A Multiomics, Molecular Atlas of Breast Cancer Survivors
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design, Recruitment, Inclusion/Exclusion Criteria, and Informed Consent
2.2. Whole Genome Sequencing
2.3. Proteomics via Aptamer
2.4. Targeted Metabolomics
2.4.1. Acylcarnitines
2.4.2. Bile Acids
2.4.3. Fatty Acid Panel
2.4.4. Targeted Metabolomics Statistical Methods
2.5. Untargeted Metabolomics
2.6. Gut Metagenomics
2.7. Quality of Life Questionnaires
3. Results
3.1. Metadata: Description of the Cohort
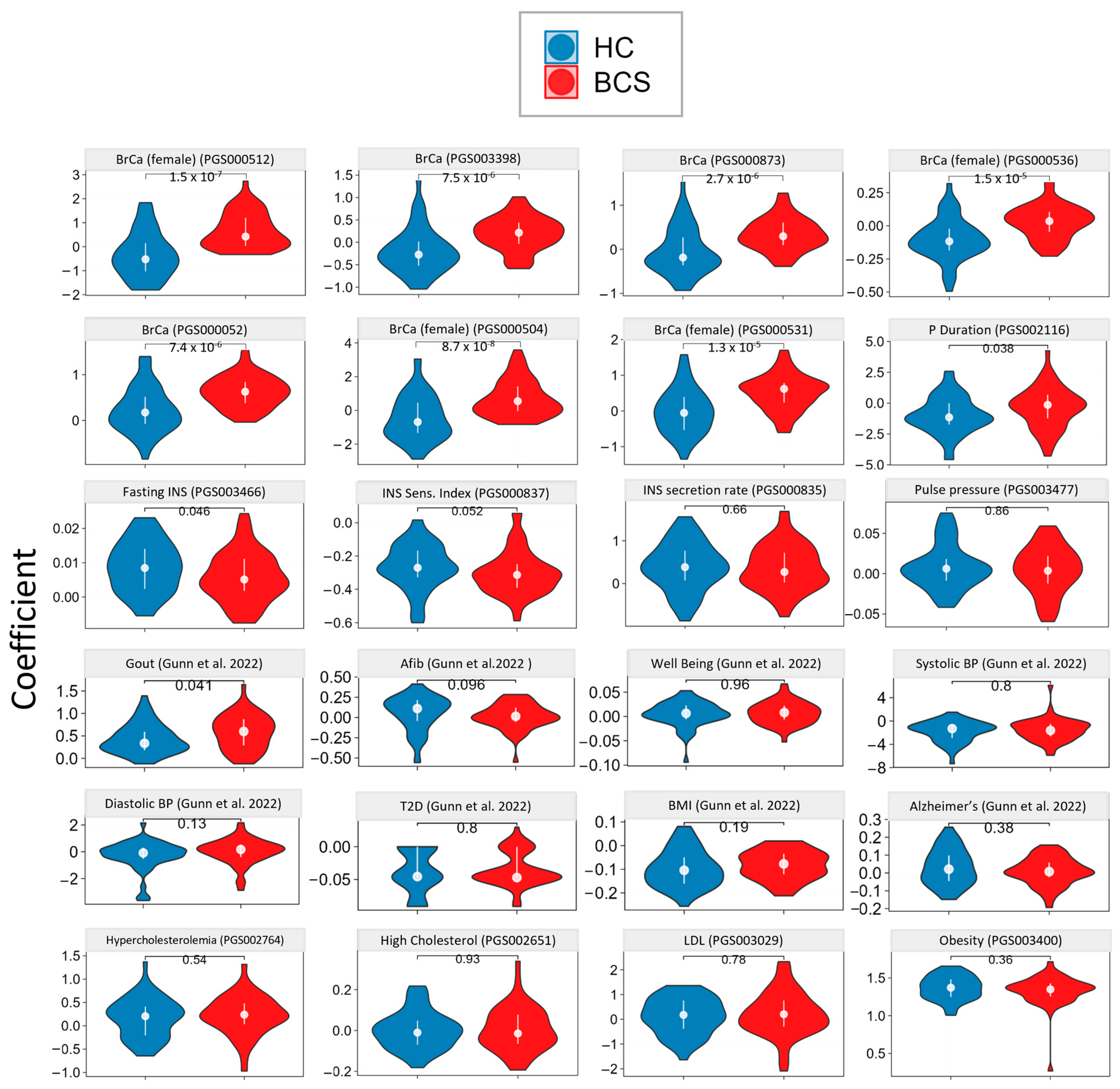
3.2. Genetic Analysis via Whole Genome Sequencing
3.3. Targeted Metabolomics
3.3.1. Acylcarnitines
3.3.2. Bile Acids
3.3.3. RBC Fatty Acids
3.4. Aptamer-Based Untargeted Proteomics
3.4.1. Primary and Exploratory Proteomic Analysis
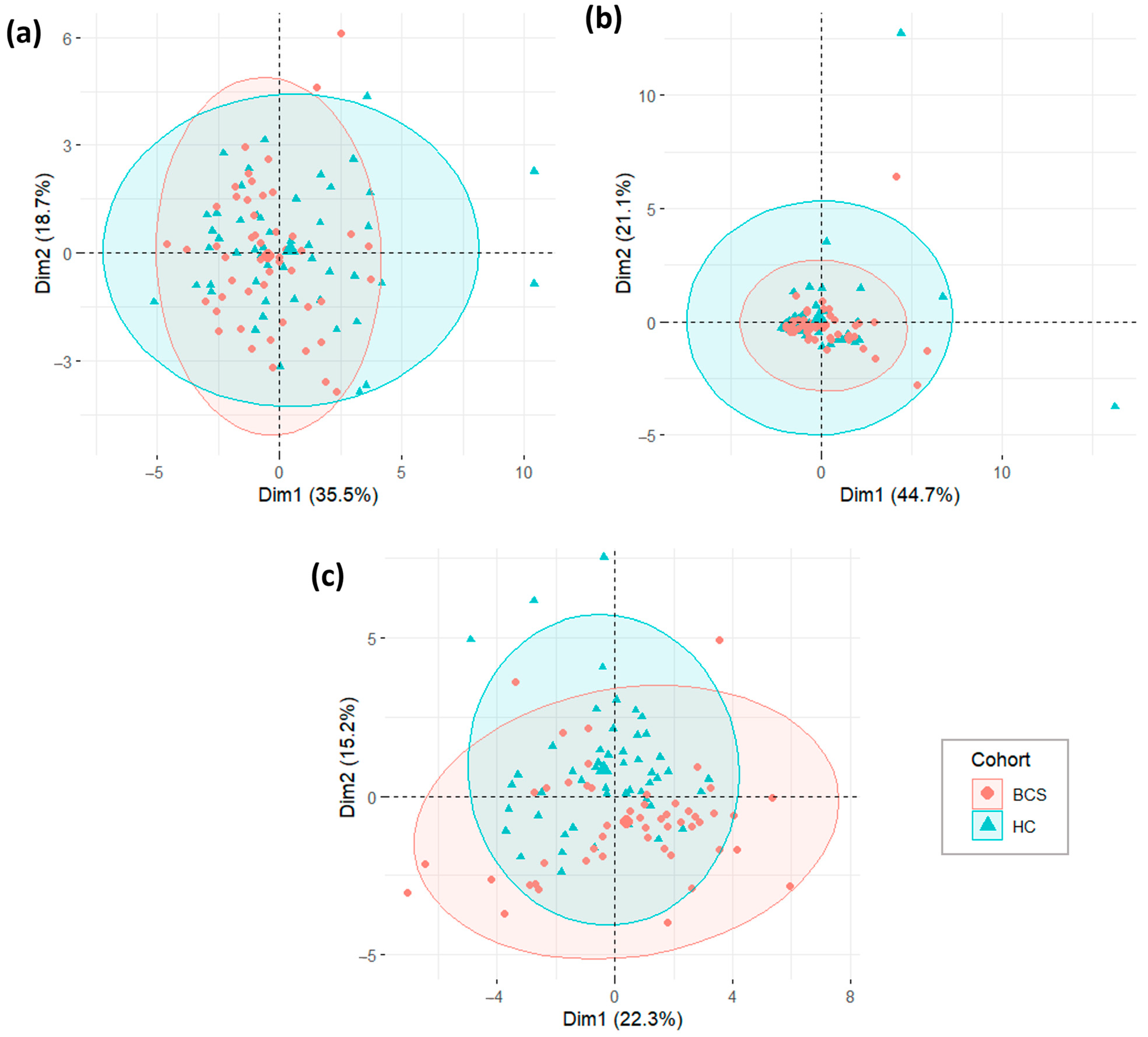
3.4.2. Univariate and Multivariate Proteomic Analysis
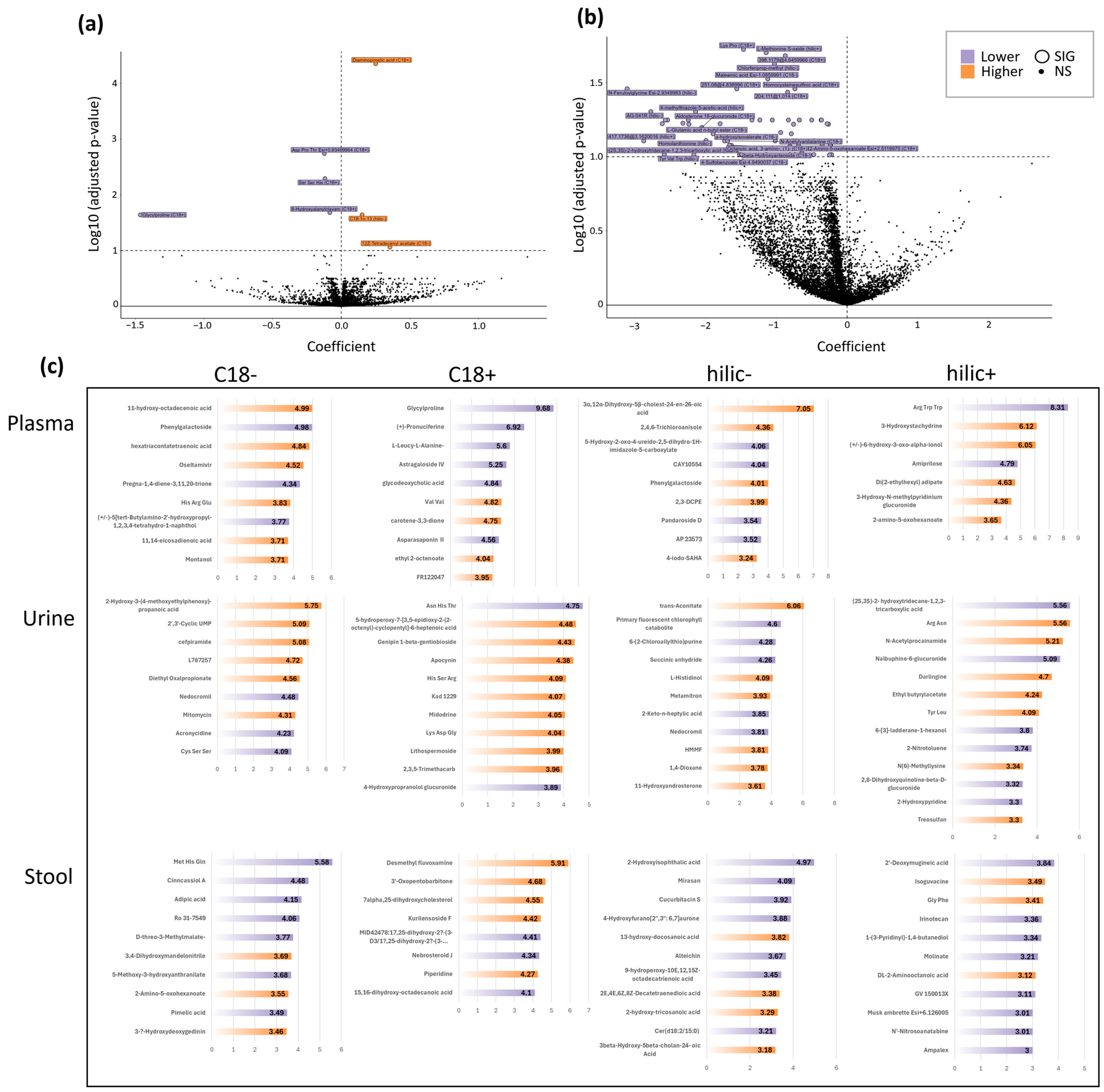
3.5. Untargeted Metabolomics
3.5.1. Untargeted Plasma Metabolomics
3.5.2. Untargeted Urine & Stool Metabolomics
3.5.3. VIP Plots and Metabolite Type Annotation
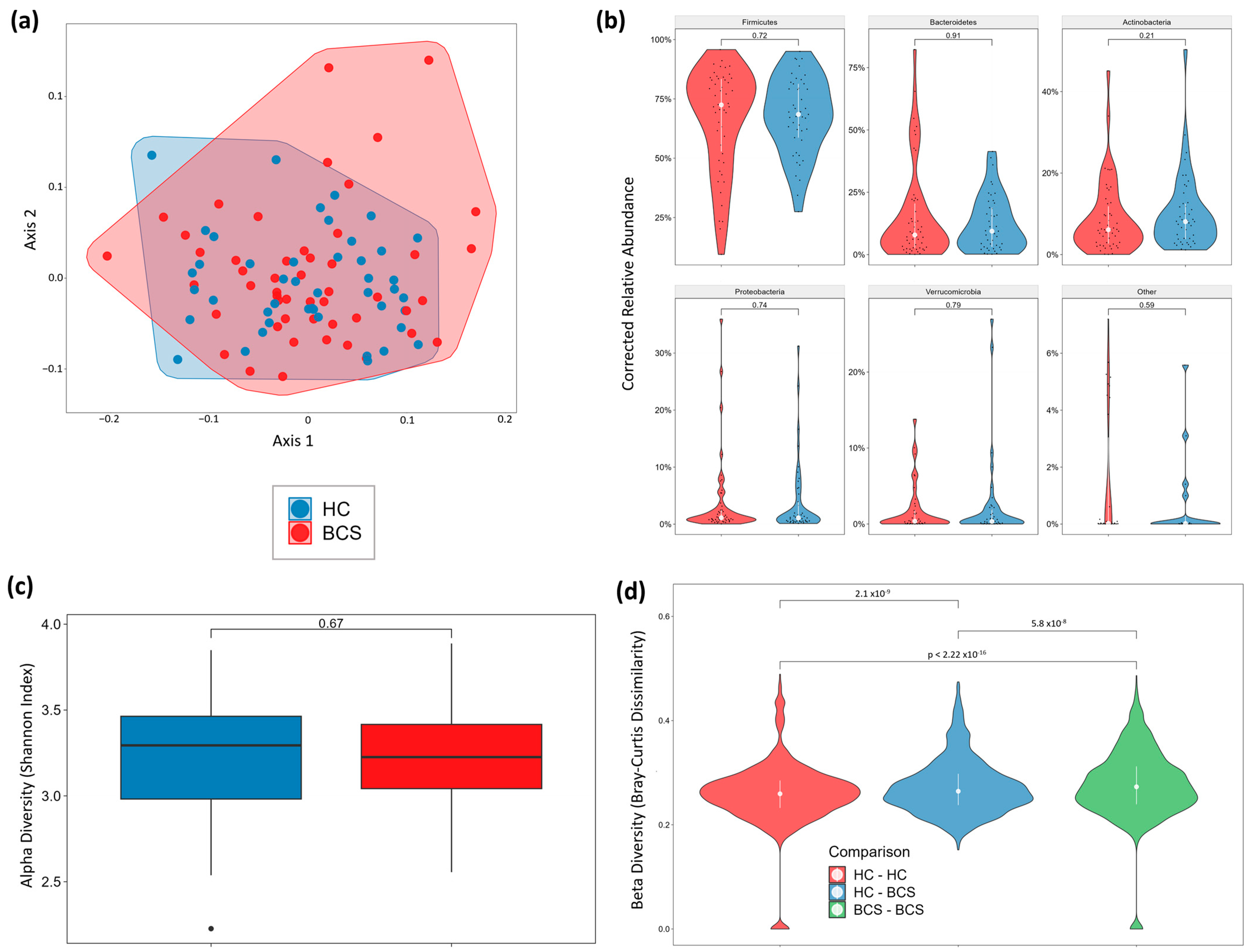
3.6. Gut Microbiome Metagenomics
Stool Microbiome
4. Discussion
4.1. Cohort
4.2. Genetics
4.3. Untargeted Proteomics
4.4. Targeted Metabolomics
4.4.1. Acylcarnitines
4.4.2. Bile Acids
4.4.3. RBC Fatty Acids
4.5. Untargeted Metabolomics
4.6. Metagenomics
5. Conclusions, Future Directions, and Limitations
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A

Appendix B
| PGSa | Wilcox.p.Value | lm.Estimate | lm.p.Value | Wilcox.q.Value | lm.q.Value | Reported.Trait | Mapped.Trait.s...EFO.Label. |
|---|---|---|---|---|---|---|---|
| PGS000511 | 1.3 × 10−7 | 0.136170 | 3.4 × 10−7 | 4.7 × 10−6 | 4.9 × 10−5 | Breast cancer (female) | breast carcinoma |
| PGS000512 | 2.4 × 10−7 | 0.503786 | 1.7 × 10−7 | 7.0 × 10−8 | 4.9 × 10−5 | Breast cancer (female) | breast carcinoma |
| PGS000510 | 2.0 × 10−7 | 0.624025 | 6.3 × 10−7 | 5.8 × 10−6 | 6.0 × 10−5 | Breast cancer (female) | breast carcinoma |
| PGS000507 | 4.7 × 10−7 | 0.144283 | 1.5 × 10−6 | 1.2 × 10−5 | 1.1 × 10−4 | Breast cancer (female) | breast carcinoma |
| PGS000008 | 5.6 × 10−7 | 0.220445 | 8.7 × 10−6 | 1.3 × 10−5 | 5.0 × 10−4 | ER-positive Breast Cancer | estrogen-receptor positive breast cancer |
| PGS000504 | 3.1 × 10−8 | 0.570125 | 1.2 × 10−5 | 1.5 × 10−6 | 5.9 × 10−4 | Breast cancer (female) | breast carcinoma |
| PGS000007 | 3.6 × 10−6 | 0.199725 | 1.8 × 10−5 | 6.5 × 10−5 | 7.4 × 10−4 | Breast Cancer | breast carcinoma |
| PGS000536 | 7.4 × 10−9 | 0.065889 | 2.4 × 10−5 | 5.3 × 10−7 | 8.6 × 10−4 | Breast cancer (female) | breast carcinoma |
| PGS003398 | 1.2 × 10−9 | 0.197326 | 3.0 × 10−5 | 1.7 × 10−7 | 9.7 × 10−4 | Breast Cancer | breast carcinoma |
| PGS000531 | 9.2 × 10−8 | 0.263083 | 3.7 × 10−5 | 3.7 × 10−6 | 1.0 × 10−3 | Breast cancer (female) | breast carcinoma |
| PGS000503 | 6.9 × 10−6 | 0.157900 | 5.0 × 10−5 | 9.0 × 10−5 | 1.3 × 10−3 | Breast cancer (female) | breast carcinoma |
| PGS000539 | 1.5 × 10−7 | 0.059395 | 5.4 × 10−5 | 4.7 × 10−6 | 1.3 × 10−3 | Breast cancer (female) | breast carcinoma |
| PGS000509 | 5.9 × 10−7 | 0.624194 | 6.9 × 10−5 | 1.3 × 10−5 | 1.4 × 10−3 | Breast cancer (female) | breast carcinoma |
| PGS000528 | 1.8 × 10−6 | 0.081497 | 6.8 × 10−5 | 3.8 × 10−5 | 1.4 × 10−3 | Breast cancer (female) | breast carcinoma |
| PGS000508 | 6.0 × 10−6 | 0.149773 | 7.4 × 10−5 | 9.0 × 10−5 | 1.4 × 10−3 | Breast cancer (female) | breast carcinoma |
| PGS000873 | 1.8 × 10−9 | 0.193709 | 1.0 × 10−4 | 1.7 × 10−7 | 1.8 × 10−3 | Breast cancer | breast carcinoma |
| PGS000052 | 1.6 × 10−8 | 0.201100 | 2.4 × 10−4 | 9.3 × 10−7 | 4.2 × 10−3 | Breast cancer | breast carcinoma |
| PGS000535 | 9.8 × 10−5 | 0.061701 | 3.5 × 10−4 | 7.6 × 10−4 | 5.4 × 10−3 | Breast cancer (female) | breast carcinoma |
| PGS003380 | 1.7 × 10−5 | 0.389549 | 3.4 × 10−4 | 2.0 × 10−4 | 5.4 × 10−3 | Breast cancer | breast carcinoma |
| PGS000527 | 3.1 × 10−5 | 0.078368 | 3.8 × 10−4 | 3.5 × 10−4 | 5.5 × 10−3 | Breast cancer (female) | breast carcinoma |
| PGS000214 | 3.0 × 10−6 | 0.269912 | 5.9 × 10−4 | 5.9 × 10−5 | 7.8 × 10−3 | Breast cancer intrinsic-like subtype (luminal B-like) | luminal B breast carcinoma |
| PGS000497 | 6.2 × 10−5 | 0.321878 | 7.0 × 10−4 | 5.5 × 10−4 | 7.8 × 10−3 | Breast cancer (female) | breast carcinoma |
| PGS000498 | 6.2 × 10−5 | 0.321878 | 7.0 × 10−4 | 5.5 × 10−4 | 7.8 × 10−3 | Breast cancer (female) | breast carcinoma |
| PGS000502 | 8.0 × 10−5 | 0.489076 | 6.6 × 10−4 | 6.6 × 10−4 | 7.8 × 10−3 | Breast cancer (female) | breast carcinoma |
| PGS000505 | 4.1 × 10−6 | 0.383257 | 7.0 × 10−4 | 6.6 × 10−5 | 7.8 × 10−3 | Breast cancer (female) | breast carcinoma |
| PGS000506 | 4.1 × 10−6 | 0.383257 | 7.0 × 10−4 | 6.6 × 10−5 | 7.8 × 10−3 | Breast cancer (female) | breast carcinoma |
