Sex-Specific Effects of Polystyrene Microplastic and Lead(II) Co-Exposure on the Gut Microbiome and Fecal Metabolome in C57BL/6 Mice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Design
2.2. Sequencing of Internal Transcribed Spacer (ITS) and 16S rRNA Gene
2.3. Metagenomic Sequencing
2.4. qRT-PCR Analysis of Tight-Junction Proteins
2.5. Untargeted Metabolomic Profiling
2.6. Statistical Analysis
3. Results
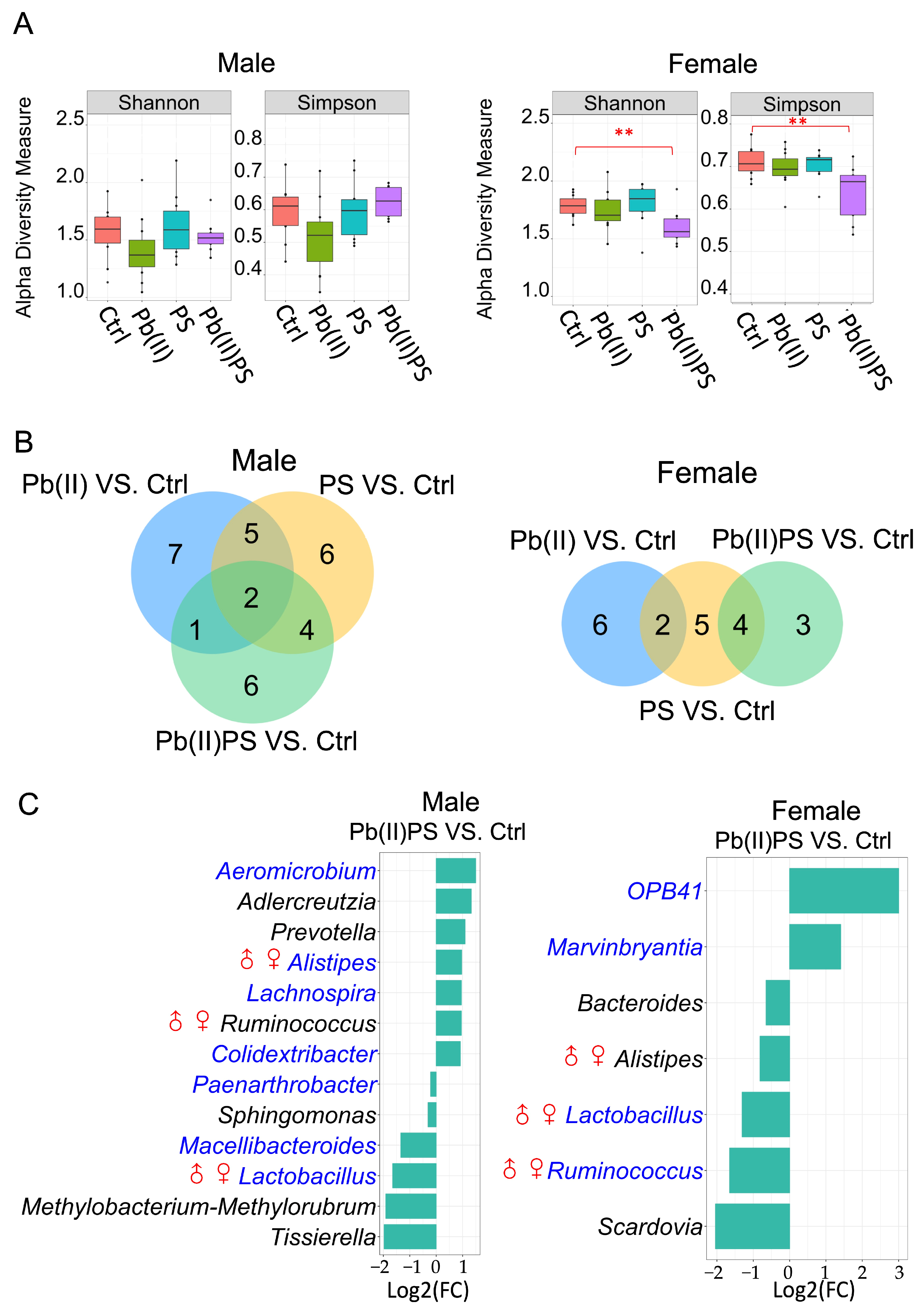
3.1. PS and Pb(II) Co-Exposure Altered the Gut Bacterial Composition
3.2. PS and Pb(II) Co-Exposure Altered the Gut Fungal Composition
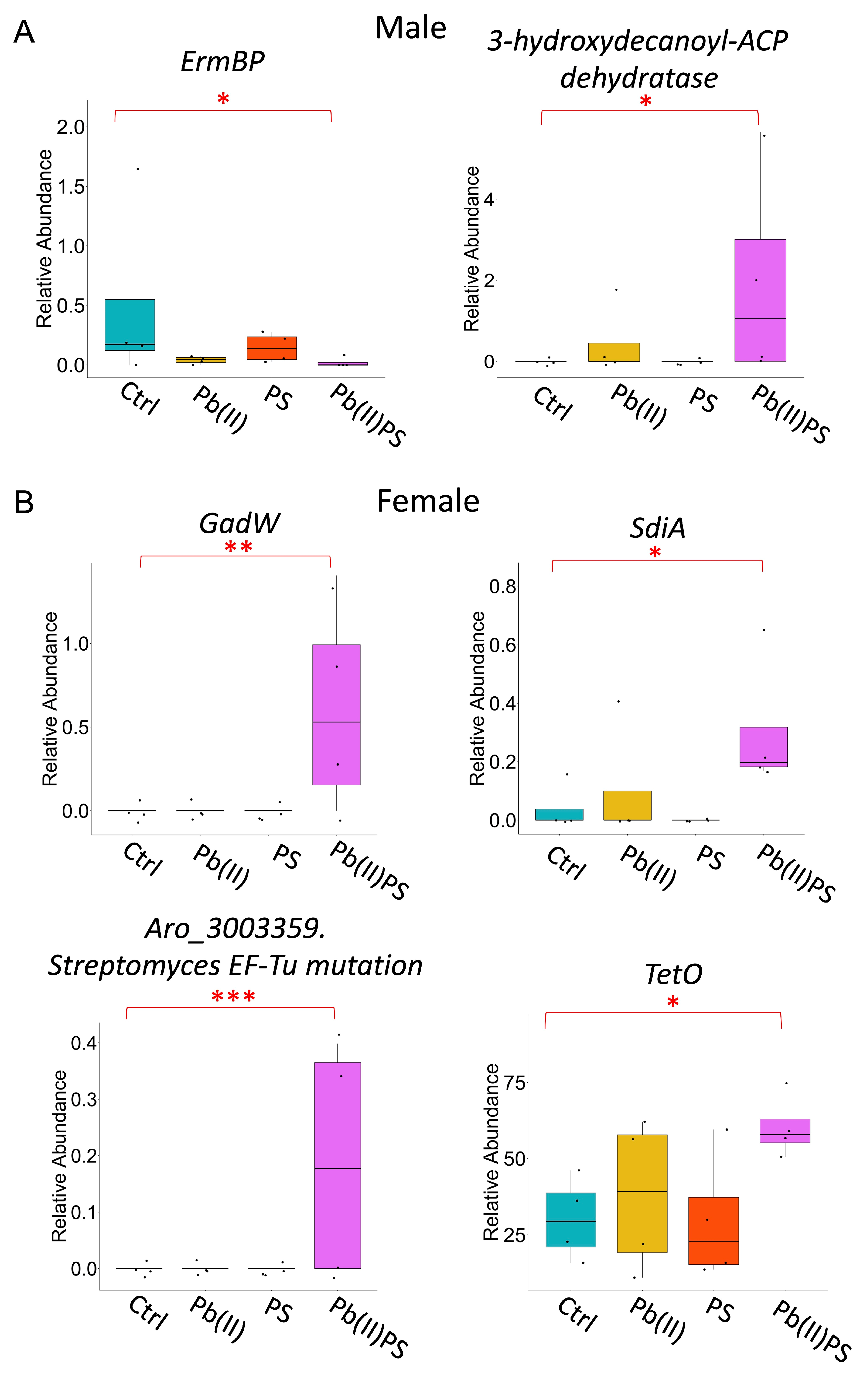
3.3. PS and Pb(II) Co-Exposure Altered the Gut Microbial Metabolic Pathways
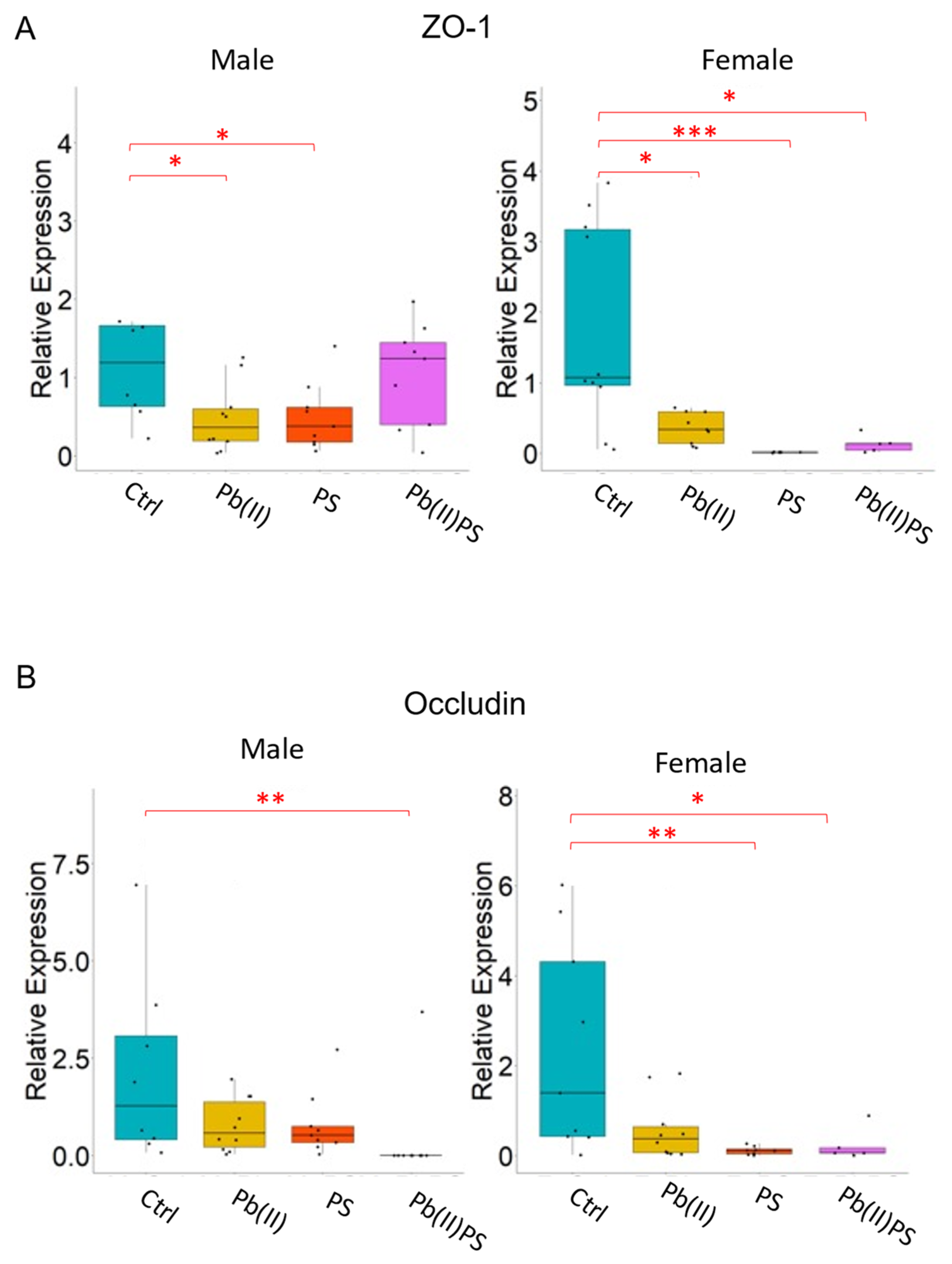
3.4. PS and Pb(II) Co-Exposure Increased the Intestinal Permeability
3.5. PS and Pb(II) Co-Exposure Altered the Fecal Metabolites
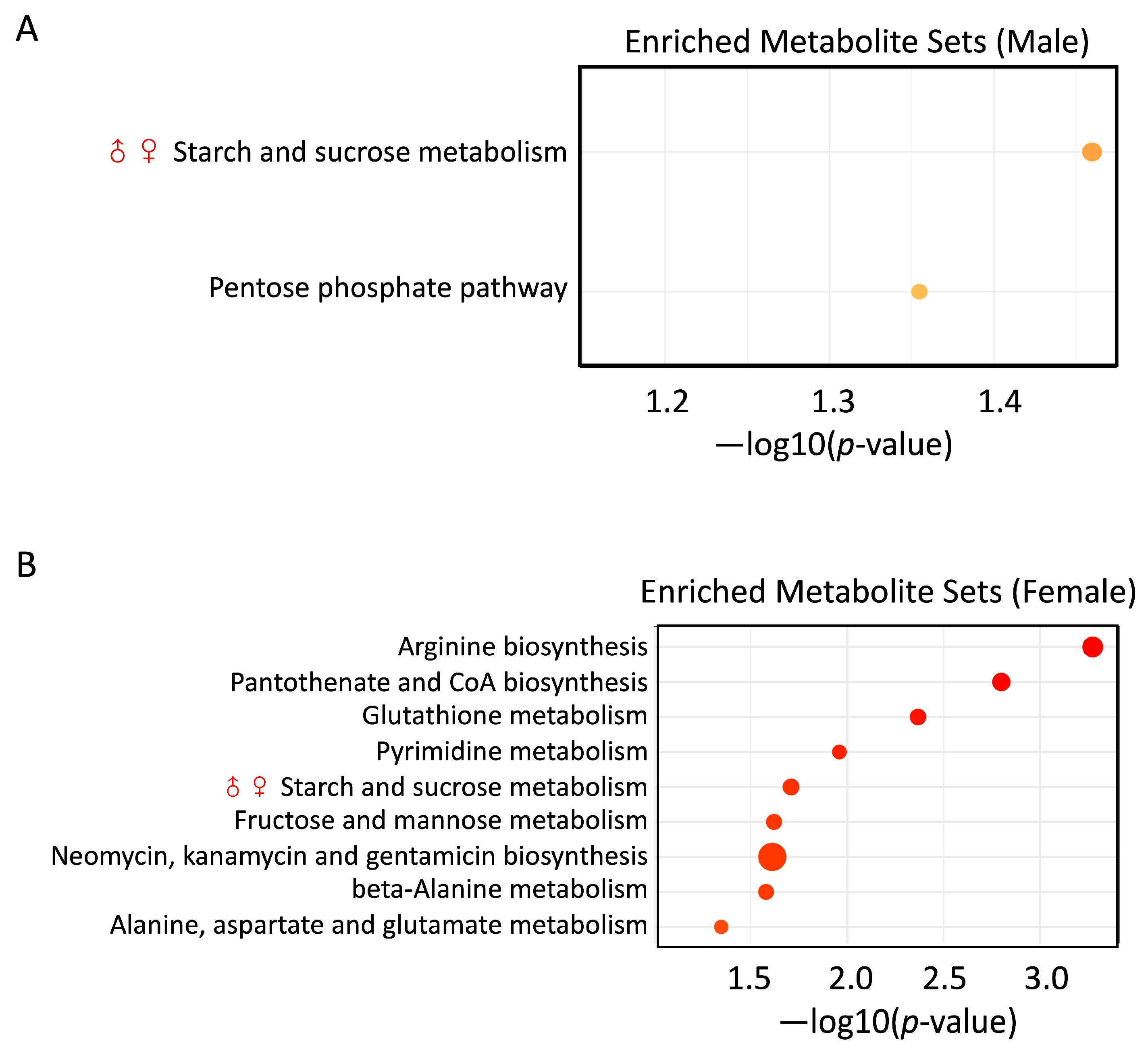
3.6. PS and Pb(II) Co-Exposure Enriched Metabolite Sets
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wu, P.; Huang, J.; Zheng, Y.; Yang, Y.; Zhang, Y.; He, F.; Chen, H.; Quan, G.; Yan, J.; Li, T.; et al. Environmental Occurrences, Fate, and Impacts of Microplastics. Ecotoxicol. Environ. Saf. 2019, 184, 109612. [Google Scholar] [CrossRef] [PubMed]
- Rochman, C.M.; Hoh, E.; Kurobe, T.; Teh, S.J. Ingested Plastic Transfers Hazardous Chemicals to Fish and Induces Hepatic Stress. Sci. Rep. 2013, 3, 3263. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Jia, Z. Recent Insights into Uptake, Toxicity, and Molecular Targets of Microplastics and Nanoplastics Relevant to Human Health Impacts. iScience 2023, 26, 106061. [Google Scholar] [CrossRef] [PubMed]
- Razanajatovo, R.M.; Ding, J.; Zhang, S.; Jiang, H.; Zou, H. Sorption and Desorption of Selected Pharmaceuticals by Polyethylene Microplastics. Mar. Pollut. Bull. 2018, 136, 516–523. [Google Scholar] [CrossRef] [PubMed]
- Brennecke, D.; Duarte, B.; Paiva, F.; Caçador, I.; Canning-Clode, J. Microplastics as Vector for Heavy Metal Contamination from the Marine Environment. Estuar. Coast. Shelf Sci. 2016, 178, 189–195. [Google Scholar] [CrossRef]
