Multi-Capillary Column-Ion Mobility Spectrometry of Volatile Metabolites Emitted by Saccharomyces Cerevisiae
Abstract
:1. Introduction
| Compound | Compound class | PubChem ID | Included in | Reference # | |
|---|---|---|---|---|---|
| Yeast 7.11 | SGD | ||||
| (2-phenylcyclopropyl) methanol | alcohols | 317540 | no | no | [35] |
| 1,2-benzenedicarboxylic acid | acids | 1017 | no | no | [29,36] |
| 1,3-butanediol | alcohols | 6440 | no | no | [35] |
| 1-butanol | alcohols | 263 | no | no | [35] |
| 1-heptanol | alcohols | 8129 | no | no | [35] |
| 1-hexanol | alcohols | 8103 | no | no | [23,24,25,37] |
| 1-propanol | alcohols | 1031 | no | no | [23,24,25,37,38] |
| 2,3-butanediol | alcohols | 262 | yes | yes | [23,24] |
| 2,5-dimethylpyrazine * | pyrazines | 31252 | no | no | [29,36] |
| 2-ethyl-1-hexanol | alcohols | 7720 | no | no | [29,36,37] |
| 2-furfuraldehyde | aldehydes | 7362 | no | no | [25] |
| 2-hexanol | alcohols | 12297 | no | no | [35] |
| 2-methyl-2-butanol * | alcohols | 6405 | no | no | [24,25,37] |
| 2-methylbutanal * | aldehydes | 7284 | yes | no | [29,36] |
| 2-methylbutanoic acid | acids | 8314 | no | no | [29,36] |
| 2-methylbutanol | alcohols | 8723 | yes | yes | [24,25,37] |
| 2-pentanone | ketones | 7895 | no | no | [29,36] |
| 2-phenylethanol * | benzenoids | 6054 | yes | yes | [23,24,25,26,29,36] |
| 2-phenylethyl acetate | esters | 7654 | no | no | [25,26] |
| 2-propane | alkanes | 6334 | no | no | [37] |
| 2-propanol | alcohols | 3776 | no | no | [29,36,37,38] |
| 2-xylene | benzenoids | 7237 | no | no | [29,36] |
| 3-methylbutanal * | aldehydes | 11552 | yes | yes | [37] |
| 3-methylbutanoic acid * | acids | 10430 | no | no | [24,25,29,36] |
| 3-methylheptyl acetate | esters | 537686 | no | no | [37] |
| 5-methyl-2-furfuraldehyde | aldehydes | 12097 | no | no | [25] |
| acetaldehyde | aldehydes | 177 | yes | yes | [24,25,37,38] |
| acetaldehyde diethylacetal | ethers | 7765 | no | no | [25] |
