Pharmacometabolomics Enables Real-World Drug Metabolism Sciences
Abstract
1. Introduction
2. Materials and Methods
2.1. A Literature Study of CsA Metabolites
2.2. Clinical Samples
2.3. LC-SWATH/MS-Based Pharmacometabolomics Analyses
2.4. Data Processing
3. Results and Discussion
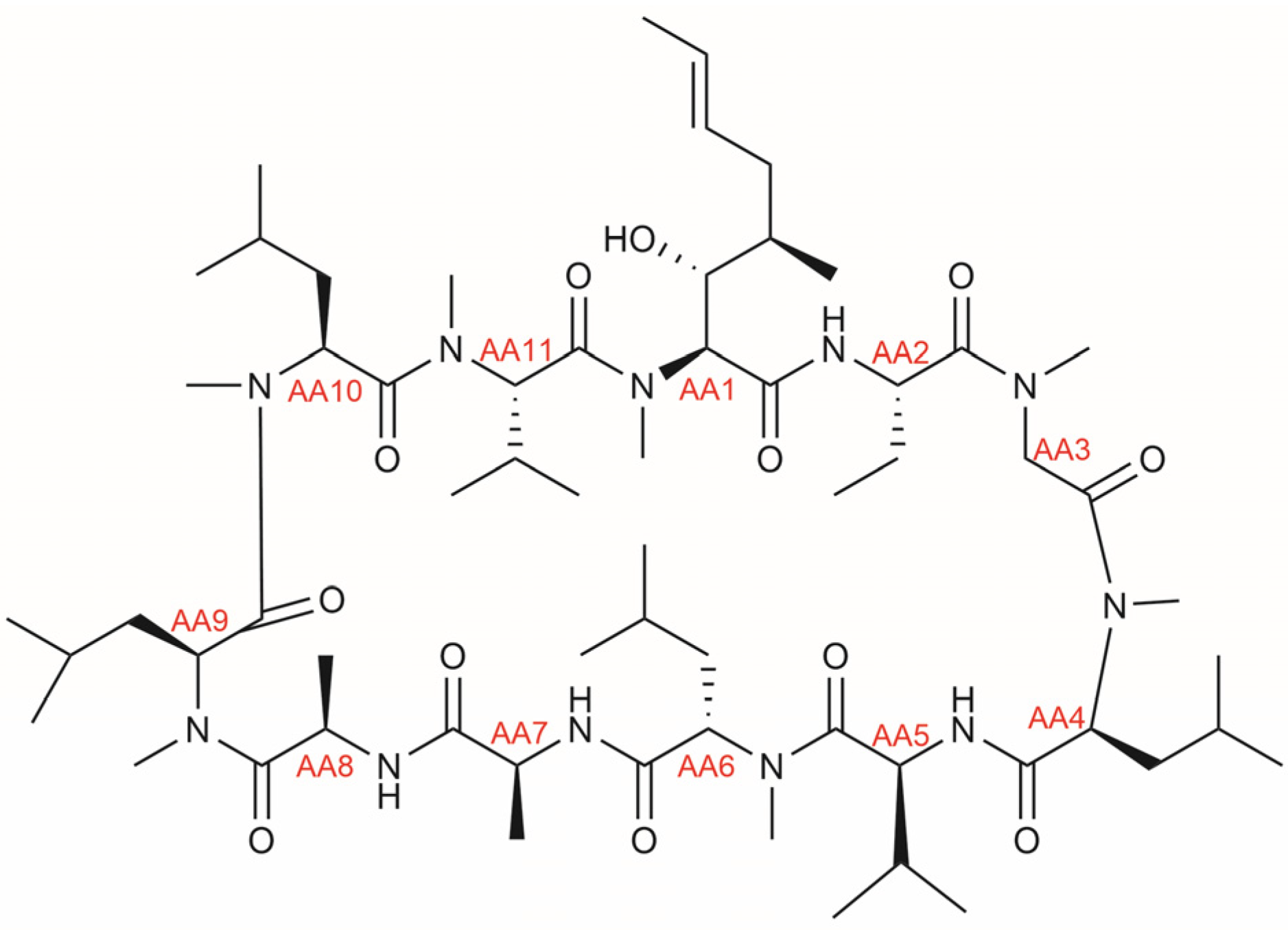
3.1. CsA Metabolites in Literature
| Substance Code | Molecular Formula | Monoisotopic Mass | Modification Reported on Position | Detected in | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 4 | 6 | 9 | Unknown | Bile | Blood | Urine | |||
| AM4N | C61H109N11O12 | 1187.83 | -CH3 | [31,33] | [28,29] | [26] | ||||
| CsA | C62H111N11O12 | 1201.84 | [30,31] | [25,28,29] | [26] | |||||
| AM4N9 | C61H109N11O13 | 1203.82 | -CH3 | +OH | [33] | [29] | [26] | |||
| AM14N | C61H109N11O13 | 1203.82 | +OH | -CH3 | [33] | [29] | ||||
| UM32.8min | C61H109N11O13 | 1203.82 | +OH | -CH3 | [33] | |||||
| AM1AL | C62H109N11O13 | 1215.82 | ald. | [33] | ||||||
| AM1 | C62H111N11O13 | 1217.84 | +OH | [31,33] | [28,29] | [26] | ||||
| AM1c | C62H111N11O13 | 1217.84 | cycl. | [31,33] | [29] | [26] | ||||
| AM9 | C62H111N11O13 | 1217.84 | +OH | [31,33] | [28,29] | [26] | ||||
| AM4N69 | C61H109N11O14 | 1219.82 | -CH3 | +OH | +OH | [33] | [26] | |||
| UM21.2min | C61H109N11O14 | 1219.82 | +OH | -CH3 | +OH | [33] | ||||
| AM1DI | C62H113N11O13 | 1219.85 | +OH +sat. | [31] | [27] | [27] | ||||
| AM1A | C62H109N11O14 | 1231.82 | carbox. | [30,33] | [29] | |||||
| AM1Ac | C62H109N11O14 | 1231.82 | carbox. cycl. | [30] | ||||||
| AM19 | C62H111N11O14 | 1233.83 | +OH | +OH | [31,33] | [29] | [26] | |||
| AM1c9 | C62H111N11O14 | 1233.83 | cycl. | +OH | [33] | [29] | ||||
| AM49 | C62H111N11O14 | 1233.83 | +OH | +OH | [31,33] | [26] | ||||
| AM69 | C62H111N11O14 | 1233.83 | +OH | +OH | [31,33] | [26] | ||||
| AM11d | C62H111N11O14 | 1233.83 | +2×OH | [33] | ||||||
| UM19.8min | C61H109N11O15 | 1235.81 | +OH | -CH3 | +2×OH | [33] | ||||
| UM23.0min | C61H109N11O15 | 1235.81 | -CH3 | +3×OH | [33] | |||||
| AM1DI9 | C62H113N11O14 | 1235.85 | +OH +sat. | +OH | [31] | |||||
| UM26.0min | C62H109N11O15 | 1247.81 | carbox. | +OH | [33] | |||||
| UM20.6min | C62H111N11O15 | 1249.83 | +OH | +2×OH | [33] | |||||
| UM22.4min | C62H111N11O15 | 1249.83 | +OH | +2×OH | [33] | |||||
