Penzo, M.; Fulle, S.; Finn, P.W.; Blackman, M.J.; Jirgensons, A. Substrate derived peptidic α-ketoamides as inhibitors of the malarial protease PfSUB1. Bioorganic Med. Chem. Lett. 2014, 24, 4486–4489. [Google Scholar] [CrossRef]
- Wu, S.; Qi, W.; Su, R.; Li, T.; Lu, D.; He, Z. CoMFA and CoMSIA analysis of ACE-inhibitory, antimicrobial and bitter-tasting peptides. Eur. J. Med. Chem. 2014, 84, 100–106. [Google Scholar] [CrossRef]
- Li, Q.; Yan, J.; Zhang, L.; Chen, P. Antifibrotic effects of N-acetyl-seryl-aspartyl-lysyl-proline mediated by the regulation of MCP-1 and ED-1 expression on rats with silicosis. Wei Sheng Yan Jiu 2008, 7, 666–670. (In Chinese) [Google Scholar]
- Mayer, R.; Picard, I.; Lawton, P.; Grellier, P.; Barrault, C.; Monsigny, M.; Schrével, J. Peptide derivatives specific for a Plasmodium falciparum proteinase inhibit the human erythrocyte invasion by merozoites. J. Med. Chem. 1991, 34, 3029–3035. [Google Scholar] [CrossRef]
- Georgsson, J.; Sköld, C.; Botros, M.; Lindeberg, G.; Nyberg, F.; Karlén, A.; Hallberg, A.; Larhed, M. Synthesis of a new class of druglike angiotensin II C-terminal mimics with affinity for the AT2 receptor. J. Med. Chem. 2007, 50, 1711–1715. [Google Scholar] [CrossRef] [PubMed]
- LeBeau, A.M.; Banerjee, S.R.; Pomper, M.G.; Mease, R.C.; Denmeade, S.R. Optimization of peptide-based inhibitors of prostate-specific antigen (PSA) as targeted imaging agents for prostate cancer. Bioorganic Med. Chem. Lett. 2009, 17, 4888–4893. [Google Scholar] [CrossRef] [PubMed]
- Kong, J.S.; Venkatraman, S.; Furness, K.; Nimkar, S.; Shepherd, T.A.; Wang, Q.M.; Aubé, J.; Hanzlik, R.P. Synthesis and evaluation of peptidyl Michael acceptors that inactivate human rhinovirus 3C protease and inhibit virus replication. J. Med. Chem. 1998, 41, 2579–2587. [Google Scholar] [CrossRef] [PubMed]
- Wood, S.G.; Lynch, M.; Plaut, A.G.; Burton, J. Tetrapeptide inhibitors of the IgA1 proteinases from type I Neisseria gonorrhoeae. J. Med. Chem. 1989, 32, 2407–2411. [Google Scholar] [CrossRef]
- Marlowe, C.K.; Sinha, U.; Gunn, A.C.; Scarborough, R.M. Design, synthesis and structure-activity relationship of a series of arginine aldehyde factor Xa inhibitors. Part 1: Structures based on the (D)-Arg-Gly-Arg tripeptide sequence. Bioorganic Med. Chem. Lett. 2003, 10, 13–16. [Google Scholar] [CrossRef]
- Noever, D. Naturally occurring protease inhibitors potent against the human immunodeficiency virus. Biochem. Biophys. Res. Commun. 1996, 227, 125–130. [Google Scholar] [CrossRef]
- Yin, Z.; Kelso, M.J.; Beck, J.L.; Oakley, A.J. Structural and thermodynamic dissection of linear motif recognition by the E. coli sliding clamp. J. Med. Chem. 2013, 56, 8665–8673. [Google Scholar] [CrossRef] [PubMed]
- Chapelat, J.; Berst, F.; Marzinzik, A.L.; Moebitz, H.; Drueckes, P.; Trappe, J.; Fabbro, D.; Seebach, D. The Substrate-Activity-Screening methodology applied to receptor tyrosine kinases: A proof-of-concept study. Eur. J. Med. Chem. 2012, 57, 1–9. [Google Scholar] [CrossRef]
- Zin, N.M.; Baba, M.S.; Zainal-Abidin, A.H.; Latip, J.; Mazlan, N.W.; Edrada-Ebel, R. Gancidin W, a potential low-toxicity antimalarial agent isolated from an endophytic Streptomyces SUK10. Drug Des. Dev. Ther. 2017, 11, 351–363. [Google Scholar] [CrossRef]
- Dragovich, P.S.; Webber, S.E.; Babine, R.E.; Fuhrman, S.A.; Patick, A.K.; Matthews, D.A.; Reich, S.H.; Marakovits, J.T.; Prins, T.J.; Zhou, R.; et al. Structure-based design, synthesis, and biological evaluation of irreversible human rhinovirus 3C protease inhibitors. 2. Peptide structure-activity studies. J. Med. Chem. 1998, 41, 2819–2834. [Google Scholar] [CrossRef] [PubMed]
- Llinàs-Brunet, M.; Bailey, M.D.; Ghiro, E.; Gorys, V.; Halmos, T.; Poirier, M.; Rancourt, J.; Goudreau, N. A systematic approach to the optimization of substrate-based inhibitors of the hepatitis C virus NS3 protease: Discovery of potent and specific tripeptide inhibitors. J. Med. Chem. 2004, 47, 6584–6594. [Google Scholar] [CrossRef]
- Zhang, X.; Fang, H.; Zhu, H.; Wang, X.; Zhang, L.; Li, M.; Li, Q.; Yuan, Y.; Xu, W. Novel aminopeptidase N (APN/CD13) inhibitors derived from 3-phenylalanyl-N′-substituted-2,6-piperidinedione. Bioorganic Med. Chem. Lett. 2010, 18, 5981–5987. [Google Scholar] [CrossRef]