| acetic acid | acids | 176 | yes | yes | [23,29,36] |
| acetic acid 2-propenyl ester | esters | 11584 | no | no | [37] |
| acetic acid ethenyl ester | esters | 7904 | no | no | [29,36] |
| acetoin | ketones | 179 | yes | yes | [25] |
| acetone | ketones | 180 | no | no | [29,36] |
| acetophenone | ketones | 7410 | no | no | [25] |
| benzaldehyde | aldehydes | 240 | no | no | [25] |
| benzyl acetate | alcohols | 8455 | no | no | [25] |
| benzyl alcohol * | alcohols | 244 | no | yes | [25] |
| butanal | aldehydes | 261 | no | no | [37] |
| butanone * | ketones | 6569 | no | no | [29,36,37] |
| butyric acid | carboxylic acids | 264 | no | no | [24,26] |
| cis-3-hexen-1-ol | alcohols | 5281167 | no | no | [24,25] |
| decanoic acid | carboxylic acids | 2969 | no | no | [23,24,25,26] |
| diacetyl * | ketones | 650 | no | yes | [25,37] |
| diethyl succinate | esters | 31249 | no | no | [23,24,25] |
| dimethyl disulfide * | sulfides | 12232 | no | no | [29,36] |
| dodecanoic acid * | carboxylic acids | 3893 | no | no | [24,26] |
| ethanol | alcohols | 702 | yes | yes | [29,36] |
| ethyl 2-methylbutyrate | esters | 24020 | no | no | [25] |
| ethyl acetate | esters | 8857 | yes | no | [24,25,29,36,37,38] |
| ethyl benzoate | esters | 7165 | no | no | [25] |
| ethyl butyrate | esters | 7762 | no | no | [23,25,26] |
| ethyl caproate | esters | 31265 | no | no | [23,24,25,26,37] |
| ethyl caprylate | esters | 7799 | no | no | [23,24,25,26,37] |
| ethyl decanoate | esters | 8048 | no | no | [23,24,25,26] |
| ethyl furoate | esters | 11980 | no | no | [25] |
| ethyl heptanoate | esters | 7797 | no | no | [25] |
| ethyl isobutyrate | esters | 7342 | no | no | [25,37] |
| ethyl isovalerate | esters | 7945 | no | no | [25,37] |
| ethyl lactate | esters | 7344 | no | no | [24,25] |
| ethyl pehnylacetate | esters | 7590 | no | no | [25] |
| ethyl propanoate | esters | 7749 | no | no | [25,37] |
| ethyl pyruvate | esters | 12041 | no | no | [24] |
| ethyl valerate | esters | 10882 | no | no | [25] |