| PGS000472 | 3.7 × 10−4 | 0.007524 | 7.5 × 10−4 | 2.3 × 10−3 | 8.0 × 10−3 | Breast cancer (female) | breast carcinoma |
| PGS000335 | 1.0 × 10−5 | 0.148391 | 8.3 × 10−4 | 1.2 × 10−4 | 8.5 × 10−3 | Breast cancer | breast carcinoma |
| PGS000499 | 6.9 × 10−5 | 0.163507 | 1.2 × 10−3 | 6.0 × 10−4 | 0.011942 | Breast cancer (female) | breast carcinoma |
| PGS000501 | 5.7 × 10−5 | 0.491264 | 1.2 × 10−3 | 5.5 × 10−4 | 0.011942 | Breast cancer (female) | breast carcinoma |
| PGS000002 | 6.8 × 10−6 | 0.141149 | 1.3 × 10−3 | 9.0 × 10−5 | 0.012108 | ER-positive Breast Cancer | estrogen-receptor positive breast cancer |
| PGS000540 | 1.2 × 10−5 | 0.030463 | 1.3 × 10−3 | 1.4 × 10−4 | 0.012108 | Breast cancer (female) | breast carcinoma |
| PGS000212 | 7.8 × 10−5 | 0.267451 | 1.3 × 10−3 | 6.6 × 10−4 | 0.012123 | Breast cancer intrinsic-like subtype (luminal A-like) | luminal A breast carcinoma |
| PGS000213 | 6.3 × 10−6 | 0.217897 | 2.0 × 10−3 | 9.0 × 10−5 | 0.017031 | Breast cancer intrinsic-like subtype (luminal B/HER2-negative-like) | HER2-negative breast carcinoma |
| PGS000001 | 4.2 × 10−4 | 0.128408 | 2.3 × 10−3 | 2.5 × 10−3 | 0.017187 | Breast Cancer | breast carcinoma |
| PGS000004 | 2.9 × 10−3 | 0.192750 | 2.5 × 10−3 | 1.4 × 10−2 | 0.017187 | Breast Cancer | breast carcinoma |
| PGS000005 | 2.0 × 10−3 | 0.204576 | 2.2 × 10−3 | 1.0 × 10−2 | 0.017187 | ER-positive Breast Cancer | estrogen-receptor positive breast cancer |
| PGS000332 | 4.3 × 10−4 | 0.136928 | 2.1 × 10−3 | 2.5 × 10−3 | 0.017187 | Breast cancer | breast carcinoma |
| PGS000344 | 5.8 × 10−5 | 0.190588 | 2.5 × 10−3 | 5.5 × 10−4 | 0.017187 | Breast cancer | breast carcinoma |
| PGS000473 | 1.2 × 10−4 | 0.038474 | 2.5 × 10−3 | 9.3 × 10−4 | 0.017187 | Breast cancer (female) | breast carcinoma |
| PGS000480 | 1.0 × 10−3 | 0.007254 | 2.5 × 10−3 | 5.5 × 10−3 | 0.017187 | Breast cancer (female) | breast carcinoma |
| PGS000533 | 5.1 × 10−5 | 0.200274 | 2.3 × 10−3 | 5.3 × 10−4 | 0.017187 | Breast cancer (female) | breast carcinoma |
| PGS000534 | 5.1 × 10−5 | 0.200274 | 2.3 × 10−3 | 5.3 × 10−4 | 0.017187 | Breast cancer (female) | breast carcinoma |
| PGS000347 | 1.5 × 10−4 | 0.197233 | 2.6 × 10−3 | 1.1 × 10−3 | 0.017434 | Estrogen receptor-positive breast cancer | estrogen-receptor positive breast cancer |
| PGS000009 | 4.4 × 10−4 | 0.143606 | 2.8 × 10−3 | 2.5 × 10−3 | 0.018270 | ER-negative Breast Cancer | estrogen-receptor-negative breast cancer |
| PGS000538 | 1.3 × 10−4 | 0.182262 | 2.9 × 10−3 | 1.0 × 10−3 | 0.018555 | Breast cancer (female) | breast carcinoma |
| PGS000500 | 2.4 × 10−4 | 0.172722 | 3.8 × 10−3 | 1.6 × 10−3 | 0.023371 | Breast cancer (female) | breast carcinoma |
| PGS000046 | 8.0 × 10−4 | 0.132093 | 5.6 × 10−3 | 4.5 × 10−3 | 0.033573 | Estrogen receptor [ER]-positive breast cancer | estrogen-receptor positive breast cancer |
| PGS000045 | 1.8 × 10−4 | 0.117902 | 6.0 × 10−3 | 1.2 × 10−3 | 0.034689 | Breast cancer | breast carcinoma |
| PGS003399 | 2.1 × 10−4 | 0.192520 | 6.0 × 10−3 | 1.4 × 10−3 | 0.034689 | Breast Cancer | breast carcinoma |
Appendix C
Appendix D

Appendix E

Appendix F
| Metabolite (MS Mode) | Variable | Estimate | p.Value | Qval |
|---|---|---|---|---|
| Lys Pro (C18+) | TxChemoY | −1.466 | 2.11 × 10−06 | 0.0189 |
| L-Methionine S-oxide (hilic+) | TxChemoY | −1.145 | 4.43 × 10−06 | 0.0199 |
| Chlorfenprop-methyl (hilic−) | TxChemoY | −1.028 | 1.06 × 10−05 | 0.0237 |
| Maleamic acid (C18−) | TxChemoY | −1.123 | 1.67 × 10−05 | 0.0299 |
| Homocysteinesulfinic acid (C18+) | TxChemoY | −0.739 | 2.72 × 10−05 | 0.0349 |
| N-Feruloylglycine (hilic−) | TxChemoY | −3.114 | 2.75 × 10−05 | 0.0349 |
| AG-041R (hilic−) | TxChemoY | −2.777 | 5.92 × 10−05 | 0.0497 |
| 4-methylthiazole-5-acetic-acid (hilic+) | TxChemoY | −2.145 | 6.09 × 10−05 | 0.0497 |
| 4-Sulfobenzoate (C18−) | TxChemoY | −1.236 | 8.45 × 10−05 | 0.0566 |
| Caprylic acid (C18−) | TxChemoY | −1.347 | 9.57 × 10−05 | 0.0566 |
| SC-58125 (C18−) | TxChemoY | −2.573 | 1.02 × 10−04 | 0.0566 |
| N-Methyl-2-oxoglutaramate (C18−) | TxChemoY | −0.363 | 1.17 × 10−04 | 0.0566 |
| Lewis a trisaccharide (C18−) | TxChemoY | −0.367 | 1.18 × 10−04 | 0.0566 |
| Lovastatin (hilic+) | TxChemoY | −2.541 | 1.24 × 10−04 | 0.0566 |
| N-Acetylneuraminic Acid (hilic−) | TxChemoY | −2.248 | 1.28 × 10−04 | 0.0566 |
| 3-Keto-scyllo-inosamine (C18+) | TxChemoY | −0.676 | 1.31 × 10−04 | 0.0566 |
| Carboxymethyloxysuccinate (C18+) | TxChemoY | −0.831 | 1.36 × 10−04 | 0.0566 |
| 4-(Trimethylammonio)but-2-enoate (hilic+) | TxChemoY | −0.500 | 1.39 × 10−04 | 0.0566 |
| 9alpha-Fluoro-6alpha-methylprednisolone 21-acetate (C18+) | TxChemoY | −2.617 | 1.65 × 10−04 | 0.0599 |
| 5-Ethyl-5-(1-methyl-3-carboxypropyl)barbituric acid (hilic+) | TxChemoY | −0.283 | 1.68 × 10−04 | 0.0599 |
