Identification and Characterization of Two Se6OMTs from Stephania epigaea Offer Novel Insights into the Biosynthetic Pathway of Cepharanthine
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials, Chemicals, and Strains
2.2. RNA Extraction and Transcriptome Sequencing
2.3. Analysis and Cloning of St6OMTs Genes
2.4. Expression in E. coli BL21(DE3) and Recombinant Protein Purification
2.5. Functional Characterization of the Recombinant St6OMTs
2.6. UPLC-QTOF-MS Analysis
3. Results
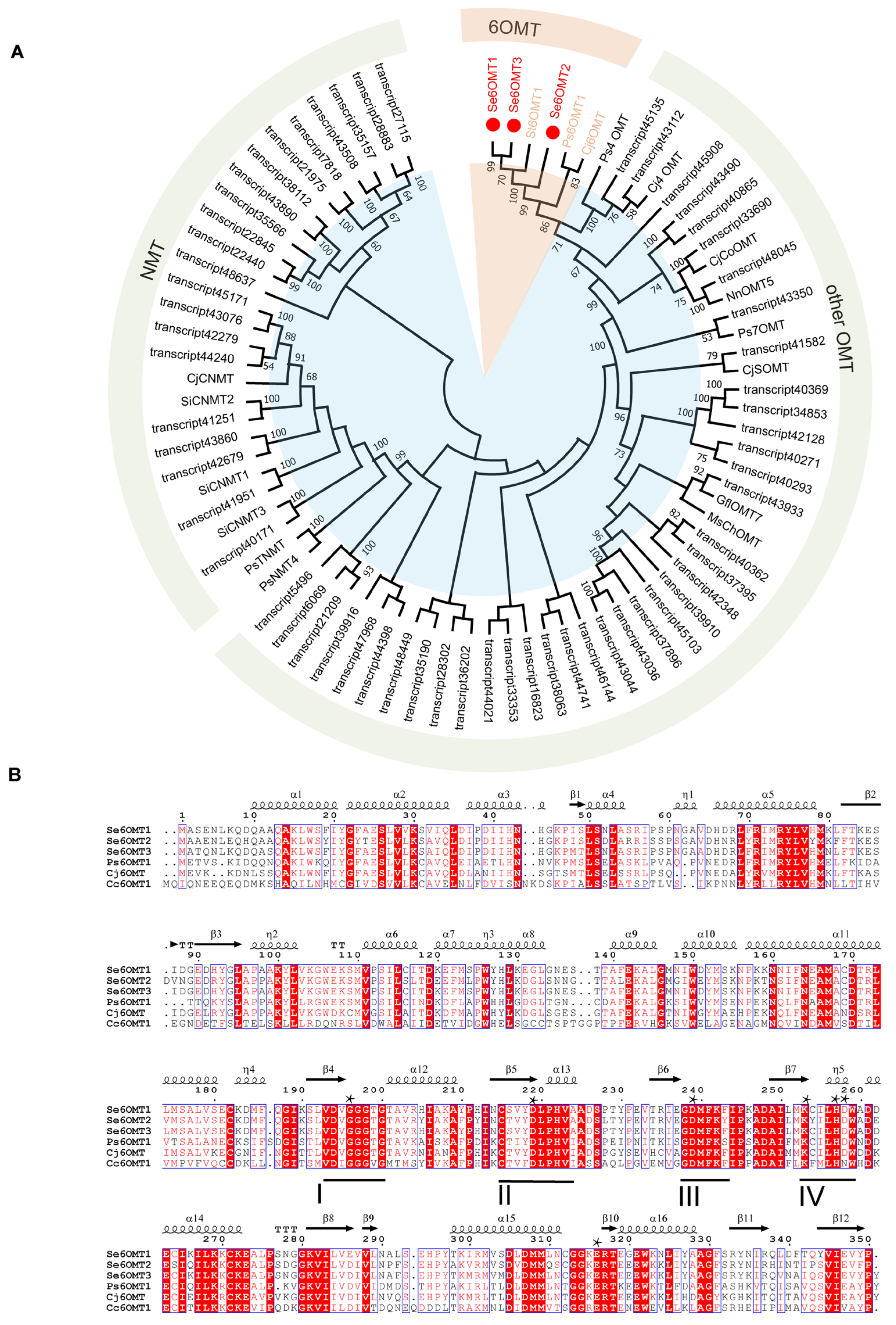
3.1. Screening, Cloning, and Analysis of 6OMTs from the Transcriptome of S. epigaea
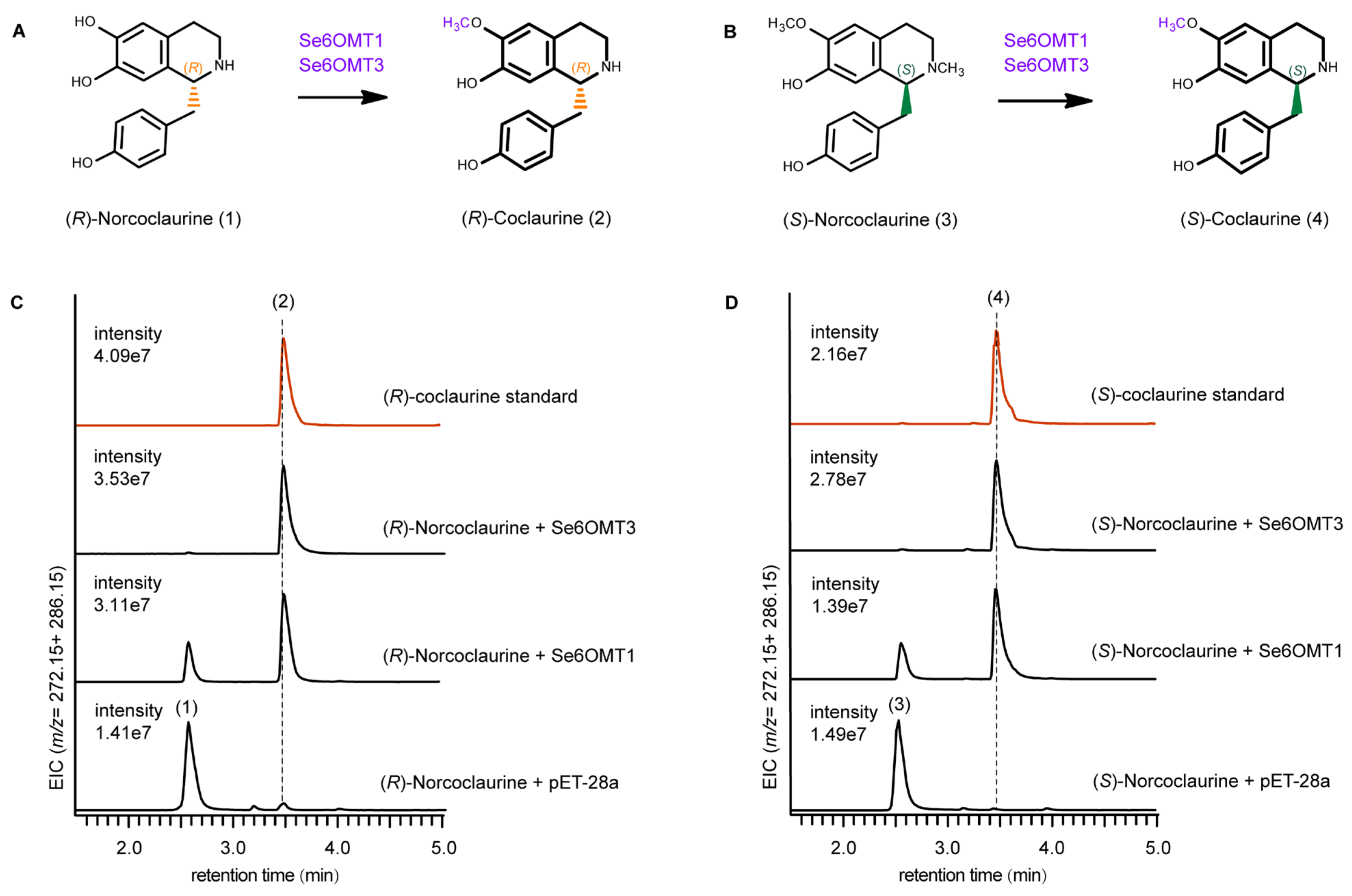
3.2. Functional Characterization of Se6OMTs Towards (R)- and (S)-Norcoclaurine
3.3. Functional Characterization of Se6OMT3 on Other Seven 1-BIAs Substrates
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wu, Z.; Raven, P.H.; Deyuan, H. Missouri Botanical Garden. Flora of China; Science Press: Beijing, China, 1996; Volume 30, p. 53. [Google Scholar]
- Administration, F.P.F.a.D. Fujian Province Processing Standards for Traditional Chinese Medicine Pieces; Fujian Science and Technology Press: Fuzhou, China, 2012; p. 82. [Google Scholar]
- Administration, Y.P.F.a.D. Yunnan Province Traditional Chinese Medicine Standards 2005 Edition; Yunnan Fine Arts Publishing House: Kunming, China, 2005; p. 19. [Google Scholar]
- Li, X.; Liu, L.; Li, G.; Dong, J. Progress in Chemical Constituents, Pharmacology and Fermentation Modification of Radix Stephaniae epigaeae. Shandong Chem. Ind. 2023, 52, 89–94. [Google Scholar]
- Li, X.; Li, M.; Mao, Z.; Du, Y.; Brown, S.; Min, X.; Zhang, R.; Zhong, Y.; Dong, Y.; Liu, Z.; et al. Identification of Bioactive Components of Stephania epigaea Lo and Their Potential Therapeutic Targets by UPLC-MS/MS and Network Pharmacology. Evid. Based Complement. Alternat. Med. 2022, 2022, 3641586. [Google Scholar] [CrossRef] [PubMed]
- Yang, L. Research on the Chemical Constituents and Biological Activities of Sinomenine and Stephania epigaea; Yunnan University: Kunming, China, 2019. [Google Scholar]
- Rogosnitzky, M.; Okediji, P.; Koman, I. Cepharanthine: A review of the antiviral potential of a Japanese-approved alopecia drug in COVID-19. Pharmacol. Rep. 2020, 72, 1509–1516. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.; Liu, K.; Hong, B.; He, S.; Han, P.; Li, M.; Wang, S.; Tong, Y. Progress in the study of antiviral activity of cepharanthine against SARS-CoV-2. J. South. Med. Univ. 2022, 42, 955–957. [Google Scholar]
