Chemical Diversity of UK-Grown Tea Explored Using Metabolomics and Machine Learning
Abstract
:1. Introduction
2. Materials and Methods
2.1. Tea Plants
2.2. Fresh Leaf Sample Collection
2.3. Sample Preparation and Extraction
2.4. Sample Preparation for Analysis
2.5. Flow Infusion Electrospray Ionisation Mass Spectrometry (FIE-MS)
2.6. Data Analysis
3. Results
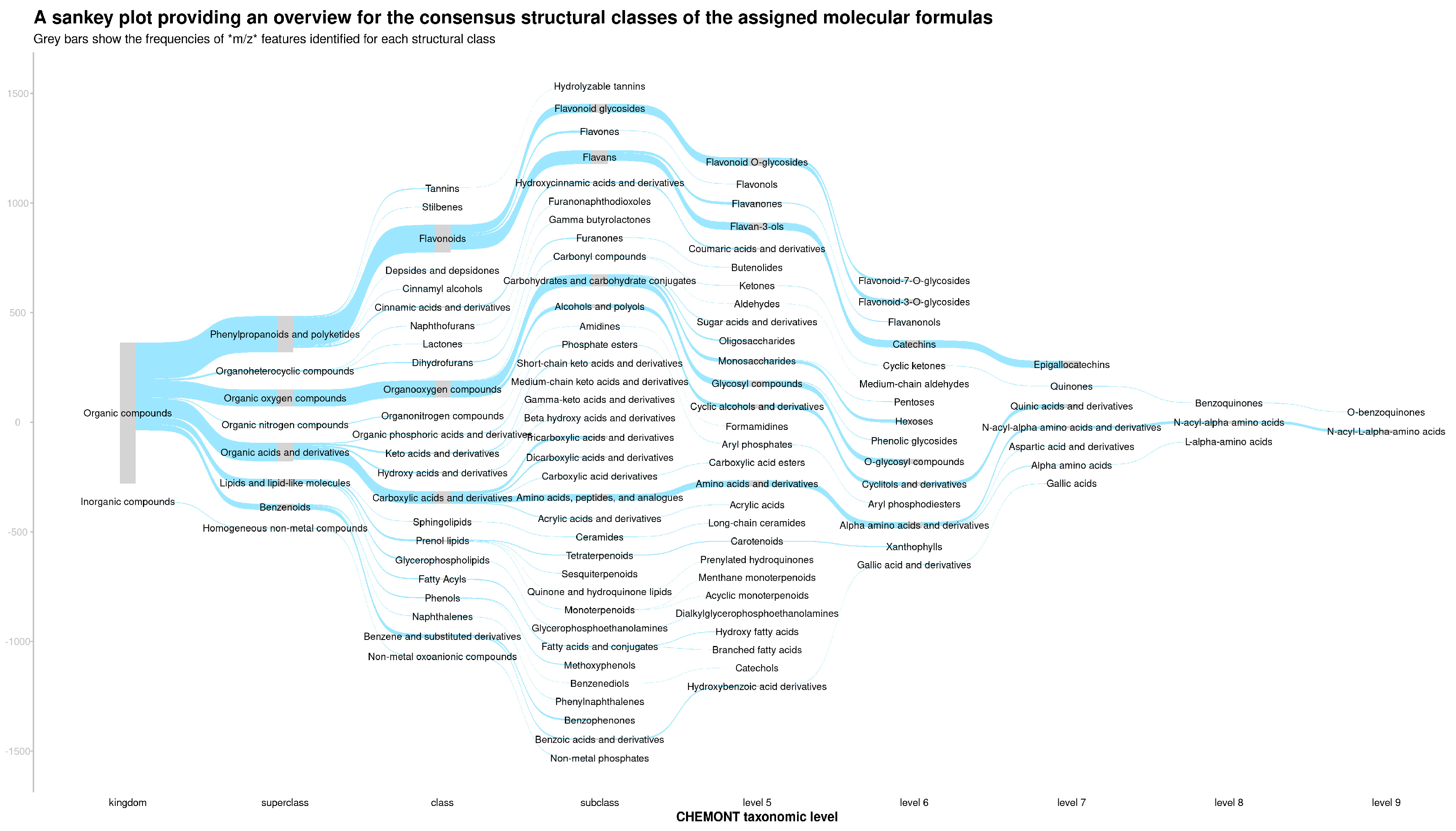
3.1. Structural Composition of the Tea Samples
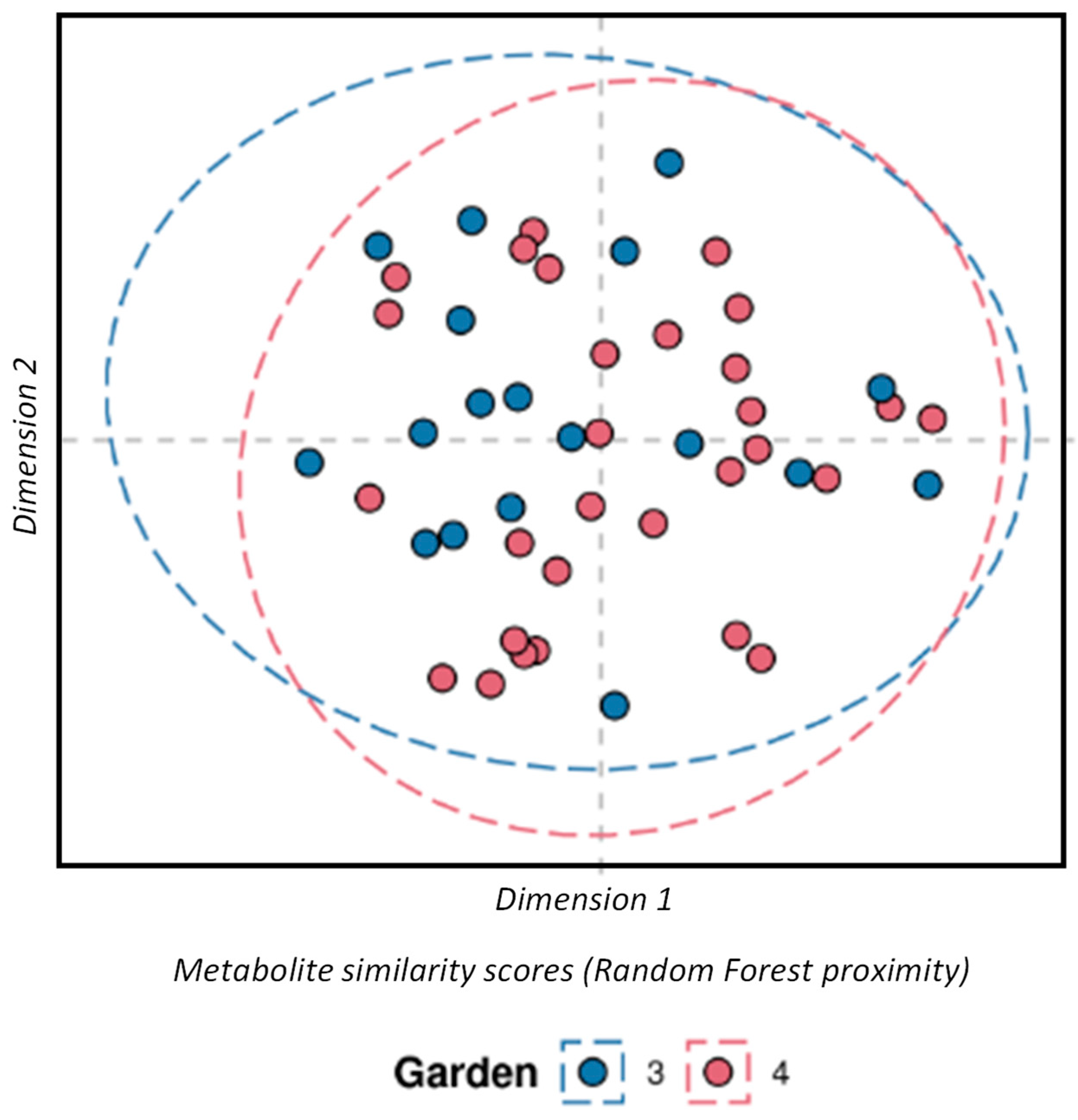
3.2. Variability Within the Georgian Tea
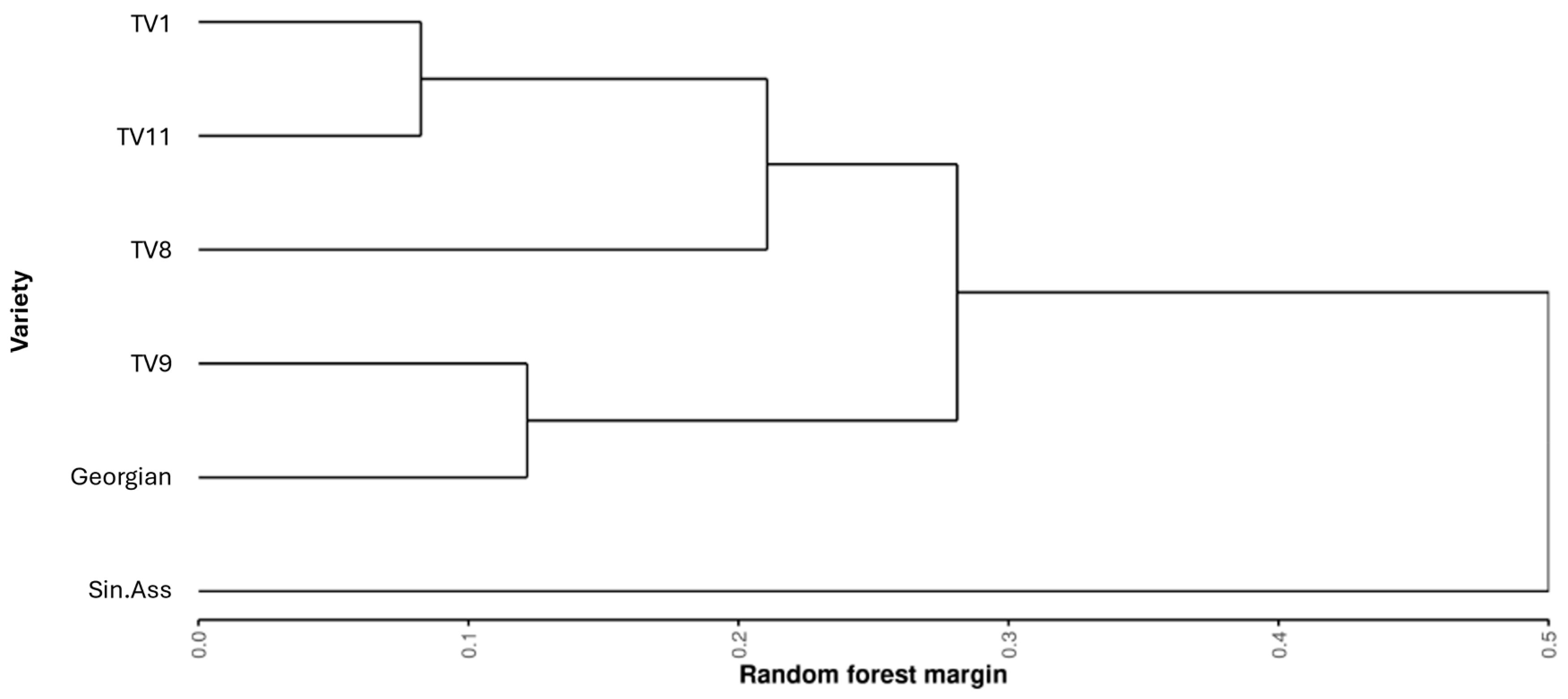
3.3. Variability Within the Tocklai Tri-Clonal Variants
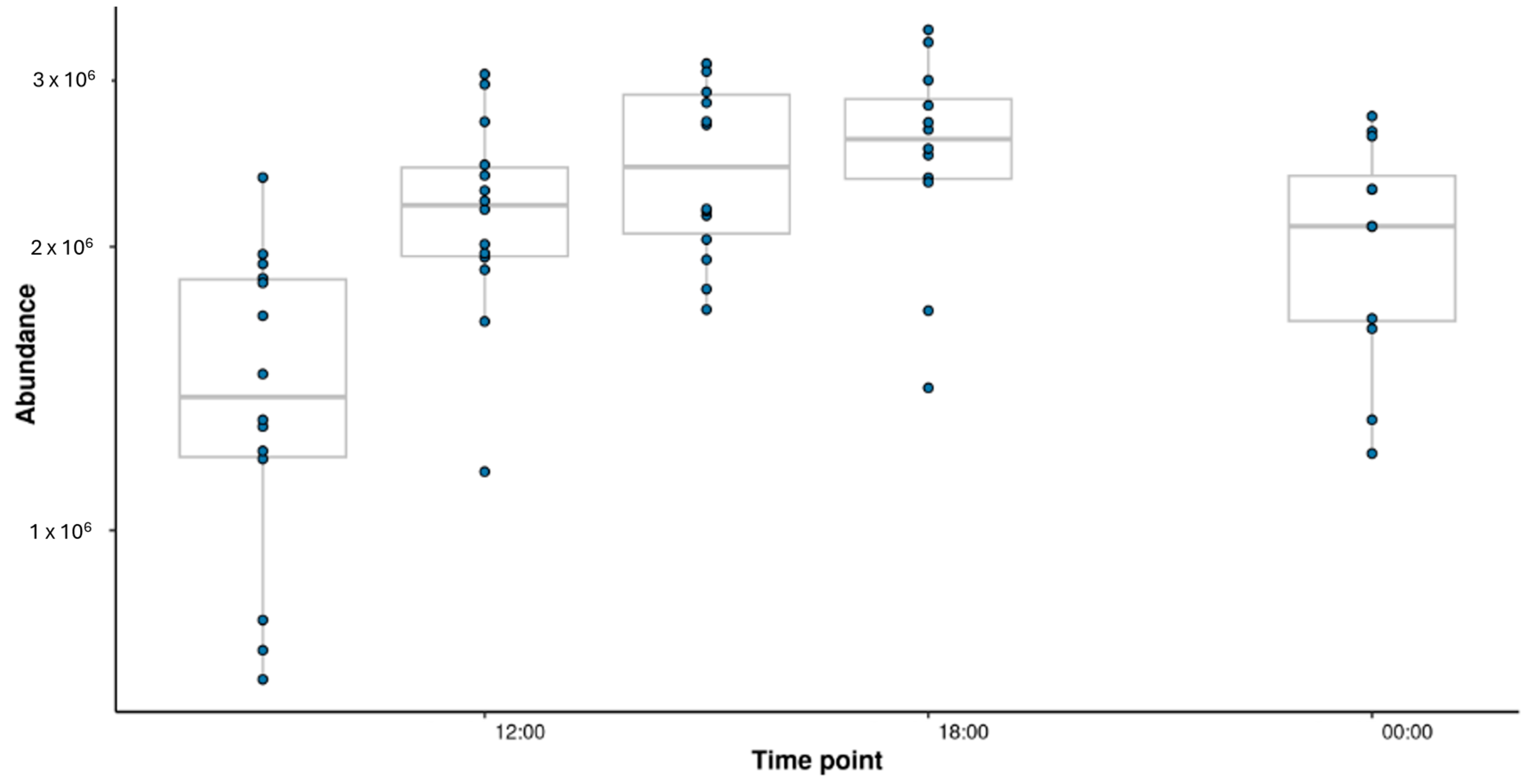
3.4. Metabolic Changes over the Day in One Variant
4. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Huda, H.-S.-A.; Majid, N.B.A.; Chen, Y.; Adnan, M.; Ashraf, S.A.; Roszko, M.; Bryła, M.; Kieliszek, M.; Sasidharan, S. Exploring the ancient roots and modern global brews of tea and herbal beverages: A comprehensive review of origins, types, health benefits, market dynamics, and future trends. Food Sci. Nutr. 2024, 12, 6938–6955. [Google Scholar] [CrossRef]
- Samanta, S. Potential Bioactive Components and Health Promotional Benefits of Tea (Camellia sinensis). J. Am. Nutr. Assoc. 2022, 41, 65–93. [Google Scholar] [CrossRef] [PubMed]
- Cabrera, C.; Artacho, R.; Giménez, R. Beneficial effects of green tea—A review. J. Am. Coll. Nutr. 2006, 25, 79–99. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, S.; Feng, Y.; Jiang, Y.; Yuan, H.; Shan, X.; Zhang, Q.; Niu, L.; Wang, S.; Zhou, Q.; et al. Seasonal variation in non-volatile flavor substances of fresh tea leaves (Camellia sinensis) by integrated lipidomics and metabolomics using UHPLC-Q-Exactive mass spectrometry. Food Chem. 2025, 462, 140986. [Google Scholar] [CrossRef]
- Tang, H.; Zhang, M.; Liu, J.; Cai, J. Metabolomic and Transcriptomic Analyses Reveal the Characteristics of Tea Flavonoids and Caffeine Accumulation and Regulation between Chinese Varieties (Camellia sinensis var. sinensis) and Assam Varieties (C. sinensis var. assamica). Genes 2022, 13, 1994. [Google Scholar] [CrossRef]