| Pivalic acid (C18−) | TxChemoY | −0.758 | 1.91 × 10−04 | 0.0604 |
| Lys-Trp-OH (C18−) | TxChemoY | −0.758 | 1.99 × 10−04 | 0.0604 |
| 4-Sulfobenzoate (hilic−) | TxChemoY | −0.263 | 2.02 × 10−04 | 0.0604 |
| Aldosterone 18-glucuronide (C18+) | TxChemoY | −2.057 | 2.20 × 10−04 | 0.0636 |
| L-Glutamic acid n-butyl ester (C18−) | TxChemoY | −1.897 | 2.63 × 10−04 | 0.0700 |
| Dopamine 3-O-sulfate (C18−) | TxChemoY | −0.790 | 2.65 × 10−04 | 0.0700 |
| Homolanthionine (hilic−) | TxChemoY | −1.997 | 3.03 × 10−04 | 0.0777 |
| N-Acetylvanilalanine (C18−) | TxChemoY | −1.691 | 3.32 × 10−04 | 0.0782 |
| 5-aminosalicyluric acid (hilic−) | TxChemoY | −1.017 | 3.37 × 10−04 | 0.0782 |
| a-hydroxyisovalerate (C18−) | TxChemoY | −1.734 | 3.51 × 10−04 | 0.0782 |
| Propanoic acid, 2-hydroxy-3-[(4-hydroxy-1-naphthalenyl)oxy]- (C18−) | TxChemoY | −0.763 | 3.62 × 10−04 | 0.0782 |
| 8-Hydroxyondansetron glucuronide (hilic+) | TxChemoY | −0.724 | 3.64 × 10−04 | 0.0782 |
| 2-Amino-5-oxohexanoate (C18+) | TxChemoY | −1.639 | 4.07 × 10−04 | 0.0837 |
| Octanoic acid, 3-amino-, (1)- (C18+) | TxChemoY | −1.664 | 4.30 × 10−04 | 0.0837 |
| Cardiogenol C (hilic+) | TxChemoY | −0.238 | 4.36 × 10−04 | 0.0837 |
| Betaine (hilic+) | TxChemoY | −0.234 | 4.37 × 10−04 | 0.0837 |
| 2,4-Dichlorophenoxybutyric Acid (C18−) | TxChemoY | −0.353 | 4.39 × 10−04 | 0.0837 |
| 2-Napthyloxyacetic acid (hilic−) | TxChemoY | −0.687 | 4.51 × 10−04 | 0.0843 |
| Ripazepam (C18+) | TxChemoY | −0.809 | 4.64 × 10−04 | 0.0850 |
| L-glycyl-L-hydroxyproline (hilic+) | TxChemoY | −0.641 | 5.25 × 10−04 | 0.0942 |
| 2,3-Dihydroxynaphthalene (C18+) | TxChemoY | −0.809 | 5.52 × 10−04 | 0.0968 |
| Clitidine (hilic+) | TxChemoY | −0.221 | 5.62 × 10−04 | 0.0968 |
| N2-Acetyl-L-aminoadipate (hilic+) | TxChemoY | −0.738 | 5.90 × 10−04 | 0.0968 |
| (2S,3S)-2-hydroxytridecane-1,2,3-tricarboxylic acid (hilic+) | TxChemoY | −2.588 | 5.93 × 10−04 | 0.0968 |
| Ursodeoxycholic acid 3-sulfate (C18−) | TxChemoY | −1.143 | 6.04 × 10−04 | 0.0968 |
| beta-Hydroxyacteoside (C18−) | TxChemoY | −1.497 | 6.13 × 10−04 | 0.0968 |
| L-prolyl-L-proline (C18+) | TxChemoY | −0.471 | 6.21 × 10−04 | 0.0968 |
| Tyr Val Trp (hilic−) | TxChemoY | −2.168 | 6.36 × 10−04 | 0.0968 |
| 4-Sulfobenzoate (C18−) | TxChemoY | −1.522 | 6.37 × 10−04 | 0.0968 |
| DL-Cycloserine (hilic+) | TxChemoY | −0.248 | 6.50 × 10−04 | 0.0971 |
Appendix G

References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA A Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer Statistics, 2023. CA A Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef] [PubMed]
- An, R.; Yu, H.; Wang, Y.; Lu, J.; Gao, Y.; Xie, X.; Zhang, J. Integrative Analysis of Plasma Metabolomics and Proteomics Reveals the Metabolic Landscape of Breast Cancer. Cancer Metab. 2022, 10, 13. [Google Scholar] [CrossRef] [PubMed]
- Barupal, D.K.; Gao, B.; Budczies, J.; Phinney, B.S.; Perroud, B.; Denkert, C.; Fiehn, O. Prioritization of Metabolic Genes as Novel Therapeutic Targets in Estrogen-Receptor Negative Breast Tumors Using Multi-Omics Data and Text Mining. Oncotarget 2019, 10, 3894–3909. [Google Scholar] [CrossRef] [PubMed]
- Bellerba, F.; Chatziioannou, A.C.; Jasbi, P.; Robinot, N.; Keski-Rahkonen, P.; Trolat, A.; Vozar, B.; Hartman, S.J.; Scalbert, A.; Bonanni, B.; et al. Metabolomic Profiles of Metformin in Breast Cancer Survivors: A Pooled Analysis of Plasmas from Two Randomized Placebo-Controlled Trials. J. Transl. Med. 2022, 20, 629. [Google Scholar] [CrossRef] [PubMed]
- Debik, J.; Euceda, L.R.; Lundgren, S.; von der Lippe Gythfeldt, H.; Garred, Ø.; Borgen, E.; Engebraaten, O.; Bathen, T.F.; Giskeødegård, G.F. Assessing Treatment Response and Prognosis by Serum and Tissue Metabolomics in Breast Cancer Patients. J. Proteome Res. 2019, 18, 3649–3660. [Google Scholar] [CrossRef] [PubMed]
- Dowling, P.; Henry, M.; Meleady, P.; Clarke, C.; Gately, K.; O’Byrne, K.; Connolly, E.; Lynch, V.; Ballot, J.; Gullo, G.; et al. Metabolomic and Proteomic Analysis of Breast Cancer Patient Samples Suggests That Glutamate and 12-HETE in Combination with CA15-3 May Be Useful Biomarkers Reflecting Tumour Burden. Metabolomics 2015, 11, 620–635. [Google Scholar] [CrossRef]
- Hassan, M.A.; Al-Sakkaf, K.; Shait Mohammed, M.R.; Dallol, A.; Al-Maghrabi, J.; Aldahlawi, A.; Ashoor, S.; Maamra, M.; Ragoussis, J.; Wu, W.; et al. Integration of Transcriptome and Metabolome Provides Unique Insights to Pathways Associated With Obese Breast Cancer Patients. Front. Oncol. 2020, 10, 804. [Google Scholar] [CrossRef] [PubMed]
- Haukaas, T.H.; Euceda, L.R.; Giskeødegård, G.F.; Lamichhane, S.; Krohn, M.; Jernström, S.; Aure, M.R.; Lingjærde, O.C.; Schlichting, E.; Garred, Ø.; et al. Metabolic Clusters of Breast Cancer in Relation to Gene- and Protein Expression Subtypes. Cancer Metab. 2016, 4, 12. [Google Scholar] [CrossRef]