- Dobaradaran, S.; Schmidt, T.C.; Nabipour, I.; Khajeahmadi, N.; Tajbakhsh, S.; Saeedi, R.; Javad Mohammadi, M.; Keshtkar, M.; Khorsand, M.; Faraji Ghasemi, F. Characterization of Plastic Debris and Association of Metals with Microplastics in Coastline Sediment along the Persian Gulf. Waste Manag. 2018, 78, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Yuan, W.; Zhou, Y.; Chen, Y.; Liu, X.; Wang, J. Toxicological Effects of Microplastics and Heavy Metals on the Daphnia Magna. Sci. Total Environ. 2020, 746, 141254. [Google Scholar] [CrossRef]
- de Freitas, C.U.; De Capitani, E.M.; Gouveia, N.; Simonetti, M.H.; de Paula E Silva, M.R.; Kira, C.S.; Sakuma, A.M.; de Fátima Henriques Carvalho, M.; Duran, M.C.; Tiglea, P.; et al. Lead Exposure in an Urban Community: Investigation of Risk Factors and Assessment of the Impact of Lead Abatement Measures. Environ. Res. 2007, 103, 338–344. [Google Scholar] [CrossRef] [PubMed]
- Qu, X.; Alvarez, P.J.J.; Li, Q. Applications of Nanotechnology in Water and Wastewater Treatment. Water Res. 2013, 47, 3931–3946. [Google Scholar] [CrossRef]
- Tang, S.; Lin, L.; Wang, X.; Feng, A.; Yu, A. Pb(II) Uptake onto Nylon Microplastics: Interaction Mechanism and Adsorption Performance. J. Hazard. Mater. 2020, 386, 121960. [Google Scholar] [CrossRef]
- Lu, X.; Zeng, F.; Wei, S.; Gao, R.; Abdurahman, A.; Wang, H.; Liang, W. Effects of Humic Acid on Pb2+ Adsorption onto Polystyrene Microplastics from Spectroscopic Analysis and Site Energy Distribution Analysis. Sci. Rep. 2022, 12, 8932. [Google Scholar] [CrossRef] [PubMed]
- Schoeler, M.; Caesar, R. Dietary Lipids, Gut Microbiota and Lipid Metabolism. Rev. Endocr. Metab. Disord. 2019, 20, 461–472. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Liu, L.; Cao, Z.; Li, W.; Li, H.; Lu, C.; Yang, X.; Liu, Y. Gut Microbiota as an “Invisible Organ” That Modulates the Function of Drugs. Biomed. Pharmacother. Biomed. Pharmacother. 2020, 121, 109653. [Google Scholar] [CrossRef]
- Tu, P.; Chi, L.; Bodnar, W.; Zhang, Z.; Gao, B.; Bian, X.; Stewart, J.; Fry, R.; Lu, K. Gut Microbiome Toxicity: Connecting the Environment and Gut Microbiome-Associated Diseases. Toxics 2020, 8, 19. [Google Scholar] [CrossRef]
- Dominianni, C.; Sinha, R.; Goedert, J.J.; Pei, Z.; Yang, L.; Hayes, R.B.; Ahn, J. Sex, Body Mass Index, and Dietary Fiber Intake Influence the Human Gut Microbiome. PLoS ONE 2015, 10, e0124599. [Google Scholar] [CrossRef]