- Zou, D.; Yin, X.-L.; Gu, H.-W.; Peng, Z.-X.; Ding, B.; Li, Z.; Hu, X.-C.; Long, W.; Fu, H.; She, Y. Insight into the effect of cultivar and altitude on the identification of EnshiYulu tea grade in untargeted metabolomics analysis. Food Chem. 2024, 436, 137768. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.; Griffin, T.S.; Kraner, D.; Schaffner, M.K.; Sharma, D.; Hazel, M.; Leitch, A.R.; Orians, C.M.; Han, W.; Stepp, J.R.; et al. Environmental Factors Variably Impact Tea Secondary Metabolites in the Context of Climate Change. Front. Plant Sci. 2019, 10, 939. [Google Scholar] [CrossRef]
- Li, H.; Song, K.; Zhang, X.; Wang, D.; Dong, S.; Liu, Y.; Yang, L. Application of Multi-Perspectives in Tea Breeding and the Main Directions. Int. J. Mol. Sci. 2023, 24, 12643. [Google Scholar] [CrossRef]
- Ma, D.; Pang, Y.; Xie, R.; Luo, J.; Xiao, S.; Wang, J.; Yang, R.; Wang, B. Unveiling metabolite network dynamics during Pu-erh tea storage via non-targeted metabolomics. LWT 2024, 209, 116789. [Google Scholar] [CrossRef]
- Aaqil, M.; Peng, C.; Kamal, A.; Nawaz, T.; Zhang, F.; Gong, J. Tea Harvesting and Processing Techniques and Its Effect on Phytochemical Profile and Final Quality of Black Tea: A Review. Foods 2023, 12, 4467. [Google Scholar] [CrossRef]
- He, Y.; Zhang, Q.; Inostroza, A.C.; Kierszniowska, S.; Liu, L.; Li, Y.; Ruan, J. Application of metabolic fingerprinting in tea quality evaluation. Food Control 2024, 160, 110361. [Google Scholar] [CrossRef]
- Draper, J.; Lloyd, A.J.; Goodacre, R.; Beckmann, M. Flow infusion electrospray ionisation mass spectrometry for high throughput, non-targeted metabolite fingerprinting: A review. Metabolomics 2013, 9, 4–29. [Google Scholar] [CrossRef]
- Fang, X.; Xue, R.; Xiao, J.; Pu, Q.; Wang, Y.; Yuan, Y.; Liu, B.; Sui, M.; Jiang, G.; Niaz, R.; et al. Effects of different fermentation modes on tea leaves: Revealing the metabolites modification by quasi-targeted metabolomics. Food Biosci. 2024, 62, 105223. [Google Scholar] [CrossRef]