- Huang, S.; Chong, N.; Lewis, N.E.; Jia, W.; Xie, G.; Garmire, L.X. Novel Personalized Pathway-Based Metabolomics Models Reveal Key Metabolic Pathways for Breast Cancer Diagnosis. Genome Med. 2016, 8, 34. [Google Scholar] [CrossRef]
- Luo, X.; Yu, H.; Song, Y.; Sun, T. Integration of Metabolomic and Transcriptomic Data Reveals Metabolic Pathway Alteration in Breast Cancer and Impact of Related Signature on Survival. J. Cell. Physiol. 2019, 234, 13021–13031. [Google Scholar] [CrossRef] [PubMed]
- Smith, K.S.; Tissier, A.; Bail, J.R.; Novak, J.R.; Morrow, C.D.; Demark-Wahnefried, W.; Frugé, A.D. Health-Related Quality of Life Is Associated with Fecal Microbial Composition in Breast Cancer Survivors. Support. Care Cancer 2022, 31, 10. [Google Scholar] [CrossRef] [PubMed]
- Starodubtseva, N.L.; Tokareva, A.O.; Rodionov, V.V.; Brzhozovskiy, A.G.; Bugrova, A.E.; Chagovets, V.V.; Kometova, V.V.; Kukaev, E.N.; Soares, N.C.; Kovalev, G.I.; et al. Integrating Proteomics and Lipidomics for Evaluating the Risk of Breast Cancer Progression: A Pilot Study. Biomedicines 2023, 11, 1786. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Lin, C.-C.; Spasojevic, I.; Iversen, E.S.; Chi, J.-T.; Marks, J.R. A Joint Analysis of Metabolomics and Genetics of Breast Cancer. Breast Cancer Res. 2014, 16, 415. [Google Scholar] [CrossRef] [PubMed]
- Terunuma, A.; Putluri, N.; Mishra, P.; Mathé, E.A.; Dorsey, T.H.; Yi, M.; Wallace, T.A.; Issaq, H.J.; Zhou, M.; Killian, J.K.; et al. MYC-Driven Accumulation of 2-Hydroxyglutarate Is Associated with Breast Cancer Prognosis. J. Clin. Investig. 2014, 124, 398–412. [Google Scholar] [CrossRef] [PubMed]
- Walker, J.; Joy, A.A.; Vos, L.J.; Stenson, T.H.; Mackey, J.R.; Jovel, J.; Kao, D.; Madsen, K.L.; Wong, G.K.-S. Chemotherapy-Induced Weight Gain in Early-Stage Breast Cancer: A Prospective Matched Cohort Study Reveals Associations with Inflammation and Gut Dysbiosis. BMC Med. 2023, 21, 178. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Ma, D.; Yang, Y.-S.; Yang, F.; Ding, J.-H.; Gong, Y.; Jiang, L.; Ge, L.-P.; Wu, S.-Y.; Yu, Q.; et al. Comprehensive Metabolomics Expands Precision Medicine for Triple-Negative Breast Cancer. Cell Res. 2022, 32, 477–490. [Google Scholar] [CrossRef] [PubMed]
- Meydan, C.; Afshinnekoo, E.; Rickard, N.; Daniels, G.; Kunces, L.; Hardy, T.; Lili, L.; Pesce, S.; Jacobson, P.; Mason, C.E.; et al. Improved Gastrointestinal Health for Irritable Bowel Syndrome with Metagenome-Guided Interventions. Precis. Clin. Med. 2020, 3, 136–146. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and Accurate Short Read Alignment with Burrows-Wheeler Transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Kendig, K.I.; Baheti, S.; Bockol, M.A.; Drucker, T.M.; Hart, S.N.; Heldenbrand, J.R.; Hernaez, M.; Hudson, M.E.; Kalmbach, M.T.; Klee, E.W.; et al. Sentieon DNASeq Variant Calling Workflow Demonstrates Strong Computational Performance and Accuracy. Front. Genet. 2019, 10, 736. [Google Scholar] [CrossRef]
- Freed, D.; Pan, R.; Chen, H.; Li, Z.; Hu, J.; Aldana, R. DNAscope: High Accuracy Small Variant Calling Using Machine Learning. bioRxiv 2022. [Google Scholar] [CrossRef]
- Mills, R.E.; Luttig, C.T.; Larkins, C.E.; Beauchamp, A.; Tsui, C.; Pittard, W.S.; Devine, S.E. An Initial Map of Insertion and Deletion (INDEL) Variation in the Human Genome. Genome Res. 2006, 16, 1182–1190. [Google Scholar] [CrossRef]
- Sherry, S.T.; Ward, M.; Sirotkin, K. dbSNP-Database for Single Nucleotide Polymorphisms and Other Classes of Minor Genetic Variation. Genome Res. 1999, 9, 677–679. [Google Scholar] [PubMed]
- Lambert, S.A.; Gil, L.; Jupp, S.; Ritchie, S.C.; Xu, Y.; Buniello, A.; McMahon, A.; Abraham, G.; Chapman, M.; Parkinson, H.; et al. The Polygenic Score Catalog as an Open Database for Reproducibility and Systematic Evaluation. Nat. Genet. 2021, 53, 420–425. [Google Scholar] [CrossRef] [PubMed]
- Wainberg, M.; Magis, A.T.; Earls, J.C.; Lovejoy, J.C.; Sinnott-Armstrong, N.; Omenn, G.S.; Hood, L.; Price, N.D. Multiomic Blood Correlates of Genetic Risk Identify Presymptomatic Disease Alterations. Proc. Natl. Acad. Sci. USA 2020, 117, 21813–21820. [Google Scholar] [CrossRef] [PubMed]
- Gold, L.; Ayers, D.; Bertino, J.; Bock, C.; Bock, A.; Brody, E.N.; Carter, J.; Dalby, A.B.; Eaton, B.E.; Fitzwater, T.; et al. Aptamer-Based Multiplexed Proteomic Technology for Biomarker Discovery. PLoS ONE 2010, 5, e15004. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.H.; Tworoger, S.S.; Stampfer, M.J.; Dillon, S.T.; Gu, X.; Sawyer, S.J.; Chan, A.T.; Libermann, T.A.; Eliassen, A.H. Stability and Reproducibility of Proteomic Profiles Measured with an Aptamer-Based Platform. Sci. Rep. 2018, 8, 8382. [Google Scholar] [CrossRef] [PubMed]