- Ding, T.; Schloss, P.D. Dynamics and Associations of Microbial Community Types across the Human Body. Nature 2014, 509, 357–360. [Google Scholar] [CrossRef]
- Hokanson, K.C.; Hernández, C.; Deitzler, G.E.; Gaston, J.E.; David, M.M. Sex Shapes Gut-Microbiota-Brain Communication and Disease. Trends Microbiol. 2024, 32, 151–161. [Google Scholar] [CrossRef] [PubMed]
- Markle, J.G.M.; Frank, D.N.; Mortin-Toth, S.; Robertson, C.E.; Feazel, L.M.; Rolle-Kampczyk, U.; von Bergen, M.; McCoy, K.D.; Macpherson, A.J.; Danska, J.S. Sex Differences in the Gut Microbiome Drive Hormone-Dependent Regulation of Autoimmunity. Science 2013, 339, 1084–1088. [Google Scholar] [CrossRef]
- Flores, R.; Shi, J.; Fuhrman, B.; Xu, X.; Veenstra, T.D.; Gail, M.H.; Gajer, P.; Ravel, J.; Goedert, J.J. Fecal Microbial Determinants of Fecal and Systemic Estrogens and Estrogen Metabolites: A Cross-Sectional Study. J. Transl. Med. 2012, 10, 253. [Google Scholar] [CrossRef]
- Calcaterra, V.; Rossi, V.; Massini, G.; Regalbuto, C.; Hruby, C.; Panelli, S.; Bandi, C.; Zuccotti, G. Precocious Puberty and Microbiota: The Role of the Sex Hormone-Gut Microbiome Axis. Front. Endocrinol. 2022, 13, 1000919. [Google Scholar] [CrossRef]
- López de Las Hazas, M.-C.; Boughanem, H.; Dávalos, A. Untoward Effects of Micro- and Nanoplastics: An Expert Review of Their Biological Impact and Epigenetic Effects. Adv. Nutr. 2022, 13, 1310–1323. [Google Scholar] [CrossRef]
- Li, B.; Ding, Y.; Cheng, X.; Sheng, D.; Xu, Z.; Rong, Q.; Wu, Y.; Zhao, H.; Ji, X.; Zhang, Y. Polyethylene Microplastics Affect the Distribution of Gut Microbiota and Inflammation Development in Mice. Chemosphere 2020, 244, 125492. [Google Scholar] [CrossRef] [PubMed]
- Qiao, R.; Sheng, C.; Lu, Y.; Zhang, Y.; Ren, H.; Lemos, B. Microplastics Induce Intestinal Inflammation, Oxidative Stress, and Disorders of Metabolome and Microbiome in Zebrafish. Sci. Total Environ. 2019, 662, 246–253. [Google Scholar] [CrossRef]