| UM24.4min | C62H111N11O15 | 1249.83 | +OH | +2×OH | [33] | |||||
| UM25.5min | C62H111N11O15 | 1249.83 | +OH | +2×OH | [33] | |||||
| AM1S | C62H111N11O15S | 1281.80 | +sul. | [32] | [32] | |||||
| AM1c-Glc | C68H119N11O19 | 1393.87 | cycl. +glu. | [33] | ||||||
3.2. Sample Analysis
3.3. Characteristics of Kidney and Liver Transplant Recipients
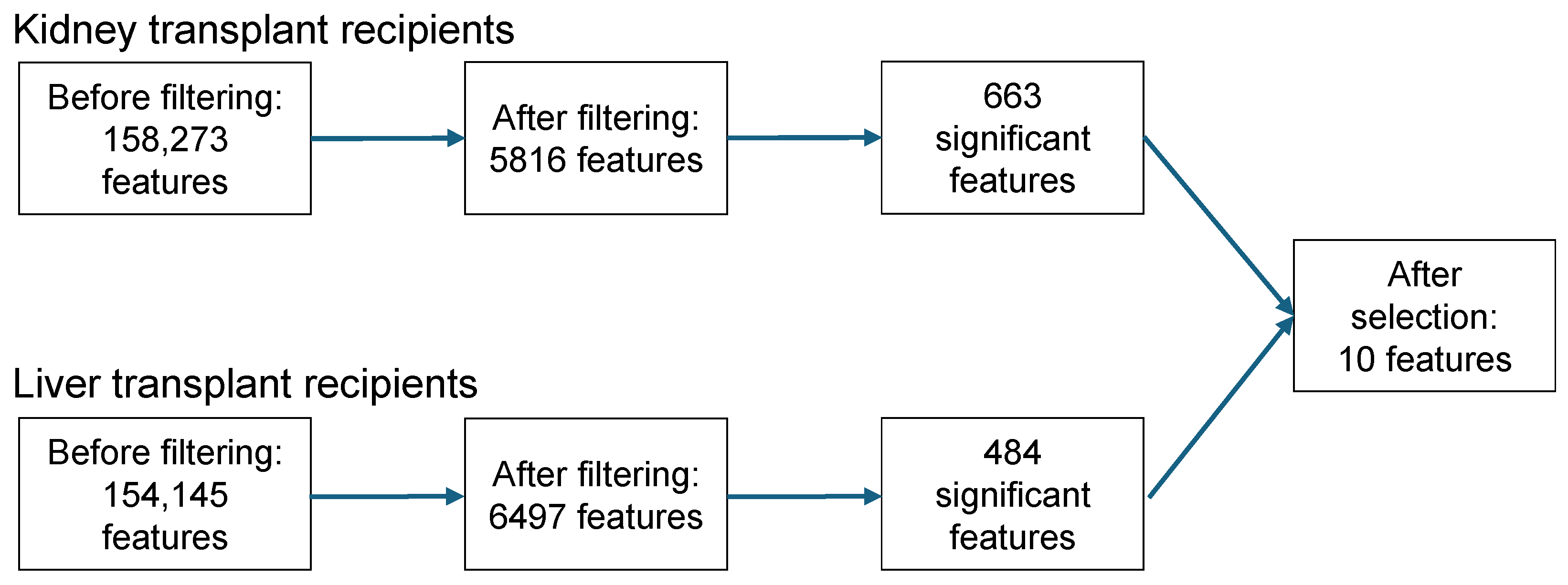
3.4. Feature Selection
3.5. Metabolite Identification (As Level 3 “Putatively Characterized Compound Classes”, According to the Metabolomics Standards Initiative, MSI [22])
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, R.C.; Wang, Z. Precision Medicine: Disease Subtyping and Tailored Treatment. Cancers 2023, 15, 3837. [Google Scholar] [CrossRef] [PubMed]
- Kantae, V.; Krekels, E.H.J.; Esdonk, M.J.V.; Lindenburg, P.; Harms, A.C.; Knibbe, C.A.J.; Van der Graaf, P.H.; Hankemeier, T. Integration of Pharmacometabolomics with Pharmacokinetics and Pharmacodynamics: Towards Personalized Drug Therapy. Metabolomics 2017, 13, 9. [Google Scholar] [CrossRef]
- Relling, M.V.; Klein, T.E. CPIC: Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network. Clin. Pharmacol. Ther. 2011, 89, 464–467. [Google Scholar] [CrossRef] [PubMed]
- Swen, J.J.; Huizinga, T.W.; Gelderblom, H.; De Vries, E.G.E.; Assendelft, W.J.J.; Kirchheiner, J.; Guchelaar, H.J. Translating Pharmacogenomics: Challenges on the Road to the Clinic. PLoS ONE 2007, 4, e209. [Google Scholar] [CrossRef]
- Gervasini, G.; Benítez, J.; Carrillo, J.A. Pharmacogenetic Testing and Therapeutic Drug Monitoring Are Complementary Tools for Optimal Individualization of Drug Therapy. Eur. J. Clin. Pharmacol. 2010, 66, 755–774. [Google Scholar] [CrossRef]
- Ramamoorthy, A.; Bende, G.; Chow, E.C.Y.; Dimova, H.; Hartman, N.; Jean, D.; Pahwa, S.; Ren, Y.; Shukla, C.; Yang, Y.; et al. Human Radiolabeled Mass Balance Studies Supporting the FDA Approval of New Drugs. Clin. Transl. Sci. 2022, 15, 2567–2575. [Google Scholar] [CrossRef] [PubMed]
- Klont, F.; Stepanović, S.; Kremer, D.; Bonner, R.; Touw, D.J.; Hak, E.; Bakker, S.J.L.; Hopfgartner, G. Untargeted ‘SWATH’ Mass Spectrometry-Based Metabolomics for Studying Chronic and Intermittent Exposure to Xenobiotics in Cohort Studies. Food Chem. Toxicol. 2022, 165, 113188. [Google Scholar] [CrossRef]