- Van der Maaten, L.; Hinton, G. Visualizing Data Using T-SNE. J. Mach. Learn. Res. 2008, 9, 2579–2605. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Breitwieser, F.P.; Baker, D.N.; Salzberg, S.L. KrakenUniq: Confident and Fast Metagenomics Classification Using Unique k-Mer Counts. Genome Biol. 2018, 19, 198. [Google Scholar] [CrossRef]
- Lu, J.; Breitwieser, F.P.; Thielen, P.; Salzberg, S.L. Bracken: Estimating Species Abundance in Metagenomics Data. PeerJ Comput. Sci. 2016, 3, e104. [Google Scholar] [CrossRef]
- Beghini, F.; McIver, L.J.; Blanco-Míguez, A.; Dubois, L.; Asnicar, F.; Maharjan, S.; Mailyan, A.; Manghi, P.; Scholz, M.; Thomas, A.M.; et al. Integrating Taxonomic, Functional, and Strain-Level Profiling of Diverse Microbial Communities with bioBakery 3. eLife 2021, 10, e65088. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Bittinger, K.; Charlson, E.S.; Hoffmann, C.; Lewis, J.; Wu, G.D.; Collman, R.G.; Bushman, F.D.; Li, H. Associating Microbiome Composition with Environmental Covariates Using Generalized UniFrac Distances. Bioinformatics 2012, 28, 2106–2113. [Google Scholar] [CrossRef]
- Bray, J.R.; Curtis, J.T. An Ordination of the Upland Forest Communities of Southern Wisconsin. Ecol. Monogr. 1957, 27, 325–349. [Google Scholar] [CrossRef]
- Mallick, H.; Rahnavard, A.; McIver, L.J.; Ma, S.; Zhang, Y.; Nguyen, L.H.; Tickle, T.L.; Weingart, G.; Ren, B.; Schwager, E.H.; et al. Multivariable Association Discovery in Population-Scale Meta-Omics Studies. PLoS Comput. Biol. 2021, 17, e1009442. [Google Scholar] [CrossRef]
- Robinson, M.D.; Oshlack, A. A Scaling Normalization Method for Differential Expression Analysis of RNA-Seq Data. Genome Biol. 2010, 11, R25. [Google Scholar] [CrossRef] [PubMed]
- Janssens, Y.; Nielandt, J.; Bronselaer, A.; Debunne, N.; Verbeke, F.; Wynendaele, E.; Van Immerseel, F.; Vandewynckel, Y.-P.; De Tré, G.; De Spiegeleer, B. Disbiome Database: Linking the Microbiome to Disease. BMC Microbiol. 2018, 18, 50. [Google Scholar] [CrossRef] [PubMed]
- Gunn, S.; Wainberg, M.; Song, Z.; Andersen, S.; Boudreau, R.; Feitosa, M.F.; Tan, Q.; Montasser, M.E.; O’Connell, J.R.; Stitziel, N.; et al. Distribution of 54 Polygenic Risk Scores for Common Diseases in Long Lived Individuals and Their Offspring. Geroscience 2022, 44, 719–729. [Google Scholar] [CrossRef] [PubMed]
- Roberts, E.; Howell, S.; Evans, D.G. Polygenic Risk Scores and Breast Cancer Risk Prediction. Breast 2023, 67, 71–77. [Google Scholar] [CrossRef]
- Gjerde, J.; Geisler, J.; Lundgren, S.; Ekse, D.; Varhaug, J.E.; Mellgren, G.; Steen, V.M.; Lien, E.A. Associations between Tamoxifen, Estrogens, and FSH Serum Levels during Steady State Tamoxifen Treatment of Postmenopausal Women with Breast Cancer. BMC Cancer 2010, 10, 313. [Google Scholar] [CrossRef]
- Zhou, J.; Chen, Y.; Huang, Y.; Long, J.; Wan, F.; Zhang, S. Serum Follicle-Stimulating Hormone Level Is Associated with Human Epidermal Growth Factor Receptor Type 2 and Ki67 Expression in Post-Menopausal Females with Breast Cancer. Oncol. Lett. 2013, 6, 1128–1132. [Google Scholar] [CrossRef]
- Sherbet, G.V.; Cajone, F. Stathmin in Cell Proliferation and Cancer Progression. Cancer Genom. Proteom. 2005, 2, 227–237. [Google Scholar]
- Kuang, X.-Y.; Jiang, H.-S.; Li, K.; Zheng, Y.-Z.; Liu, Y.-R.; Qiao, F.; Li, S.; Hu, X.; Shao, Z.-M. The Phosphorylation-Specific Association of STMN1 with GRP78 Promotes Breast Cancer Metastasis. Cancer Lett. 2016, 377, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Askeland, C.; Wik, E.; Finne, K.; Birkeland, E.; Arnes, J.B.; Collett, K.; Knutsvik, G.; Krüger, K.; Davidsen, B.; Aas, T.; et al. Stathmin Expression Associates with Vascular and Immune Responses in Aggressive Breast Cancer Subgroups. Sci. Rep. 2020, 10, 2914. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Zhen, T.; Lin, Y.; Shao, N.; Kuang, X. The Prognostic Role of a Phospho-Stathmin 1 Signature in Breast Cancer Treated with Neoadjuvant Chemotherapy. Gland. Surg. 2022, 11, 1808–1816. [Google Scholar] [CrossRef] [PubMed]
- Dingjan, I.; Linders, P.T.A.; Verboogen, D.R.J.; Revelo, N.H.; ter Beest, M.; van den Bogaart, G. Endosomal and Phagosomal SNAREs. Physiol. Rev. 2018, 98, 1465–1492. [Google Scholar] [CrossRef]
- Parveen, S.; Khamari, A.; Raju, J.; Coppolino, M.G.; Datta, S. Syntaxin 7 Contributes to Breast Cancer Cell Invasion by Promoting Invadopodia Formation. J. Cell Sci. 2022, 135, jcs259576. [Google Scholar] [CrossRef] [PubMed]
- Maniam, S.; Maniam, S. Small Molecules Targeting Programmed Cell Death in Breast Cancer Cells. Int. J. Mol. Sci. 2021, 22, 9722. [Google Scholar] [CrossRef]
- Luo, H.; Qin, Y.; Reu, F.; Ye, S.; Dai, Y.; Huang, J.; Wang, F.; Zhang, D.; Pan, L.; Zhu, H.; et al. Microarray-Based Analysis and Clinical Validation Identify Ubiquitin-Conjugating Enzyme E2E1 (UBE2E1) as a Prognostic Factor in Acute Myeloid Leukemia. J. Hematol. Oncol. 2016, 9, 125. [Google Scholar] [CrossRef]