- Xiao, H.; Liu, G.; Wang, Z.; Yan, Y.; Xiao, P. Improvement on the Determination Method of Cepharanthine Tablets. Chin. Tradit. Pat. Med. 2005, 03, 118–120. [Google Scholar]
- Fan, H.-H.; Wang, L.-Q.; Liu, W.-L.; An, X.-P.; Liu, Z.-D.; He, X.-Q.; Song, L.-H.; Tong, Y.-G. Repurposing of clinically approved drugs for treatment of coronavirus disease 2019 in a 2019-novel coronavirus-related coronavirus model. Chin. Med. J. 2020, 133, 1051–1056. [Google Scholar] [CrossRef]
- Leng, L.; Xu, Z.; Hong, B.; Zhao, B.; Tian, Y.; Wang, C.; Yang, L.; Zou, Z.; Li, L.; Liu, K.; et al. Cepharanthine analogs mining and genomes of Stephania accelerate anti-coronavirus drug discovery. Nat. Commun. 2024, 15, 1537. [Google Scholar] [CrossRef]
- Li, X.; Li, X.; Zeng, Y.; Zhang, H.; Wang, J. Determination of Cepharanthine and Cycleanine in Stephania epigaea H.S.Lo by RP-HPLC. Res. Pract. Chin. Med. Mod. 2010, 24, 61–63. [Google Scholar]
- Ma, Y.; Guo, J.; Mao, Y.P. Analysis for biosynthetic pathways of active constituents in medicinal plants and its availability. Chin. J. Tradit. Chin. Med. 2017, 32, 2079–2083. [Google Scholar]
- Bhakuni, D.S.; Labroo, V.M.; Singh, A.N.; Kapil, R.S. Biosynthesis of the Bisbenzylisoquinoline Alkaloid Cocsulin. J. Chem. Soc. 1978, 1, 121–125. [Google Scholar] [CrossRef]
- Bhakuni, D.S.; Jain, S.; Singh, A.N. Biosynthesis of the bisbenzylisoquinoline alkaloid, tetrandrine. Phytochemistry 1980, 19, 2347–2350. [Google Scholar] [CrossRef]
- Kraus, P.F.; Kutchan, T.M. Molecular cloning and heterologous expression of a cDNA encoding berbamunine synthase, a C--O phenol-coupling cytochrome P450 from the higher plant Berberis stolonifera. Proc. Natl. Acad. Sci. USA 1995, 92, 2071–2075. [Google Scholar] [CrossRef] [PubMed]
- Pyne, M.E.; Gold, N.D.; Martin, V.J.J. Pathway elucidation and microbial synthesis of proaporphine and bis-benzylisoquinoline alkaloids from sacred lotus (Nelumbo nucifera). Metab. Eng. 2023, 77, 162–173. [Google Scholar] [CrossRef]
- Lin, Z.; Hu, Z.; Qu, X.; Lin, S. Research progress and challenges in microbial synthesis of benzylisoquinoline alkaloids. Synth. Biol. 2021, 2, 716–733. [Google Scholar]