- Klont, F.; Sosnowski, P.; Kremer, D.; Knobbe, T.J.; Bonner, R.; Blokzijl, H.; Weersma, R.K.; Bakker, S.J.L.; Investigators, T.L.; Hak, E.; et al. Assessing the Potential of Untargeted SWATH Mass Spectrometry-Based Metabolomics to Differentiate Closely Related Exposures in Observational Studies. Metabolites 2022, 12, 942. [Google Scholar] [CrossRef]
- Beger, R.D.; Schmidt, M.A.; Kaddurah-Daouk, R. Current Concepts in Pharmacometabolomics, Biomarker Discovery, and Precision Medicine. Metabolites 2020, 10, 129. [Google Scholar] [CrossRef] [PubMed]
- Clayton, T.A.; Lindon, J.C.; Cloarec, O.; Antti, H.; Charuel, C.; Hanton, G.; Provost, J.-P.; Le Net, J.-L.; Baker, D.; Walley, R.J.; et al. Pharmaco-Metabonomic Phenotyping and Personalized Drug Treatment. Nature 2006, 440, 1073–1077. [Google Scholar] [CrossRef]
- Tran, D.T.; Dahlin, A. Pharmacometabolomics: General Applications of Metabolomics in Drug Development and Personalized Medicine. In Metabolomics; Springer International Publishing: Cham, Switzerland, 2023; pp. 127–164. [Google Scholar]
- Emwas, A.H.; Szczepski, K.; McKay, R.T.; Asfour, H.; Chang, C.K.; Lachowicz, J.; Jaremko, M. Pharmacometabolomics: A New Horizon in Personalized Medicine. In Metabolomics—Methodology and Applications in Medical Sciences and Life Sciences; IntechOpen: London, UK, 2021. [Google Scholar] [CrossRef]
- Jian, J.; He, D.; Gao, S.; Tao, X.; Dong, X. Pharmacokinetics in Pharmacometabolomics: Towards Personalized Medication. Pharmaceuticals 2023, 16, 1568. [Google Scholar] [CrossRef]
- Balashova, E.E.; Maslov, D.L.; Lokhov, P.G. A Metabolomics Approach to Pharmacotherapy Personalization. J. Pers. Med. 2018, 8, 28. [Google Scholar] [CrossRef]
- Rattray, N.J.W.; Daouk, R.K. Pharmacometabolomics and Precision Medicine Special Issue Editorial. Metabolomics 2017, 13, 59. [Google Scholar] [CrossRef][Green Version]
- Klont, F.; Jahn, S.; Grivet, C.; König, S.; Bonner, R.; Hopfgartner, G. SWATH Data Independent Acquisition Mass Spectrometry for Screening of Xenobiotics in Biological Fluids: Opportunities and Challenges for Data Processing. Talanta 2020, 211, 120747. [Google Scholar] [CrossRef] [PubMed]