- May, J.J.; Wendrich, T.M.; Marahiel, M.A. The dhb operon of Bacillus subtilis encodes the biosynthetic template for the catecholic siderophore 2,3-dihydroxybenzoate-glycine-threonine trimeric ester bacillibactin. J. Biol. Chem. 2001, 276, 7209–7217. [Google Scholar] [CrossRef]
- Deaton, J.; Sun, J.; Holzenburg, A.; Struck, D.K.; Berry, J.; Young, R. Functional bacteriorhodopsin is efficiently solubilized and delivered to membranes by the chaperonin GroEL. Proc. Natl. Acad. Sci. USA 2004, 101, 2281–2286. [Google Scholar] [CrossRef] [PubMed]
- Hu, K.; Chung, D.D.L. Flexible graphite modified by carbon black paste for use as a thermal interface material. Carbon 2011, 49, 1075–1086. [Google Scholar] [CrossRef]
- Downs, C.A.; DiNardo, J.C.; Stien, D.; Rodrigues, A.M.; Lebaron, P. Benzophenone accumulates over time from the degradation of octocrylene in commercial sunscreen products. Chem. Res. Toxicol. 2021, 34, 1046–1054. [Google Scholar] [CrossRef]
- Dimopoulou, A.; Theologidis, I.; Benaki, D.; Koukounia, M.; Zervakou, A.; Tzima, A.; Diallinas, G.; Hatzinikolaou, D.G.; Skandalis, N. Direct Antibiotic Activity of Bacillibactin Broadens the Biocontrol Range of Bacillus amyloliquefaciens MBI600. Msphere 2021, 6, 0037621. [Google Scholar] [CrossRef] [PubMed]
- Sur, S.; Romo, T.D.; Grossfield, A. Selectivity and Mechanism of Fengycin, an Antimicrobial Lipopeptide, from Molecular Dynamics. J. Phys. Chem. B 2018, 122, 2219–2226. [Google Scholar] [CrossRef] [PubMed]
- Alizadeh, S.R.; Hashemi, S.M. Development and therapeutic potential of 2-aminothiazole derivatives in anticancer drug discovery. Med. Chem. Res. 2021, 30, 771–806. [Google Scholar] [CrossRef] [PubMed]
- Eckert, E.; Münch, F.; Göen, T.; Purbojo, A.; Müller, J.; Cesnjevar, R. Comparative study on the migration of di-2-ethylhexyl phthalate (DEHP) and tri-2-ethylhexyl trimellitate (TOTM) into blood from PVC tubing material of a heart-lung machine. Chemosphere 2016, 145, 10–16. [Google Scholar] [CrossRef]
- Chen, X.; Dai, X.; Liu, Y.; Yang, Y.; Yuan, L.; He, X.; Gong, G. Solanum nigrum Linn.: An Insight into Current Research on Traditional Uses, Phytochemistry, and Pharmacology. Front. Pharmacol. 2022, 13, 918071. [Google Scholar] [CrossRef]
- Park, S.T.; Kim, J. Trends in next-generation sequencing and a new era for whole genome sequencing. Int. Neurourol. J. 2016, 20, S76. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Hahnke, R.L.; Petersen, J.; Scheuner, C.; Michael, V.; Fiebig, A.; Rohde, C.; Rohde, M.; Fartmann, B.; Goodwin, L.A.; et al. Complete genome sequence of DSM 30083 T, the type strain (U5/41 T) of Escherichia coli, and a proposal for delineating subspecies in microbial taxonomy. Stand. Genom. Sci. 2014, 9, 1–19. [Google Scholar]
- Nalli, Y.; Singh, S.; Gajjar, A.; Mahizhaveni, B.; Dusthackeer, V.N.A.; Shinde, P.B. Bacillibactin class siderophores produced by the endophyte Bacillus subtilis NPROOT3 as antimycobacterial agents. Lett. Appl. Microbiol. 2023, 76, ovac026. [Google Scholar]
- Wu, Z.Z.; Ding, G.F.; Huang, F.F.; Yang, Z.S.; Yu, F.M.; Tang, Y.P.; Jia, Y.L.; Zheng, Y.Y.; Chen, R. Anticancer activity of Anthopleura anjunae oligopeptides in prostate cancer DU-145 cells. Mar. Drugs 2018, 16, 125. [Google Scholar] [CrossRef] [PubMed]
- Bae, Y.; Joo, C.; Kim, G.Y.; Ko, K.S.; Huh, K.M.; Han, J.; Choi, J.S. Cationic oligopeptide-functionalized mitochondria targeting sequence show mitochondria targeting and anticancer activity. Macromol. Res. 2019, 27, 1071–1080. [Google Scholar] [CrossRef]
- Gurung, A.B.; Ali, M.A.; Lee, J.; Farah, M.A.; Al-Anazi, K.M. Molecular docking and dynamics simulation study of bioactive compounds from Ficus carica L. with important anticancer drug targets. PLoS ONE 2021, 16, e0254035. [Google Scholar] [CrossRef] [PubMed]
- Cao, D.; Wang, J.; Zhou, R.; Li, Y.; Yu, H.; Hou, T. ADMET evaluation in drug discovery. 11. PharmacoKinetics Knowledge Base (PKKB): A comprehensive database of pharmacokinetic and toxic properties for drugs. J. Chem. Inf. Model. 2012, 52, 1132–1137. [Google Scholar] [CrossRef] [PubMed]
- Aaghaz, S.; Gohel, V.; Kamal, A. Peptides as potential anticancer agents. Curr. Top. Med. Chem. 2019, 19, 1491–1511. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).