- Li, J.; Wang, Y.; Suh, J.H. Multi-omics approach in tea polyphenol research regarding tea plant growth, development and tea processing: Current technologies and perspectives. Food Sci. Hum. Wellness 2022, 11, 524–536. [Google Scholar] [CrossRef]
- Lee, J.E.; Lee, B.J.; Chung, J.O.; Kim, H.N.; Kim, E.H.; Jung, S.; Lee, H.; Lee, S.J.; Hong, Y.S. Metabolomic unveiling of a diverse range of green tea (Camellia sinensis) metabolites dependent on geography. Food Chem. 2015, 174, 452–459. [Google Scholar] [CrossRef]
- Rubel Mozumder, N.H.M.; Lee, Y.-R.; Hwang, K.H.; Lee, M.-S.; Kim, E.-H.; Hong, Y.-S. Characterization of tea leaf metabolites dependent on tea (Camellia sinensis) plant age through 1H NMR-based metabolomics. Appl. Biol. Chem. 2020, 63, 10. [Google Scholar] [CrossRef]
- Patel, M.K.; Pandey, S.; Kumar, M.; Haque, M.I.; Pal, S.; Yadav, N.S. Plants Metabolome Study: Emerging Tools and Techniques. Plants 2021, 10, 2409. [Google Scholar] [CrossRef]
- Beckmann, M.; Parker, D.; Enot, D.P.; Duval, E.; Draper, J. High-throughput, nontargeted metabolite fingerprinting using nominal mass flow injection electrospray mass spectrometry. Nat. Protoc. 2008, 3, 486–504. [Google Scholar] [CrossRef] [PubMed]
- Finch, J.P.; Wilson, T.; Lyons, L.; Phillips, H.; Beckmann, M.; Draper, J. Spectral binning as an approach to post-acquisition processing of high resolution FIE-MS metabolome fingerprinting data. Metabolomics 2022, 18, 64. [Google Scholar] [CrossRef]
- Rawal, H.C.; Borchetia, S.; Rohilla, M.; Mazumder, A.; Gogoi, M.; Patel, P.K.; Ilango, R.V.J.; Das, B.; Mazumder, A.B.; Bandyopadhyay, T.; et al. First chromosome-scale genome of Indian tea (Camellia assamica Masters; syn C. sinensis var assamica) cultivar TV 1 reveals its evolution and domestication of caffeine synthesis. Ind. Crops Prod. 2024, 222, 119992. [Google Scholar] [CrossRef]
- Martens, L.; Chambers, M.; Sturm, M.; Kessner, D.; Levander, F.; Shofstahl, J.; Tang, W.H.; Römpp, A.; Neumann, S.; Pizarro, A.D.; et al. mzML—A Community Standard for Mass Spectrometry Data. Mol. Cell. Proteom. 2011, 10, R110.000133. [Google Scholar] [CrossRef] [PubMed]