- Cheung, F.; Wong, P.Y.; Loo, J.; Cole, E.H.; Levy, G.A. Identification of Cyclosporine Metabolites in Human Bile, Blood, and Urine by High-Performance Liquid Chromatography/Radioimmunoassay/Fast Atomic Bombardment Mass Spectroscopy. Transplant. Proc. 1988, 20, 602–608. [Google Scholar] [PubMed]
- Christians, U.; Schlitt, H.J.; Bleck, J.S.; Schiebel, H.M.; Kownatzki, R.; Maurer, G.; Strohmeyer, S.S.; Schottmann, R.; Wonigeit, K.; Pichlmayr, R.; et al. Measurement of Cyclosporine and 18 Metabolites in Blood, Bile, and Urine by High-Performance Liquid Chromatography Extraction for Semipreparative Isolation. Transplant. Proc. 1988, 20, 609–613. [Google Scholar]
- Yatscoff, R.W.; Rosano, T.G.; Bowers, L.D. The Clinical Significance of Cyclosporine Metabolites. Clin. Biochem. 1991, 24, 23–35. [Google Scholar] [CrossRef]
- Klont, F.; Nijdam, F.B.; Bakker, S.J.L.; Keski-Rahkonen, P.; Hopfgartner, G.; Investigators, T. High-Abundance Peaks and Peak Clusters Associate with Pharmaceutical Polymers and Excipients in Urinary Untargeted Clinical Metabolomics Data: Exploration of Their Origin and Possible Impact on Label-Free Quantification. Analyst 2024, 149, 1061–1067. [Google Scholar] [CrossRef]
- Eisenga, M.F.; Gomes-Neto, A.W.; Van Londen, M.; Ziengs, A.L.; Douwes, R.M.; Stam, S.P.; Osté, M.C.J.; Knobbe, T.J.; Hessels, N.R.; Buunk, A.M.; et al. Rationale and Design of TransplantLines: A Prospective Cohort Study and Biobank of Solid Organ Transplant Recipients. BMJ Open 2018, 8, e024502. [Google Scholar] [CrossRef]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.M.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed Minimum Reporting Standards for Chemical Analysis: Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef] [PubMed]
- The EndNote Team. EndNote, Version 20; Clarivate: Philadelphia, PA, USA, 2013.
- Klont, F.; Kremer, D.; Gomes Neto, A.W.; Berger, S.P.; Touw, D.J.; Hak, E.; Bonner, R.; Bakker, S.J.L.; Hopfgartner, G. Metabolomics Data Complemented Drug Use Information in Epidemiological Databases: Pilot Study of Potential Kidney Donors. J. Clin. Epidemiol. 2021, 135, 10–16. [Google Scholar] [CrossRef]
- Yee, G.C.; Gmur, D.J.; Kennedy, M.S. Liquid-Chromatographic Determination of Cyclosporine in Serum with Use of a Rapid Extraction Procedure. Clin. Chem. 1982, 28, 2269–2271. [Google Scholar] [CrossRef]