- Robin, A.Y.; Giustini, C.; Graindorge, M.; Matringe, M.; Dumas, R. Crystal structure of norcoclaurine-6-O-methyltransferase, a key rate-limiting step in the synthesis of benzylisoquinoline alkaloids. Plant J. 2016, 87, 641–653. [Google Scholar] [CrossRef]
- Li, Q.; Bu, J.; Ma, Y.; Yang, J.; Hu, Z.; Lai, C.; Xu, Y.; Tang, J.; Cui, G.; Wang, Y.; et al. Characterization of O-methyltransferases involved in the biosynthesis of tetrandrine in Stephania tetrandra. J. Plant Physiol. 2020, 250, 153181. [Google Scholar] [CrossRef]
- Noel, J.P.; Dixon, R.A.; Pichersky, E.; Zubieta, C.; Ferrer, J.-L. Chapter two Structural, functional, and evolutionary basis for methylation of plant small molecules. In Recent Advances in Phytochemistry; Romeo, J.T., Ed.; Elsevier: Amsterdam, The Netherlands, 2003; pp. 37–58. [Google Scholar]
- Inui, T.; Tamura, K.-I.; Fujii, N.; Morishige, T.; Sato, F. Overexpression of Coptis japonica norcoclaurine 6-O-methyltransferase overcomes the rate-limiting step in Benzylisoquinoline alkaloid biosynthesis in cultured Eschscholzia californica. Plant Cell Physiol. 2007, 48, 252–262. [Google Scholar] [CrossRef]
- Morishige, T.; Tsujita, T.; Yamada, Y.; Sato, F. Molecular Characterization of the S-Adenosyl-l-methionine:3′-Hydroxy-N-methylcoclaurine 4′-O-Methyltransferase Involved in Isoquinoline Alkaloid Biosynthesis in Coptis japonica. J. Biol. Chem. 2000, 275, 23398–23405. [Google Scholar] [CrossRef]
- Li, K.; Chen, X.; Zhang, J.; Wang, C.; Xu, Q.; Hu, J.; Kai, G.; Feng, Y. Transcriptome Analysis of Stephania tetrandra and Characterization of Norcoclaurine-6-O-Methyltransferase Involved in Benzylisoquinoline Alkaloid Biosynthesis. Front. Plant Sci. 2022, 13, 874583. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S.; Kawashima, S.; Okuno, Y.; Hattori, M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004, 32, D277–D280. [Google Scholar] [CrossRef]
- Apweiler, R.; Bairoch, A.; Wu, C.H.; Barker, W.C.; Boeckmann, B.; Ferro, S.; Gasteiger, E.; Huang, H.; Lopez, R.; Magrane, M.; et al. UniProt: The Universal Protein knowledgebase. Nucleic Acids Res. 2004, 32, D115–D119. [Google Scholar] [CrossRef] [PubMed]
- Tatusov, R.L.; Fedorova, N.D.; Jackson, J.D.; Jacobs, A.R.; Kiryutin, B.; Koonin, E.V.; Krylov, D.M.; Mazumder, R.; Mekhedov, S.L.; Nikolskaya, A.N.; et al. The COG database: An updated version includes eukaryotes. BMC Bioinform. 2003, 4, 41. [Google Scholar] [CrossRef]