- Chambers, M.C.; Maclean, B.; Burke, R.; Amodei, D.; Ruderman, D.L.; Neumann, S.; Gatto, L.; Fischer, B.; Pratt, B.; Egertson, J.; et al. A Cross-platform Toolkit for Mass Spectrometry and Proteomics. Nat. Biotechnol. 2012, 30, 918–920. [Google Scholar] [CrossRef]
- Draper, J.; Enot, D.P.; Parker, D.; Beckmann, M.; Snowdon, S.; Lin, W.; Zubair, H. Metabolite signal identification in accurate mass metabolomics data with MZedDB, an interactive m/z annotation tool utilising predicted ionisation behaviour ‘rules’. BMC Bioinform. 2009, 10, 227. [Google Scholar] [CrossRef]
- Beckmann, M.; Lloyd, A.J.; Wilson, T.; Torres, D.P.M.; Goios, A.C.L.; Willis, N.D.; Lyons, L.; Phillips, H.; Mathers, J.C.; Nash, R.J.; et al. Calystegines are Potential Urine Biomarkers for Dietary Exposure to Potato Products. Mol. Nutr. Food Res. 2020, 64, 2000515. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, A.J.; Wilson, T.; Willis, N.D.; Lyons, L.; Phillips, H.; Janssen, H.G.; Stiegler, M.; Xie, L.; Tailliart, K.; Beckmann, M.; et al. Developing community-based urine sampling methods to deploy biomarker technology for assessment of dietary exposure. Public Health Nutr. 2020, 23, 3081–3092. [Google Scholar] [CrossRef]
- Djoumbou Feunang, Y.; Eisner, R.; Knox, C.; Chepelev, L.; Hastings, J.; Owen, G.; Fahy, E.; Steinbeck, C.; Subramanian, S.; Bolton, E.; et al. ClassyFire: Automated chemical classification with a comprehensive, computable taxonomy. J. Cheminform. 2016, 8, 61. [Google Scholar] [CrossRef]
- Picart-Armada, S.; Fernández-Albert, F.; Vinaixa, M.; Rodríguez, M.A.; Aivio, S.; Stracker, T.H.; Yanes, O.; Perera-Lluna, A. Null diffusion-based enrichment for metabolomics data. PLoS ONE 2017, 12, e0189012. [Google Scholar] [CrossRef] [PubMed]
- Picart-Armada, S.; Fernández-Albert, F.; Vinaixa, M.; Yanes, O.; Perera-Lluna, A. FELLA: An R package to enrich metabolomics data. BMC Bioinform. 2018, 19, 538. [Google Scholar] [CrossRef]