- Maurer, G.; Loosli, H.A.; Schreier, E.; Keller, B. Disposition of Cyclosporine in Several Animal Species and Man: I. Structural Elucidation of Its Metabolites. Drug Metab. Dispos. 1984, 12, 120–126. [Google Scholar] [PubMed]
- Meier, G.P.; Park, S.B.; Yee, G.C.; Gmur, D.J. Isolation and Identification of a Novel Human Metabolite of Cyclosporin A: Dihydro-CsA M17. Drug Metab. Dispos. 1990, 18, 68–71. [Google Scholar] [PubMed]
- Rosano, T.; Freed, B.; Cerilli, J.; Lempert, N. Immunosuppressive Metabolites of Cyclosporine in the Blood of Renal Allograft Recipients. Transplantation 1986, 42, 262–267. [Google Scholar] [CrossRef] [PubMed]
- Lensmeyer, G.; Wiebe, D.; Carlson, I. Identification and Analysis of Nine Metabolites of Cyclosporine in Whole Blood by Liquid Chromatography. 1: Purification of Analytical Standards and Optimization of the Assay. Clin. Chem. 1987, 33, 1841–1850. [Google Scholar] [CrossRef] [PubMed]
- Hartman, N.; Trimble, L.; Vederas, J.; Jardine, I. An Acid Metabolite of Cyclosporine. Biochem. Biophys. Res. Commun. 1985, 133, 964–971. [Google Scholar] [CrossRef]
- Wang, C.P.; Hartman, N.R.; Venkataramanan, R.; Jardine, I.; Lin, F.-T.; Knapp, J.E.; Starzl, T.E.; Clinical, G.J.B.; Laboratory, P. Isolation of 10 Cyclosporine Metabolites from Human Bile. Drug Metab. Dispos. 1989, 17, 292–296. [Google Scholar] [PubMed]
- Henricsson, S. A Sulfate Conjugate of Cyclosporin. Pharmacol. Toxicol. 1990, 66, 53–55. [Google Scholar] [CrossRef]
- Christians, U.; Strohmeyer, S.; Kownatzki, R.; Schiebel, H.; Bleck, J.; Greipel, J.; Kohlhaw, K.; Schottmann, R.; Sewing, K. Investigations on the Metabolic Pathways of Cyclosporine: I. Excretion of Cyclosporine and Its Metabolites in Human Bile—Isolation of 12 New Cyclosporine Metabolites. Xenobiotica 1991, 21, 1185–1198. [Google Scholar] [CrossRef]
- Trevor, G.R.; Lim, Y.J.; Urquhart, B.L. Pharmacometabolomics in Drug Disposition, Toxicity, and Precision Medicine. Drug Metab. Dispos. 2024, 52, 1187–1195. [Google Scholar] [CrossRef] [PubMed]
- Saigusa, D.; Matsukawa, N.; Hishinuma, E.; Koshiba, S. Identification of Biomarkers to Diagnose Diseases and Find Adverse Drug Reactions by Metabolomics. Drug Metab. Pharmacokinet. 2021, 37, 100373. [Google Scholar] [CrossRef] [PubMed]