- Deng, Y.; Li, J.; Wu, S.; Zhu, Y.; Chen, Y.; He, F. Integrated nr Database in Protein Annotation System and Its Localization. Comput. Eng. 2006, 32, 71–73. [Google Scholar]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: The protein families database. Nucleic Acids Res. 2014, 42, D222–D230. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Jiao, X.; Li, X.; Shi, W.; Ma, Y.; Tan, X.; Gan, J.; Liu, J.; Yang, J.; Wang, J.; et al. Identification of the cytochrome P450s responsible for the biosynthesis of two types of aporphine alkaloids and their de novo biosynthesis in yeast. J. Integr. Plant Biol. 2024, 66, 1703–1717. [Google Scholar] [CrossRef]
- Liscombe, D.K.; Louie, G.V.; Noel, J.P. Architectures, mechanisms and molecular evolution of natural product methyltransferases. Nat. Prod. Rep. 2012, 29, 1238–1250. [Google Scholar] [CrossRef] [PubMed]
- Samanani, N.; Alcantara, J.; Bourgault, R.; Zulak, K.G.; Facchini, P.J. The role of phloem sieve elements and laticifers in the biosynthesis and accumulation of alkaloids in opium poppy. Plant J. 2006, 47, 547–563. [Google Scholar] [CrossRef]
- He, S.-M.; Liang, Y.-L.; Cong, K.; Chen, G.; Zhao, X.; Zhao, Q.-M.; Zhang, J.-J.; Wang, X.; Dong, Y.; Yang, J.-L.; et al. Identification and Characterization of Genes Involved in Benzylisoquinoline Alkaloid Biosynthesis in Coptis Species. Front. Plant Sci. 2018, 9, 731. [Google Scholar] [CrossRef] [PubMed]
- Payne, J.T.; Valentic, T.R.; Smolke, C.D. Complete biosynthesis of the bisbenzylisoquinoline alkaloids guattegaumerine and berbamunine in yeast. Proc. Natl. Acad. Sci. USA 2021, 118, e2112520118. [Google Scholar] [CrossRef] [PubMed]
- Bu, J.; Zhang, X.; Li, Q.; Ma, Y.; Hu, Z.; Yang, J.; Liu, X.; Wang, R.; Jiao, X.; Chen, T.; et al. Catalytic promiscuity of O-methyltransferases from Corydalis yanhusuo leading to the structural diversity of benzylisoquinoline alkaloids. Hortic. Res. 2022, 9, uhac152. [Google Scholar] [CrossRef] [PubMed]
- Ziegler, J.; Facchini, P.J. Alkaloid biosynthesis: Metabolism and trafficking. Annu. Rev. Plant. Biol. 2008, 59, 735–769. [Google Scholar] [CrossRef]