- Desai, S.D.; Reed, R.E.; Burks, J.; Wood, L.M.; Pullikuth, A.K.; Haas, A.L.; Liu, L.F.; Breslin, J.W.; Meiners, S.; Sankar, S. ISG15 Disrupts Cytoskeletal Architecture and Promotes Motility in Human Breast Cancer Cells. Exp. Biol. Med. 2012, 237, 38–49. [Google Scholar] [CrossRef]
- Du, X.; Song, H.; Shen, N.; Hua, R.; Yang, G. The Molecular Basis of Ubiquitin-Conjugating Enzymes (E2s) as a Potential Target for Cancer Therapy. Int. J. Mol. Sci. 2021, 22, 3440. [Google Scholar] [CrossRef] [PubMed]
- Voutsadakis, I.A. Ubiquitin- and Ubiquitin-like Proteins-Conjugating Enzymes (E2s) in Breast Cancer. Mol. Biol. Rep. 2013, 40, 2019–2034. [Google Scholar] [CrossRef] [PubMed]
- Picon-Ruiz, M.; Morata-Tarifa, C.; Valle-Goffin, J.J.; Friedman, E.R.; Slingerland, J.M. Obesity and Adverse Breast Cancer Risk and Outcome: Mechanistic Insights and Strategies for Intervention. CA Cancer J. Clin. 2017, 67, 378–397. [Google Scholar] [CrossRef] [PubMed]
- Mihalik, S.J.; Goodpaster, B.H.; Kelley, D.E.; Chace, D.H.; Vockley, J.; Toledo, F.G.S.; DeLany, J.P. Increased Levels of Plasma Acylcarnitines in Obesity and Type 2 Diabetes and Identification of a Marker of Glucolipotoxicity. Obesity 2010, 18, 1695–1700. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Yan, L.; Liu, S.; Ambrosone, C.B.; Zhao, H. Plasma Metabolomic Profiles in Breast Cancer Patients and Healthy Controls: By Race and Tumor Receptor Subtypes. Transl. Oncol. 2013, 6, 757–765. [Google Scholar] [CrossRef] [PubMed]
- Ozmen, H.K.; Erdemci, B.; Askin, S.; Sezen, O. Carnitine and Adiponectin Levels in Breast Cancer after Radiotherapy. Open Med. 2017, 12, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Režen, T.; Rozman, D.; Kovács, T.; Kovács, P.; Sipos, A.; Bai, P.; Mikó, E. The Role of Bile Acids in Carcinogenesis. Cell. Mol. Life Sci. 2022, 79, 243. [Google Scholar] [CrossRef] [PubMed]
- Giordano, C.; Catalano, S.; Panza, S.; Vizza, D.; Barone, I.; Bonofiglio, D.; Gelsomino, L.; Rizza, P.; Fuqua, S.A.W.; Andò, S. Farnesoid X Receptor Inhibits Tamoxifen-Resistant MCF-7 Breast Cancer Cell Growth through Downregulation of HER2 Expression. Oncogene 2011, 30, 4129–4140. [Google Scholar] [CrossRef] [PubMed]
- Journe, F.; Durbecq, V.; Chaboteaux, C.; Rouas, G.; Laurent, G.; Nonclercq, D.; Sotiriou, C.; Body, J.-J.; Larsimont, D. Association between Farnesoid X Receptor Expression and Cell Proliferation in Estrogen Receptor-Positive Luminal-like Breast Cancer from Postmenopausal Patients. Breast Cancer Res. Treat. 2009, 115, 523–535. [Google Scholar] [CrossRef]
- Luo, C.; Zhang, X.; He, Y.; Chen, H.; Liu, M.; Wang, H.; Tang, L.; Tu, G.; Ding, M. A Pseudo-Targeted Metabolomics Study Based on Serum Bile Acids Profiling for the Differential Diagnosis of Benign and Malignant Breast Lesions. Steroids 2021, 175, 108914. [Google Scholar] [CrossRef]
- Costarelli, V.; Sanders, T.A.B. Plasma Deoxycholic Acid Concentration Is Elevated in Postmenopausal Women with Newly Diagnosed Breast Cancer. Eur. J. Clin. Nutr. 2002, 56, 925–927. [Google Scholar] [CrossRef][Green Version]
- Wang, N.; Yang, J.; Han, W.; Han, M.; Liu, X.; Jiang, L.; Cao, H.; Jing, M.; Sun, T.; Xu, J. Identifying Distinctive Tissue and Fecal Microbial Signatures and the Tumor-Promoting Effects of Deoxycholic Acid on Breast Cancer. Front. Cell. Infect. Microbiol. 2022, 12. [Google Scholar] [CrossRef]
- Tang, W.; Putluri, V.; Ambati, C.R.; Dorsey, T.H.; Putluri, N.; Ambs, S. Liver- and Microbiome-Derived Bile Acids Accumulate in Human Breast Tumors and Inhibit Growth and Improve Patient Survival. Clin. Cancer Res. 2019, 25, 5972–5983. [Google Scholar] [CrossRef] [PubMed]
- Cala, M.P.; Aldana, J.; Medina, J.; Sánchez, J.; Guio, J.; Wist, J.; Meesters, R.J.W. Multiplatform Plasma Metabolic and Lipid Fingerprinting of Breast Cancer: A Pilot Control-Case Study in Colombian Hispanic Women. PLoS ONE 2018, 13, e0190958. [Google Scholar] [CrossRef]
- Pakiet, A.; Jędrzejewska, A.; Duzowska, K.; Wacławska, A.; Jabłońska, P.; Zieliński, J.; Mika, A.; Śledziński, T.; Słomińska, E. Serum Fatty Acid Profiles in Breast Cancer Patients Following Treatment. BMC Cancer 2023, 23, 433. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Chen, T.; Dong, L.; Li, T.; Xue, H.; Gao, B.; Ding, X.; Wang, H.; Li, H. Fatty Acid Synthase Promotes Breast Cancer Metastasis by Mediating Changes in Fatty Acid Metabolism. Oncol. Lett. 2020, 21, 1. [Google Scholar] [CrossRef]
- Hidaka, B.H.; Li, S.; Harvey, K.E.; Carlson, S.E.; Sullivan, D.K.; Kimler, B.F.; Zalles, C.M.; Fabian, C.J. Omega-3 and Omega-6 Fatty Acids in Blood and Breast Tissue of High-Risk Women and Association with Atypical Cytomorphology. Cancer Prev. Res. 2015, 8, 359–364. [Google Scholar] [CrossRef] [PubMed]