- Kelly, R.S.; Cote, M.F.; Begum, S.; Lasky-Su, J. Pharmacometabolomics of Asthma as a Road Map to Precision Medicine. In Metabolomics and Its Impact on Health and Diseases; Ghini, V., Stringer, K.A., Luchinat, C., Eds.; Springer International Publishing: Cham, Switzerland, 2022; pp. 247–273. [Google Scholar]
- Amaro, F.; Carvalho, M.; Bastos, M.d.L.; de Pinho, P.G.; Pinto, J. Pharmacometabolomics Applied to Personalized Medicine in Urological Cancers. Pharmaceuticals 2022, 15, 295. [Google Scholar] [CrossRef] [PubMed]
- Gianazza, E.; Brioschi, M.; Iezzi, A.; Paglia, G.; Banfi, C. Pharmacometabolomics for the Study of Lipid-Lowering Therapies: Opportunities and Challenges. Int. J. Mol. Sci. 2023, 24, 3291. [Google Scholar] [CrossRef] [PubMed]
- Everett, J.R. Pharmacometabonomics: The Prediction of Drug Effects Using Metabolic Profiling. In Concepts and Principles of Pharmacology; Barrett, J., Page, C., Michel, M., Eds.; Springer International Publishing: Cham, Switzerland, 2019; Volume 260, pp. 263–299. [Google Scholar]
- Hissong, R.; Evans, K.R.; Evans, C.R. Compound Identification Strategies in Mass Spectrometry-Based Metabolomics and Pharmacometabolomics. In Metabolomics and Its Impact on Health and Diseases; Ghini, V., Stringer, K.A., Luchinat, C., Eds.; Springer International Publishing: Cham, Switzerland, 2022; pp. 43–71. [Google Scholar]
- Vignoli, A.; Takis, P.; Montuschi, P. Editorial: Pharmacometabolomics: Biomarker Discovery, Precision Medicine, Technical Advances, Perspectives and Future Applications in Respiratory Diseases. Front. Mol. Biosci. 2023, 10, 1268001. [Google Scholar] [CrossRef] [PubMed]
| Kidney Transplant Recipients | Liver Transplant Recipients | |||||
|---|---|---|---|---|---|---|
| Characteristic As Median (IQR) or n (%) | CsA Users n = 126 | CsA Nonusers n = 606 | p-Value | CsA Users n = 38 | CsA Nonusers n = 312 | p-Value |
| Age (years) | 59 (51, 66) | 58 (48, 66) | 0.09 | 60 (47, 66) | 58 (46, 66) | 0.95 |
| Female sex | 62 (49%) | 230 (38%) | 0.03 | 18 (47%) | 134 (43%) | 0.73 |
| BMI (kg/m2) | 26.7 (24.0, 29.6) | 26.5 (23.9, 30.1) | 0.68 | 25.7 (22.5, 27.5) | 25.9 (23.3, 29.6) | 0.34 |
| Smoking status | 0.92 | 0.27 | ||||
| Current | 11 (11%) | 48 (12%) | 1 (3.3%) | 31 (12%) | ||
| Former | 38 (39%) | 147 (37%) | 12 (40%) | 79 (30%) | ||
| Never | 48 (49%) | 202 (51%) | 17 (57%) | 154 (58%) | ||