- Piyasena, K.N.P.; Hettiarachchi, L.S.K.; Jayawardhane, S.A.D.P.S.; Edirisinghe, E.N.U.; Jayasinghe, W.S. Evaluation of inherent fructose, glucose and sucrose concentrations in tea leaves (Camellia sinensis L.) and in black tea. Appl. Food Res. 2022, 2, 100100. [Google Scholar] [CrossRef]
- Gong, A.-D.; Lian, S.-B.; Wu, N.-N.; Zhou, Y.-J.; Zhao, S.-Q.; Zhang, L.-M.; Cheng, L.; Yuan, H.-Y. Integrated transcriptomics and metabolomics analysis of catechins, caffeine and theanine biosynthesis in tea plant (Camellia sinensis) over the course of seasons. BMC Plant Biol. 2020, 20, 294. [Google Scholar] [CrossRef]
- Fu, Z.; Chen, L.; Zhou, S.; Hong, Y.; Zhang, X.; Chen, H. Analysis of differences in the accumulation of tea compounds under various processing techniques, geographical origins, and harvesting seasons. Food Chem. 2024, 430, 137000. [Google Scholar] [CrossRef]
- Wen, M.; Zhu, M.; Han, Z.; Ho, C.T.; Granato, D.; Zhang, L. Comprehensive applications of metabolomics on tea science and technology: Opportunities, hurdles, and perspectives. Compr. Rev. Food Sci. Food Saf. 2023, 22, 4890–4924. [Google Scholar] [CrossRef]
- Su, S.; Long, P.; Zhang, Q.; Wen, M.; Han, Z.; Zhou, F.; Ke, J.; Wan, X.; Ho, C.-T.; Zhang, L. Chemical, sensory and biological variations of black tea under different drying temperatures. Food Chem. 2024, 446, 138827. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, A.J.; Finch, J.; Warren-Walker, A.; Watson, A.; Lyons, L.; Martin, P.M.; Wilson, T.; Beckmann, M. Chemical attributes of UK grown tea and identifying health-promoting chemistry in tea after different processing methods using metabolomics and machine learning. Metabolomics 2025, Submitted. [Google Scholar]
- Catchpole, G.S.; Beckmann, M.; Enot, D.P.; Mondhe, M.; Zywicki, B.; Taylor, J.; Hardy, N.; Smith, A.; King, R.D.; Kell, D.B.; et al. Hierarchical metabolomics demonstrates substantial compositional similarity between genetically modified and conventional potato crops. Proc. Natl. Acad. Sci. USA 2005, 102, 14458–14462. [Google Scholar] [CrossRef]