- Sun, J.; Fang, R.; Wang, H.; Xu, D.-X.; Yang, J.; Huang, X.; Cozzolino, D.; Fang, M.; Huang, Y. A Review of Environmental Metabolism Disrupting Chemicals and Effect Biomarkers Associating Disease Risks: Where Exposomics Meets Metabolomics. Environ. Int. 2022, 158, 106941. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.; Zeng, Z.; Wang, Q.; Liang, W.; Guo, Y.; Huo, X. Alterations of the Gut Microbiota and Metabolomics in Children with E-Waste Lead Exposure. J. Hazard. Mater. 2022, 434, 128842. [Google Scholar] [CrossRef]
- Feng, Y.; Yuan, H.; Wang, W.; Xu, Y.; Zhang, J.; Xu, H.; Fu, F. Co-Exposure to Polystyrene Microplastics and Lead Aggravated Ovarian Toxicity in Female Mice via the PERK/eIF2α Signaling Pathway. Ecotoxicol. Environ. Saf. 2022, 243, 113966. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Lin, S.; Tang, J.; Li, Y.; Wang, X.; Jiang, Y.; Zhang, H.; Wang, B. Effects of Microplastics and Lead Exposure on Gut Oxidative Stress and Intestinal Inflammation in Common Carp (Cyprinus carpio L.). Environ. Pollut. 2023, 327, 121528. [Google Scholar] [CrossRef]
- Gao, B.; Shi, X.; Li, S.; Xu, W.; Gao, N.; Shan, J.; Shen, W. Size-Dependent Effects of Polystyrene Microplastics on Gut Metagenome and Antibiotic Resistance in C57BL/6 Mice. Ecotoxicol. Environ. Saf. 2023, 254, 114737. [Google Scholar] [CrossRef] [PubMed]
- Gao, B.; Lue, H.-W.; Podolak, J.; Fan, S.; Zhang, Y.; Serawat, A.; Alumkal, J.J.; Fiehn, O.; Thomas, G.V. Multi-Omics Analyses Detail Metabolic Reprogramming in Lipids, Carnitines, and Use of Glycolytic Intermediates between Prostate Small Cell Neuroendocrine Carcinoma and Prostate Adenocarcinoma. Metabolites 2019, 9, 82. [Google Scholar] [CrossRef]
- Tsugawa, H.; Cajka, T.; Kind, T.; Ma, Y.; Higgins, B.; Ikeda, K.; Kanazawa, M.; VanderGheynst, J.; Fiehn, O.; Arita, M. MS-DIAL: Data-Independent MS/MS Deconvolution for Comprehensive Metabolome Analysis. Nat. Methods 2015, 12, 523–526. [Google Scholar] [CrossRef]
- Mallick, H.; Rahnavard, A.; McIver, L.J.; Ma, S.; Zhang, Y.; Nguyen, L.H.; Tickle, T.L.; Weingart, G.; Ren, B.; Schwager, E.H.; et al. Multivariable Association Discovery in Population-Scale Meta-Omics Studies. PLoS Comput. Biol. 2021, 17, e1009442. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.; Chong, J.; Zhou, G.; de Lima Morais, D.A.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.-É.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the Gap between Raw Spectra and Functional Insights. Nucleic Acids Res. 2021, 49, W388–W396. [Google Scholar] [CrossRef] [PubMed]