| Alcohol units per week | 0.5 (0.0, 3.5) | 1.2 (0.0, 6.2) | 0.13 | 0.0 (0.0, 0.9) | 0.0 (0.0, 0.5) | 0.20 |
| eGFR (mL/min/1.73 m2) | 48 (37, 62) | 55 (41, 66) | 0.009 | 75 (61, 97) | 73 (57, 92) | 0.37 |
| Serum albumin (g/L) | 43 (41, 45) | 44 (42, 46) | 0.02 | 44 (42, 47) | 44 (42, 46) | 0.83 |
| ALT (U/L) | 17 (13, 22) | 19 (14, 24) | 0.03 | 26 (18, 31) | 25 (18, 35) | 0.96 |
| Serum CRP (mg/L) | 1.8 (0.7, 5.0) | 1.8 (0.8, 4.6) | 0.88 | 1.8 (0.7, 4.2) | 2.0 (0.9, 4.7) | 0.41 |
| Serum glucose (mmol/L) | 5.6 (5.1, 6.5) | 5.5 (5.0, 6.3) | 0.28 | 5.6 (5.0, 6.4) | 5.6 (5.2, 6.7) | 0.80 |
| Urinary albumin (mg/24 h) | 41 (11, 179) | 32 (11, 147) | 0.50 | 14 (9, 53) | 13 (7, 49) | 0.41 |
| Time since transplantation (years) | 9 (6, 17) | 5 (2, 11) | <0.001 | 14 (10, 22) | 8 (3, 17) | <0.001 |
| Tacrolimus use | 0 (0%) | 451 (74%) | <0.001 | 2 (5.3%) | 209 (67%) | <0.001 |
| Mycophenolate use | 75 (60%) | 463 (76%) | <0.001 | 4 (11%) | 86 (28%) | 0.04 |
| Azathioprine use | 16 (13%) | 65 (11%) | 0.63 | 15 (39%) | 74 (24%) | 0.06 |
| mTOR inhibitor use | 1 (0.8%) | 23 (3.8%) | 0.15 | 1 (2.6%) | 48 (15%) | 0.06 |
| Prednisolone use | 125 (99%) | 577 (95%) | 0.07 | 23 (61%) | 104 (33%) | 0.002 |
| Histamine H2-receptor antagonist use | 3 (2.4%) | 25 (4.1%) | 0.50 | 1 (2.6%) | 26 (8.3%) | 0.36 |
| Calcium channel blocker use | 29 (23%) | 238 (39%) | <0.001 | 8 (21%) | 53 (17%) | 0.69 |
| ACE inhibitor use | 40 (32%) | 151 (25%) | 0.14 | 3 (7.9%) | 55 (18%) | 0.20 |
| Statin use | 71 (56%) | 336 (55%) | 0.93 | 7 (18%) | 70 (22%) | 0.72 |
| Kidney Transplant Recipients | Liver Transplant Recipients | ||||
|---|---|---|---|---|---|
| m/z 1 | RT (min) | Rel. Median (%) 2 | p-Value | Rel. Median (%) 2 | p-Value |
| 594.92 | 15.0 | 2.2 | 4.3 × 10−152 | 2.6 | 4.2 × 10−63 |
| 601.92 | 15.2 | 1.9 | 1.4 × 10−150 | 2.0 | 3.3 × 10−60 |
| 602.92 | 14.6 | 4.0 | 1.1 × 10−150 | 3.9 | 3.9 × 10−48 |
| 608.92 | 14.6 | 3.4 | 2.8 × 10−138 | - | n.s. |
| 608.92 | 14.8 | - | n.s. | 3.2 | 4.1 × 10−66 |
| 609.92 | 14.8 | 100.0 | 6.5 × 10−142 | 100.0 | 2.7 × 10−40 |
| 616.92 | 14.6 | 2.0 | 3.9 × 10−154 | 3.2 | 1.4 × 10−67 |
| 617.92 | 14.2 | 30.3 | 1.1 × 10−150 | 34.5 | 3.5 × 10−66 |
| 624.91 | 14.2 | 1.0 | 9.8 × 10−152 | - | n.s. |
| 670.43 | 14.1 | 1.4 | 3.9 × 10−154 | 2.0 | 2.9 × 10−72 |