- Ohashi, H.; Watashi, K.; Saso, W.; Shionoya, K.; Iwanami, S.; Hirokawa, T.; Shirai, T.; Kanaya, S.; Ito, Y.; Kim, K.S.; et al. Potential anti-COVID-19 agents, cepharanthine and nelfinavir, and their usage for combination treatment. iScience 2021, 24, 102367. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Shen, X.; Sun, X.; Wang, J.; Yuan, Q. Application of Methyltransferase in Microbial Synthesis of Natural Products. J. Bioeng. 2021, 37, 1869–1886. [Google Scholar]
- Chang, L.; Hagel, J.M.; Facchini, P.J. Isolation and Characterization of O-methyltransferases Involved in the Biosynthesis of Glaucine in Glaucium flavum. Plant Physiol. 2015, 169, 1127–1140. [Google Scholar] [CrossRef]
- Morris, J.S.; Facchini, P.J. Molecular Origins of Functional Diversity in Benzylisoquinoline Alkaloid Methyltransferases. Front. Plant Sci. 2019, 10, 1058. [Google Scholar] [CrossRef]
- Valentic, T.R.; Payne, J.T.; Smolke, C.D. Structure-Guided Engineering of a Scoulerine 9-O-Methyltransferase Enables the Biosynthesis of Tetrahydropalmatrubine and Tetrahydropalmatine in Yeast. ACS Catal. 2020, 10, 4497–4509. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gan, J.; Shi, W.; Li, Q.; Li, X.; Zhao, X.; Tang, J.; Ma, Y.; Wang, J.; Gong, S.; Ma, X.; et al. Identification and Characterization of Two Se6OMTs from Stephania epigaea Offer Novel Insights into the Biosynthetic Pathway of Cepharanthine. Metabolites 2025, 15, 92. https://doi.org/10.3390/metabo15020092
Gan J, Shi W, Li Q, Li X, Zhao X, Tang J, Ma Y, Wang J, Gong S, Ma X, et al. Identification and Characterization of Two Se6OMTs from Stephania epigaea Offer Novel Insights into the Biosynthetic Pathway of Cepharanthine. Metabolites. 2025; 15(2):92. https://doi.org/10.3390/metabo15020092
Chicago/Turabian StyleGan, Jingyi, Wenlong Shi, Qishuang Li, Xinyi Li, Xingyu Zhao, Junhao Tang, Ying Ma, Jian Wang, Shukun Gong, Xiaohui Ma, and et al. 2025. "Identification and Characterization of Two Se6OMTs from Stephania epigaea Offer Novel Insights into the Biosynthetic Pathway of Cepharanthine" Metabolites 15, no. 2: 92. https://doi.org/10.3390/metabo15020092
APA StyleGan, J., Shi, W., Li, Q., Li, X., Zhao, X., Tang, J., Ma, Y., Wang, J., Gong, S., Ma, X., & Guo, J. (2025). Identification and Characterization of Two Se6OMTs from Stephania epigaea Offer Novel Insights into the Biosynthetic Pathway of Cepharanthine. Metabolites, 15(2), 92. https://doi.org/10.3390/metabo15020092