- Org, E.; Mehrabian, M.; Parks, B.W.; Shipkova, P.; Liu, X.; Drake, T.A.; Lusis, A.J. Sex Differences and Hormonal Effects on Gut Microbiota Composition in Mice. Gut Microbes 2016, 7, 313–322. [Google Scholar] [CrossRef] [PubMed]
- Yurkovetskiy, L.; Burrows, M.; Khan, A.A.; Graham, L.; Volchkov, P.; Becker, L.; Antonopoulos, D.; Umesaki, Y.; Chervonsky, A.V. Gender Bias in Autoimmunity Is Influenced by Microbiota. Immunity 2013, 39, 400–412. [Google Scholar] [CrossRef] [PubMed]
- Ai, X.; Xin, X.; Wei, W.; Xie, J.; Hong, J. Polysorbate-80 Pretreatment Contributing to Volatile Fatty Acids Production Associated Microbial Interactions via Acidogenic Fermentation of Waste Activated Sludge. Bioresour. Technol. 2022, 345, 126488. [Google Scholar] [CrossRef] [PubMed]
- Larsen, J.M. The Immune Response to Prevotella Bacteria in Chronic Inflammatory Disease. Immunology 2017, 151, 363–374. [Google Scholar] [CrossRef] [PubMed]
- Zafar, H.; Saier, M.H. Gut Bacteroides Species in Health and Disease. Gut Microbes 2021, 13, 1848158. [Google Scholar] [CrossRef]
- Khomyakova, M.A.; Zavarzina, D.G.; Merkel, A.Y.; Klyukina, A.A.; Pikhtereva, V.A.; Gavrilov, S.N.; Slobodkin, A.I. The First Cultivated Representatives of the Actinobacterial Lineage OPB41 Isolated from Subsurface Environments Constitute a Novel Order Anaerosomatales. Front. Microbiol. 2022, 13, 1047580. [Google Scholar] [CrossRef] [PubMed]
- Parker, B.J.; Wearsch, P.A.; Veloo, A.C.M.; Rodriguez-Palacios, A. The Genus Alistipes: Gut Bacteria with Emerging Implications to Inflammation, Cancer, and Mental Health. Front. Immunol. 2020, 11, 906. [Google Scholar] [CrossRef]
- Hall, A.B.; Yassour, M.; Sauk, J.; Garner, A.; Jiang, X.; Arthur, T.; Lagoudas, G.K.; Vatanen, T.; Fornelos, N.; Wilson, R.; et al. A Novel Ruminococcus Gnavus Clade Enriched in Inflammatory Bowel Disease Patients. Genome Med. 2017, 9, 103. [Google Scholar] [CrossRef]
- Novicki, T.J.; LaFe, K.; Bui, L.; Bui, U.; Geise, R.; Marr, K.; Cookson, B.T. Genetic Diversity among Clinical Isolates of Acremonium Strictum Determined during an Investigation of a Fatal Mycosis. J. Clin. Microbiol. 2003, 41, 2623–2628. [Google Scholar] [CrossRef] [PubMed]