| Substance | Molecular Formula | Monoisotopic Mass | m/z 1 | RT (min) | Median Metabolite Abundance 2 in KTR (%) | Median Metabolite Abundance 2 in LTR (%) |
|---|---|---|---|---|---|---|
| Demethylcyclosporine | C61H109N11O12 | 1187.83 | 594.92 | 15.0 | 1.41 | 1.38 |
| Cyclosporine | C62H111N11O12 | 1201.84 | 601.92 | 15.2 | 1.01 | 0.73 |
| Demethylhydroxycyclosporine | C61H109N11O13 | 1203.82 | 602.92 | 14.6 14.8 | 2.37 1.36 | 2.52 0.98 |
| Cyclosporine aldehyde | C62H109N11O13 | 1215.82 | 608.92 | 14.6 14.8 | 0.75 1.27 | 0.73 1.25 |
| Hydroxycyclosporine | C62H111N11O13 | 1217.84 | 609.92 | 14.8 | 64.01 | 60.66 |
| Demethyldihydroxycyclosporine | C62H113N11O13 | 1219.85 | 610.92 | 14.3 | 1.10 | 1.19 |
| Cyclosporine carboxylic acid | C62H109N11O14 | 1231.82 | 616.92 | 14.6 | 0.61 | 1.15 |
| Dihydroxycyclosporine | C62H111N11O14 | 1233.83 | 617.92 | 14.2 14.7 | 20.29 4.35 | 21.99 4.75 |
| Hydroxycyclosporine carboxylic acid | C62H109N11O15 | 1247.81 | 624.91 | 14.2 | 0.76 | 1.54 |
| Unknown metabolite | C65H118N12O15S | 1338.86 | 670.43 | 14.1 | 0.71 | 1.13 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nijdam, F.B.; Hof, M.A.J.; Blokzijl, H.; Bakker, S.J.L.; Hak, E.; Hopfgartner, G.; Klont, F.; on behalf of the TransplantLines Investigators. Pharmacometabolomics Enables Real-World Drug Metabolism Sciences. Metabolites 2025, 15, 39. https://doi.org/10.3390/metabo15010039
Nijdam FB, Hof MAJ, Blokzijl H, Bakker SJL, Hak E, Hopfgartner G, Klont F, on behalf of the TransplantLines Investigators. Pharmacometabolomics Enables Real-World Drug Metabolism Sciences. Metabolites. 2025; 15(1):39. https://doi.org/10.3390/metabo15010039
Chicago/Turabian StyleNijdam, Fleur B., Marieke A. J. Hof, Hans Blokzijl, Stephan J. L. Bakker, Eelko Hak, Gérard Hopfgartner, Frank Klont, and on behalf of the TransplantLines Investigators. 2025. "Pharmacometabolomics Enables Real-World Drug Metabolism Sciences" Metabolites 15, no. 1: 39. https://doi.org/10.3390/metabo15010039
APA StyleNijdam, F. B., Hof, M. A. J., Blokzijl, H., Bakker, S. J. L., Hak, E., Hopfgartner, G., Klont, F., & on behalf of the TransplantLines Investigators. (2025). Pharmacometabolomics Enables Real-World Drug Metabolism Sciences. Metabolites, 15(1), 39. https://doi.org/10.3390/metabo15010039