- Enot, D.P.; Lin, W.; Beckmann, M.; Parker, D.; Overy, D.P.; Draper, J. Preprocessing, classification modeling and feature selection using flow injection electrospray mass spectrometry metabolite fingerprint data. Nat. Protoc. 2008, 3, 446–470. [Google Scholar] [CrossRef]


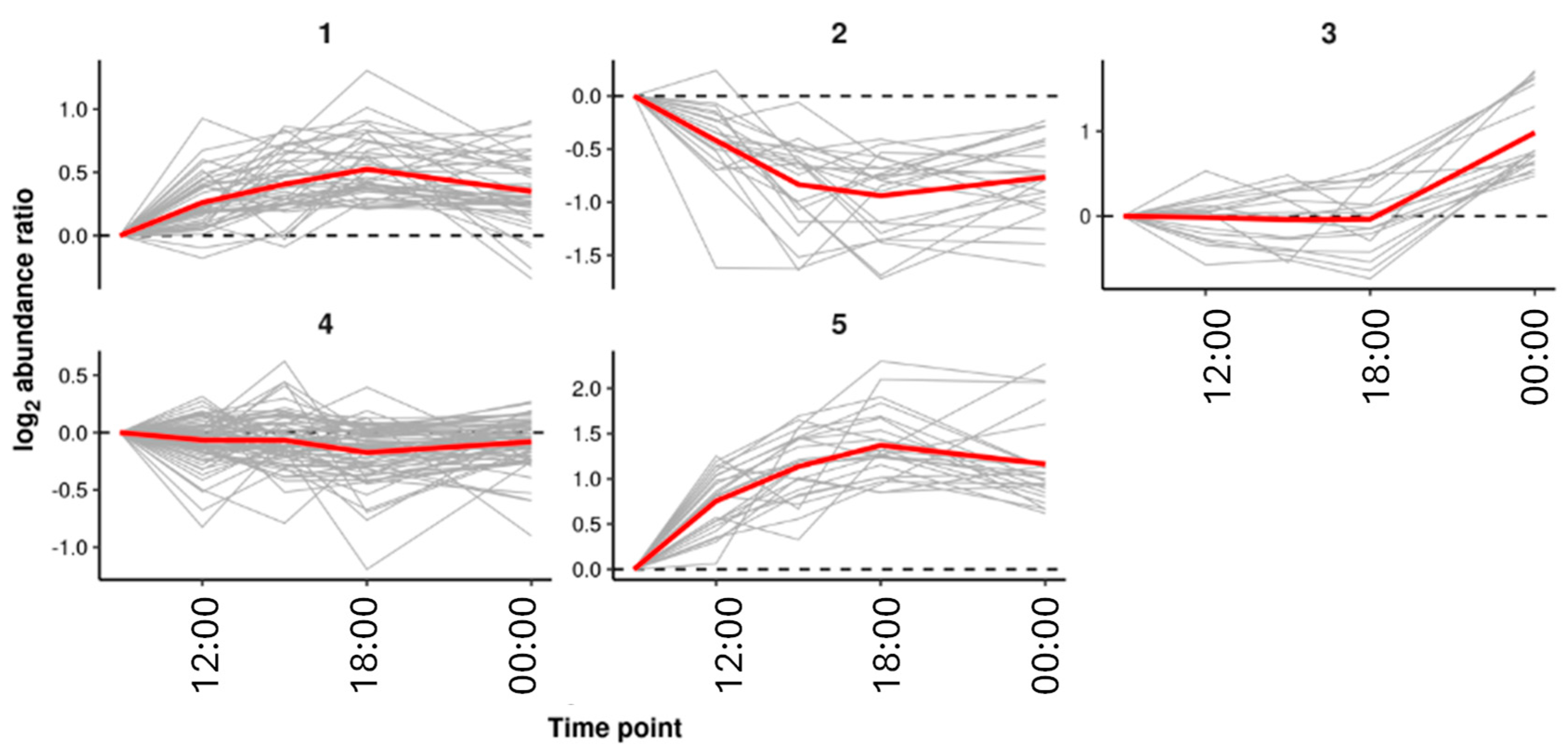
| Cluster | KEGG id | KEGG Name | p-Score |
|---|---|---|---|
| 1 | csin00020 | Citrate cycle (TCA cycle) | 0.0000 |
| 1 | csin00040 | Pentose and glucuronate interconversions | 0.0000 |
| 1 | csin00053 | Ascorbate and aldarate metabolism | 0.0000 |
| 1 | csin00500 | Starch and sucrose metabolism | 0.0000 |
| 1 | csin00600 | Sphingolipid metabolism | 0.0000 |
| 1 | csin00603 | Glycosphingolipid biosynthesis | 0.0000 |
| 1 | csin00660 | C5-Branched dibasic acid metabolism | 0.0000 |
| 1 | csin00760 | Nicotinate and nicotinamide metabolism | 0.0000 |
| 1 | csin02010 | ABC transporters | 0.0000 |
| 1 | csin04016 | MAPK signaling pathway | 0.0000 |
| 3 | csin00030 | Pentose phosphate pathway | 0.0472 |
| 3 | csin00040 | Pentose and glucuronate interconversions | 0.0054 |
| 3 | csin00240 | Pyrimidine metabolism | 0.0110 |
| 3 | csin00250 | Alanine, aspartate and glutamate metabolism | 0.0000 |
| 3 | csin00280 | Valine, leucine and isoleucine degradation | 0.0000 |
| 3 | csin00330 | Arginine and proline metabolism | 0.0001 |
| 3 | csin00410 | beta-Alanine metabolism | 0.0001 |
| 3 | csin00470 | D-Amino acid metabolism | 0.0408 |
| 3 | csin00511 | Other glycan degradation | 0.0394 |
| 3 | csin00561 | Glycerolipid metabolism | 0.0000 |
| 3 | csin00620 | Pyruvate metabolism | 0.0000 |
| 3 | csin00670 | One carbon pool by folate | 0.0000 |
| 3 | csin00740 | Riboflavin metabolism | 0.0000 |
| 3 | csin00970 | Aminoacyl-tRNA biosynthesis | 0.0000 |
| 3 | csin01200 | Carbon metabolism | 0.0000 |
| 3 | csin04148 | Efferocytosis | 0.0000 |
| 4 | csin00941 | Flavonoid biosynthesis | 0.0000 |
| 4 | csin00999 | Biosynthesis of various plant secondary metab... | 0.0001 |
| 5 | csin00030 | Pentose phosphate pathway | 0.0000 |