- Fabian, C.J.; Kimler, B.F.; Hursting, S.D. Omega-3 Fatty Acids for Breast Cancer Prevention and Survivorship. Breast Cancer Res. 2015, 17, 62. [Google Scholar] [CrossRef] [PubMed]
- Nindrea, R.D.; Aryandono, T.; Lazuardi, L.; Dwiprahasto, I. Association of Dietary Intake Ratio of N-3/n-6 Polyunsaturated Fatty Acids with Breast Cancer Risk in Western and Asian Countries: A Meta-Analysis. Asian Pac. J. Cancer Prev. 2019, 20, 1321–1327. [Google Scholar] [CrossRef]
- Yang, B.; Ren, X.-L.; Fu, Y.-Q.; Gao, J.-L.; Li, D. Ratio of N-3/n-6 PUFAs and Risk of Breast Cancer: A Meta-Analysis of 274135 Adult Females from 11 Independent Prospective Studies. BMC Cancer 2014, 14, 105. [Google Scholar] [CrossRef]
- Bel’skaya, L.V.; Gundyrev, I.A.; Solomatin, D.V. The Role of Amino Acids in the Diagnosis, Risk Assessment, and Treatment of Breast Cancer: A Review. Curr. Issues Mol. Biol. 2023, 45, 7513–7537. [Google Scholar] [CrossRef]
- Lai, H.-S.; Lee, J.-C.; Lee, P.-H.; Wang, S.-T.; Chen, W.-J. Plasma Free Amino Acid Profile in Cancer Patients. Semin. Cancer Biol. 2005, 15, 267–276. [Google Scholar] [CrossRef]
- Yang, L.; Wang, Y.; Cai, H.; Wang, S.; Shen, Y.; Ke, C. Application of Metabolomics in the Diagnosis of Breast Cancer: A Systematic Review. J. Cancer 2020, 11, 2540–2551. [Google Scholar] [CrossRef]
- Jobard, E.; Dossus, L.; Baglietto, L.; Fornili, M.; Lécuyer, L.; Mancini, F.R.; Gunter, M.J.; Trédan, O.; Boutron-Ruault, M.-C.; Elena-Herrmann, B.; et al. Investigation of Circulating Metabolites Associated with Breast Cancer Risk by Untargeted Metabolomics: A Case–Control Study Nested within the French E3N Cohort. Br. J. Cancer 2021, 124, 1734–1743. [Google Scholar] [CrossRef]
- Lécuyer, L.; Victor Bala, A.; Deschasaux, M.; Bouchemal, N.; Nawfal Triba, M.; Vasson, M.-P.; Rossary, A.; Demidem, A.; Galan, P.; Hercberg, S.; et al. NMR Metabolomic Signatures Reveal Predictive Plasma Metabolites Associated with Long-Term Risk of Developing Breast Cancer. Int. J. Epidemiol. 2018, 47, 484–494. [Google Scholar] [CrossRef]
- Stevens, V.L.; Carter, B.D.; Jacobs, E.J.; McCullough, M.L.; Teras, L.R.; Wang, Y. A Prospective Case–Cohort Analysis of Plasma Metabolites and Breast Cancer Risk. Breast Cancer Res. 2023, 25, 5. [Google Scholar] [CrossRef]
- His, M.; Viallon, V.; Dossus, L.; Gicquiau, A.; Achaintre, D.; Scalbert, A.; Ferrari, P.; Romieu, I.; Onland-Moret, N.C.; Weiderpass, E.; et al. Prospective Analysis of Circulating Metabolites and Breast Cancer in EPIC. BMC Med. 2019, 17, 178. [Google Scholar] [CrossRef]
- Catchpole, G.; Platzer, A.; Weikert, C.; Kempkensteffen, C.; Johannsen, M.; Krause, H.; Jung, K.; Miller, K.; Willmitzer, L.; Selbig, J.; et al. Metabolic Profiling Reveals Key Metabolic Features of Renal Cell Carcinoma. J. Cell. Mol. Med. 2011, 15, 109–118. [Google Scholar] [CrossRef]
- Hirayama, A.; Kami, K.; Sugimoto, M.; Sugawara, M.; Toki, N.; Onozuka, H.; Kinoshita, T.; Saito, N.; Ochiai, A.; Tomita, M.; et al. Quantitative Metabolome Profiling of Colon and Stomach Cancer Microenvironment by Capillary Electrophoresis Time-of-Flight Mass Spectrometry. Cancer Res. 2009, 69, 4918–4925. [Google Scholar] [CrossRef]
- Zareba, I.; Celinska-Janowicz, K.; Surazynski, A.; Miltyk, W.; Palka, J. Proline Oxidase Silencing Induces Proline-Dependent pro-Survival Pathways in MCF-7 Cells. Oncotarget 2018, 9, 13748–13757. [Google Scholar] [CrossRef][Green Version]
- Lewoniewska, S.; Oscilowska, I.; Forlino, A.; Palka, J. Understanding the Role of Estrogen Receptor Status in PRODH/POX-Dependent Apoptosis/Survival in Breast Cancer Cells. Biology 2021, 10, 1314. [Google Scholar] [CrossRef]
- Subramani, R.; Poudel, S.; Smith, K.D.; Estrada, A.; Lakshmanaswamy, R. Metabolomics of Breast Cancer: A Review. Metabolites 2022, 12, 643. [Google Scholar] [CrossRef]
- Nam, H.; Chung, B.C.; Kim, Y.; Lee, K.; Lee, D. Combining Tissue Transcriptomics and Urine Metabolomics for Breast Cancer Biomarker Identification. Bioinformatics 2009, 25, 3151–3157. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Jiang, J.; Hua, L.; Jiang, D.; Wang, Y.; Li, D.; Wang, R.; Zhang, X.; Li, H. Rapid Detection of Volatile Organic Metabolites in Urine by High-Pressure Photoionization Mass Spectrometry for Breast Cancer Screening: A Pilot Study. Metabolites 2023, 13, 870. [Google Scholar] [CrossRef]
- Slupsky, C.M.; Steed, H.; Wells, T.H.; Dabbs, K.; Schepansky, A.; Capstick, V.; Faught, W.; Sawyer, M.B. Urine Metabolite Analysis Offers Potential Early Diagnosis of Ovarian and Breast Cancers. Clin. Cancer Res. 2010, 16, 5835–5841. [Google Scholar] [CrossRef]
- Cala, M.; Aldana, J.; Sánchez, J.; Guio, J.; Meesters, R.J.W. Urinary Metabolite and Lipid Alterations in Colombian Hispanic Women with Breast Cancer: A Pilot Study. J. Pharm. Biomed. Anal. 2018, 152, 234–241. [Google Scholar] [CrossRef]