- Silva, S.; Negri, M.; Henriques, M.; Oliveira, R.; Williams, D.W.; Azeredo, J. Candida Glabrata, Candida Parapsilosis and Candida Tropicalis: Biology, Epidemiology, Pathogenicity and Antifungal Resistance. FEMS Microbiol. Rev. 2012, 36, 288–305. [Google Scholar] [CrossRef] [PubMed]
- Cho, K.Y. Lifestyle Modifications Result in Alterations in the Gut Microbiota in Obese Children. BMC Microbiol. 2021, 21, 10. [Google Scholar] [CrossRef] [PubMed]
- Wolin, M.J.; Zhang, Y.; Bank, S.; Yerry, S.; Miller, T.L. NMR Detection of 13CH313COOH from 3-13C-Glucose: A Signature for Bifidobacterium Fermentation in the Intestinal Tract. J. Nutr. 1998, 128, 91–96. [Google Scholar] [CrossRef] [PubMed]
- Lanigan, N.; Kelly, E.; Arzamasov, A.A.; Stanton, C.; Rodionov, D.A.; van Sinderen, D. Transcriptional Control of Central Carbon Metabolic Flux in Bifidobacteria by Two Functionally Similar, yet Distinct LacI-Type Regulators. Sci. Rep. 2019, 9, 17851. [Google Scholar] [CrossRef]
- Larson, T.J.; Ehrmann, M.; Boos, W. Periplasmic Glycerophosphodiester Phosphodiesterase of Escherichia coli, a New Enzyme of the Glp Regulon. J. Biol. Chem. 1983, 258, 5428–5432. [Google Scholar] [CrossRef]
- Kaval, K.G.; Garsin, D.A. Ethanolamine Utilization in Bacteria. mBio 2018, 9, e00066-18. [Google Scholar] [CrossRef] [PubMed]
- Maicas, S.; Ferrer, S.; Pardo, I. NAD(P)H Regeneration Is the Key for Heterolactic Fermentation of Hexoses in Oenococcus Oeni. Microbiol. Read. Engl. 2002, 148, 325–332. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Dai, X. Growth Suppression by Altered (p)ppGpp Levels Results from Non-Optimal Resource Allocation in Escherichia Coli. Nucleic Acids Res. 2019, 47, 4684–4693. [Google Scholar] [CrossRef]
- Knirel, Y.A.; Prokhorov, N.S.; Shashkov, A.S.; Ovchinnikova, O.G.; Zdorovenko, E.L.; Liu, B.; Kostryukova, E.S.; Larin, A.K.; Golomidova, A.K.; Letarov, A.V. Variations in O-Antigen Biosynthesis and O-Acetylation Associated with Altered Phage Sensitivity in Escherichia Coli 4s. J. Bacteriol. 2015, 197, 905–912. [Google Scholar] [CrossRef]