| 5 | csin00040 | Pentose and glucuronate interconversions | 0.0000 |
| 5 | csin00053 | Ascorbate and aldarate metabolism | 0.0000 |
| 5 | csin00250 | Alanine, aspartate and glutamate metabolism | 0.0000 |
| 5 | csin00270 | Cysteine and methionine metabolism | 0.0000 |
| 5 | csin00290 | Valine, leucine and isoleucine biosynthesis | 0.0000 |
| 5 | csin00330 | Arginine and proline metabolism | 0.0042 |
| 5 | csin00750 | Vitamin B6 metabolism | 0.0000 |
| 5 | csin01230 | Biosynthesis of amino acids | 0.0019 |
| 5 | csin04016 | MAPK signaling pathway | 0.0000 |
| Cluster | Classification | p-Value | Adjusted p-Value |
|---|---|---|---|
| 1 | Organic compounds | 0.000 | 0.002 |
| 1 | Organic acids and derivatives | 0.002 | 0.052 |
| 1 | O-glycosyl compounds | 0.003 | 0.056 |
| 1 | Carbohydrates and carbohydrate conjugates | 0.003 | 0.068 |
| 1 | Keto acids and derivatives | 0.003 | 0.072 |
| 1 | Glycosyl compounds | 0.004 | 0.078 |
| 1 | Organooxygen compounds | 0.010 | 0.224 |
| 1 | Organic oxygen compounds | 0.010 | 0.224 |
| 1 | Tricarboxylic acids and derivatives | 0.010 | 0.225 |
| 1 | Medium-chain keto acids and derivatives | 0.011 | 0.246 |
| 1 | Pentoses | 0.022 | 0.490 |
| 1 | Gamma-keto acids and derivatives | 0.022 | 0.490 |
| 1 | Carboxylic acids and derivatives | 0.025 | 0.554 |
| 1 | Alpha amino acids | 0.044 | 0.969 |
| 2 | Cyclic alcohols and derivatives | 0.005 | 0.126 |
| 2 | Cyclitols and derivatives | 0.005 | 0.126 |
| 2 | Quinic acids and derivatives | 0.005 | 0.126 |
| 2 | Alcohols and polyols | 0.005 | 0.126 |
| 2 | Organic compounds | 0.006 | 0.154 |
| 2 | Organooxygen compounds | 0.009 | 0.227 |
| 2 | Organic oxygen compounds | 0.009 | 0.227 |
| 2 | Sphingolipids | 0.011 | 0.286 |
| 2 | Long-chain ceramides | 0.011 | 0.286 |
| 2 | Ceramides | 0.011 | 0.286 |
| 2 | Non-metal oxoanionic compounds | 0.017 | 0.427 |
| 2 | Inorganic compounds | 0.017 | 0.427 |
| 2 | Non-metal phosphates | 0.017 | 0.427 |
| 2 | Homogeneous non-metal compounds | 0.017 | 0.427 |
| 2 | Cinnamic acids and derivatives | 0.040 | 0.986 |