- Woo, H.M.; Kim, K.M.; Choi, M.H.; Jung, B.H.; Lee, J.; Kong, G.; Nam, S.J.; Kim, S.; Bai, S.W.; Chung, B.C. Mass Spectrometry Based Metabolomic Approaches in Urinary Biomarker Study of Women’s Cancers. Clin. Chim. Acta 2009, 400, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Zhang, L.; Du, Y.; Du, W.; Liu, D.; Guo, C.; Pan, Y.; Tang, D. Enrichment and Quantitative Determination of 5-(Hydroxymethyl)-2’-Deoxycytidine, 5-(Formyl)-2’-Deoxycytidine, and 5-(Carboxyl)-2’-Deoxycytidine in Human Urine of Breast Cancer Patients by Magnetic Hyper-Cross-Linked Microporous Polymers Based on Polyionic Liquid. Anal. Chem. 2018, 90, 3906–3913. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhang, R.; Song, Y.; He, J.; Sun, J.; Bai, J.; An, Z.; Dong, L.; Zhan, Q.; Abliz, Z. RRLC-MS/MS-Based Metabonomics Combined with in-Depth Analysis of Metabolic Correlation Network: Finding Potential Biomarkers for Breast Cancer. Analyst 2009, 134, 2003–2011. [Google Scholar] [CrossRef]
- Zahran, F.; Rashed, R.; Omran, M.; Darwish, H.; Belal, A. Study on Urinary Candidate Metabolome for the Early Detection of Breast Cancer. Indian. J. Clin. Biochem. 2021, 36, 319–329. [Google Scholar] [CrossRef]
- Silva, C.L.; Olival, A.; Perestrelo, R.; Silva, P.; Tomás, H.; Câmara, J.S. Untargeted Urinary 1H NMR-Based Metabolomic Pattern as a Potential Platform in Breast Cancer Detection. Metabolites 2019, 9, 269. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Jiang, C.; Huang, S.; Gong, X.; Wang, S.; Shen, P. Analysis of Urinary Metabolites for Breast Cancer Patients Receiving Chemotherapy by CE-MS Coupled with on-Line Concentration. Clin. Biochem. 2013, 46, 1065–1073. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.J.; Takita, C.; Reis, I.M.; Yang, G.; Zhao, W.; Lee, E. Abstract 2328: Metabolomics Pathways and Biomarkers in Predicting Breast Cancer Prognosis. Cancer Res. 2022, 82, 2328. [Google Scholar] [CrossRef]





| Metric 1 | BCS | HC | |
|---|---|---|---|
| Subjects (N) | 50 | 50 | |
| Age, yrs (mean (SD)) | 62.8 (9.91) | 63.2 (9.7) | |
| BMI, kg/m2 (mean (SD)) | 28.8 (5.9) | 27.5 (5) | |
| Breast Cancer Type | Ductal: 35 (70%) Lobular: 12 (24%) Mixed: 3 (6%) | NA | |
| Treatment Type (N (%)) | Chemo: 10 (20%) Endo: 34 (68%) Radio: 32 (64%) | NA | |
| Tamoxifen Use (N (%)) | 16 (32%) | NA | |
| Breast Cancer Stage at Diagnosis (N (%)) | Stage 0: 7 (14%) Stage I: 31 (62%) Stage II: 8 (16%) Stage III: 4 (8%) | NA | |
| Type II Diabetes—Pre-Treatment | Yes: 5 (10%) No: 45 (90%) | NA | |
| Type II Diabetes—Post-Treatment | Yes: 5 (10%) 3 No: 45 (90%) | NA | |
| Type II Diabetes Health Cohort | NA | Yes: 6 (12%) No: 45 (88%) | |
| HER2 BrCa Status | Negative: 40 (80%) Positive: 3 (6%) Unknown: 7 (14%) | NA | |
| BRCA1 BrCa Status | Negative: 17 (34%) Pathogenic: 1 (2%) Not tested: 31 (62%) VUS: 1 (2%) | NA | |
| BRCA2 BrCa Status | Negative: 17 (34%) Pathogenic: 1 (2%) Not tested: 31 (62%) VUS: 1 (2%) | NA | |
| Menopause Status (N (%)) | Pre: 8 (16%) Post: 42 (84%) | Pre: 6 (12%) Post: 44 (88%) | |
| PHQ-8 Total Score (mean (SD)) | 1.58 (2.59) | 1.06 (1.62) | |
| GAD-7 Total Score (mean (SD)) | 1.16 (2.71) | 1.30 (2.70) | |
| Biotics (N (%)) | Prebiotics | 3 (6%) | 1 (2%) |
| Probiotics | 1 (2%) | 0 (0%) | |
| Antibiotics | 2 (4%) | 1 (2%) | |
| History (N (%)) | Diabetes | 5 (10%) | 6 (12%) |
| Blood Pressure | 14 (28%) | 20 (40%) | |
| Depression | 9 (18%) | 5 (10%) | |
| Anxiety 2 | 12 (24%) | 2 (4%) | |
| Pain | 7 (14%) | 3 (6%) | |
| Heart Problems | 3 (6%) | 3 (6%) | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bauer, B.A.; Schmidt, C.M.; Ruddy, K.J.; Olson, J.E.; Meydan, C.; Schmidt, J.C.; Smith, S.Y.; Couch, F.J.; Earls, J.C.; Price, N.D.; et al. A Multiomics, Molecular Atlas of Breast Cancer Survivors. Metabolites 2024, 14, 396. https://doi.org/10.3390/metabo14070396
Bauer BA, Schmidt CM, Ruddy KJ, Olson JE, Meydan C, Schmidt JC, Smith SY, Couch FJ, Earls JC, Price ND, et al. A Multiomics, Molecular Atlas of Breast Cancer Survivors. Metabolites. 2024; 14(7):396. https://doi.org/10.3390/metabo14070396
Chicago/Turabian StyleBauer, Brent A., Caleb M. Schmidt, Kathryn J. Ruddy, Janet E. Olson, Cem Meydan, Julian C. Schmidt, Sheena Y. Smith, Fergus J. Couch, John C. Earls, Nathan D. Price, and et al. 2024. "A Multiomics, Molecular Atlas of Breast Cancer Survivors" Metabolites 14, no. 7: 396. https://doi.org/10.3390/metabo14070396
APA StyleBauer, B. A., Schmidt, C. M., Ruddy, K. J., Olson, J. E., Meydan, C., Schmidt, J. C., Smith, S. Y., Couch, F. J., Earls, J. C., Price, N. D., Dudley, J. T., Mason, C. E., Zhang, B., Phipps, S. M., & Schmidt, M. A. (2024). A Multiomics, Molecular Atlas of Breast Cancer Survivors. Metabolites, 14(7), 396. https://doi.org/10.3390/metabo14070396