- Benavides, M.A.; Bosland, M.C.; da Silva, C.P.; Gomes Sares, C.T.; de Oliveira, A.M.C.; Kemp, R.; dos Reis, R.B.; Martins, V.R.; Sampaio, S.V.; Bland, K.I.; et al. L-Methionine Inhibits Growth of Human Pancreatic Cancer Cells. Anticancer Drugs 2014, 25, 200–203. [Google Scholar] [CrossRef] [PubMed]
- Lucock, M. Folic Acid: Nutritional Biochemistry, Molecular Biology, and Role in Disease Processes. Mol. Genet. Metab. 2000, 71, 121–138. [Google Scholar] [CrossRef] [PubMed]
- Mock, D.M. Marginal Biotin Deficiency Is Common in Normal Human Pregnancy and Is Highly Teratogenic in Mice. J. Nutr. 2009, 139, 154–157. [Google Scholar] [CrossRef] [PubMed]
- Delcour, J.; Ferain, T.; Deghorain, M.; Palumbo, E.; Hols, P. The Biosynthesis and Functionality of the Cell-Wall of Lactic Acid Bacteria. Antonie Van Leeuwenhoek 1999, 76, 159–184. [Google Scholar] [CrossRef] [PubMed]
- Hernández, S.B.; Dörr, T.; Waldor, M.K.; Cava, F. Modulation of Peptidoglycan Synthesis by Recycled Cell Wall Tetrapeptides. Cell Rep. 2020, 31, 107578. [Google Scholar] [CrossRef] [PubMed]
- Manna, D.; Lozano-Amado, D.; Ehrenkaufer, G.; Singh, U. The NAD+ Responsive Transcription Factor ERM-BP Functions Downstream of Cellular Aggregation and Is an Early Regulator of Development and Heat Shock Response in Entamoeba. Front. Cell. Infect. Microbiol. 2020, 10, 363. [Google Scholar] [CrossRef] [PubMed]
- Pacheco, T.; Gomes, A.É.I.; Siqueira, N.M.G.; Assoni, L.; Darrieux, M.; Venter, H.; Ferraz, L.F.C. SdiA, a Quorum-Sensing Regulator, Suppresses Fimbriae Expression, Biofilm Formation, and Quorum-Sensing Signaling Molecules Production in Klebsiella Pneumoniae. Front. Microbiol. 2021, 12, 597735. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Zhang, S.; Xu, Z.; Li, H.; Xiao, Q.; Qiu, F.; Zhang, W.; Long, Y.; Zheng, D.; Huang, B.; et al. SdiA Improves the Acid Tolerance of E. Coli by Regulating GadW and GadY Expression. Front. Microbiol. 2020, 11, 1078. [Google Scholar] [CrossRef] [PubMed]
- Camilleri, M.; Madsen, K.; Spiller, R.; Greenwood-Van Meerveld, B.; Verne, G.N. Intestinal Barrier Function in Health and Gastrointestinal Disease. Neurogastroenterol. Motil. 2012, 24, 503–512. [Google Scholar] [CrossRef]