| 2 | Coumaric acids and derivatives | 0.040 | 0.986 |
| 2 | Hydroxycinnamic acids and derivatives | 0.040 | 0.986 |
| 3 | Hydroxy acids and derivatives | 0.001 | 0.005 |
| 3 | Organic acids and derivatives | 0.006 | 0.057 |
| 3 | Aspartic acid and derivatives | 0.013 | 0.132 |
| 4 | No database hits | 0.001 | 0.009 |
| 5 | Amino acids and derivatives | 0.014 | 0.206 |
| 5 | Alpha amino acids and derivatives | 0.014 | 0.206 |
| 5 | Amino acids, peptides, and analogues | 0.014 | 0.206 |
| 5 | Hydroxy fatty acids | 0.019 | 0.280 |
| 5 | Aspartic acid and derivatives | 0.019 | 0.280 |
| 5 | Alpha amino acids | 0.025 | 0.372 |
| 5 | Fatty Acyls | 0.031 | 0.463 |
| 5 | Fatty acids and conjugates | 0.031 | 0.463 |
| 5 | Carboxylic acids and derivatives | 0.049 | 0.727 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lloyd, A.J.; Warren-Walker, A.; Finch, J.; Harper, J.; Bennet, K.; Watson, A.; Lyons, L.; Martinez Martin, P.; Wilson, T.; Beckmann, M. Chemical Diversity of UK-Grown Tea Explored Using Metabolomics and Machine Learning. Metabolites 2025, 15, 52. https://doi.org/10.3390/metabo15010052
Lloyd AJ, Warren-Walker A, Finch J, Harper J, Bennet K, Watson A, Lyons L, Martinez Martin P, Wilson T, Beckmann M. Chemical Diversity of UK-Grown Tea Explored Using Metabolomics and Machine Learning. Metabolites. 2025; 15(1):52. https://doi.org/10.3390/metabo15010052
Chicago/Turabian StyleLloyd, Amanda J., Alina Warren-Walker, Jasen Finch, Jo Harper, Kathryn Bennet, Alison Watson, Laura Lyons, Pilar Martinez Martin, Thomas Wilson, and Manfred Beckmann. 2025. "Chemical Diversity of UK-Grown Tea Explored Using Metabolomics and Machine Learning" Metabolites 15, no. 1: 52. https://doi.org/10.3390/metabo15010052
APA StyleLloyd, A. J., Warren-Walker, A., Finch, J., Harper, J., Bennet, K., Watson, A., Lyons, L., Martinez Martin, P., Wilson, T., & Beckmann, M. (2025). Chemical Diversity of UK-Grown Tea Explored Using Metabolomics and Machine Learning. Metabolites, 15(1), 52. https://doi.org/10.3390/metabo15010052