- Yin, K.; Wang, D.; Zhao, H.; Wang, Y.; Zhang, Y.; Liu, Y.; Li, B.; Xing, M. Polystyrene Microplastics Up-Regulates Liver Glutamine and Glutamate Synthesis and Promotes Autophagy-Dependent Ferroptosis and Apoptosis in the Cerebellum through the Liver-Brain Axis. Environ. Pollut. 2022, 307, 119449. [Google Scholar] [CrossRef]
- El Gazzar, W.B.; Sliem, R.E.; Bayoumi, H.; Nasr, H.E.; Shabanah, M.; Elalfy, A.; Radwaan, S.E.; Gebba, M.A.; Mansour, H.M.; Badr, A.M.; et al. Melatonin Alleviates Intestinal Barrier Damaging Effects Induced by Polyethylene Microplastics in Albino Rats. Int. J. Mol. Sci. 2023, 24, 13619. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Su, P.; Meng, S.; Aschner, M.; Cao, Y.; Luo, W.; Zheng, G.; Liu, M. Role of Matrix Metalloproteinase-2/9 (MMP2/9) in Lead-Induced Changes in an In Vitro Blood-Brain Barrier Model. Int. J. Biol. Sci. 2017, 13, 1351–1360. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Chen, L.; Wu, B. Size-Specific Effects of Microplastics and Lead on Zebrafish. Chemosphere 2023, 337, 139383. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shen, W.; Zhao, M.; Xu, W.; Shi, X.; Ren, F.; Tu, P.; Gao, N.; Shan, J.; Gao, B. Sex-Specific Effects of Polystyrene Microplastic and Lead(II) Co-Exposure on the Gut Microbiome and Fecal Metabolome in C57BL/6 Mice. Metabolites 2024, 14, 189. https://doi.org/10.3390/metabo14040189
Shen W, Zhao M, Xu W, Shi X, Ren F, Tu P, Gao N, Shan J, Gao B. Sex-Specific Effects of Polystyrene Microplastic and Lead(II) Co-Exposure on the Gut Microbiome and Fecal Metabolome in C57BL/6 Mice. Metabolites. 2024; 14(4):189. https://doi.org/10.3390/metabo14040189
Chicago/Turabian StyleShen, Weishou, Meng Zhao, Weichen Xu, Xiaochun Shi, Fangfang Ren, Pengcheng Tu, Nan Gao, Jinjun Shan, and Bei Gao. 2024. "Sex-Specific Effects of Polystyrene Microplastic and Lead(II) Co-Exposure on the Gut Microbiome and Fecal Metabolome in C57BL/6 Mice" Metabolites 14, no. 4: 189. https://doi.org/10.3390/metabo14040189
APA StyleShen, W., Zhao, M., Xu, W., Shi, X., Ren, F., Tu, P., Gao, N., Shan, J., & Gao, B. (2024). Sex-Specific Effects of Polystyrene Microplastic and Lead(II) Co-Exposure on the Gut Microbiome and Fecal Metabolome in C57BL/6 Mice. Metabolites, 14(4), 189. https://doi.org/10.3390/metabo14040189