| ethyl-2-hydroxy propionate | esters | 545098 | no | no | [23] |
| furfuryl alcohol * | alcohols | 7360 | no | no | [25] |
| guaiacol * | alcohols | 460 | no | yes | [25] |
| heptanal * | aldehydes | 8130 | no | no | [37] |
| heptanoic acid | carboxylic acids | 8094 | no | no | [25] |
| hexanal | aldehydes | 6184 | no | no | [25] |
| hexanoic acid * | carboxylic acids | 8892 | no | no | [23,24,25,26] |
| hexyl acetate | esters | 8908 | no | no | [24,25,26] |
| isoamyl acetate | esters | 31276 | yes | no | [37] |
| isoamyl alcohol | alcohols | 31260 | yes | yes | [23,24,25,26,37] |
| isobutanal * | aldehydes | 6561 | no | yes | [37] |
| isobutanol | alcohols | 6560 | yes | yes | [23,24,25,29,36,37,38] |
| isobutyl acetate | esters | 8038 | yes | no | [25,37] |
| limonene | terpenes | 22311 | no | no | [29,36] |
| linalyl propionate | esters | 61098 | no | no | [23] |
| methanol * | alcohols | 887 | no | no | [24,25] |
| methyl acetate | silanes | 76214 | no | no | [25] |
| methylpropanoic acid * | acids | 6590 | no | yes | [24,29,36] |
| monoethyl succinate | esters | 70610 | no | no | [23] |
| n-butyl acetate | esters | 31272 | no | no | [25,37] |
| nonanal | aldehydes | 31289 | no | no | [37] |
| nonanoic acid | carboxylic acids | 8158 | no | no | [25] |
| n-propyl acetate | esters | 7997 | no | no | [25,37] |
| octanoic acid * | carboxylic acids | 379 | no | no | [23,24,25,26] |
| pentanal | aldehydes | 8063 | no | no | [37] |
| propionic acid | carboxylic acids | 1032 | no | no | [24] |
| pyrazine | pyrazines | 9261 | no | no | [29,36] |
| undecane | alkanes | 14257 | no | no | [29,36] |
| α-terpineol | terpenes | 17100 | no | no | [25] |
| β-phenylethyl formate | esters | 7711 | no | no | [37] |
2. Results and Discussion
2.1. Experimental Setup
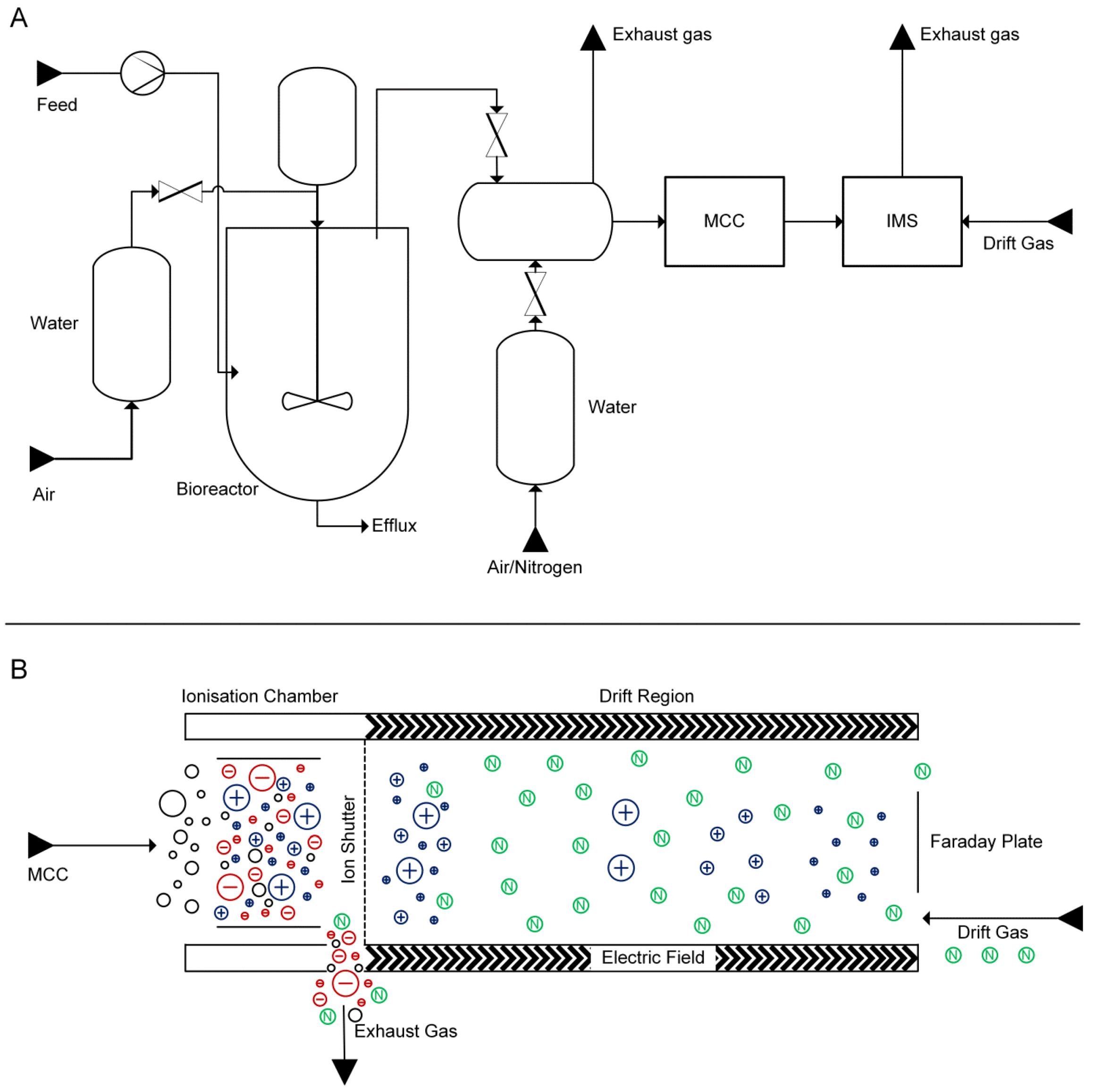
2.2. Growth-Dependent Production of Volatile Metabolites
2.2.1. MCC-IMS Monitoring of Batch Cultures




2.2.2. MCC-IMS Monitoring of Glucose-Limited Chemostat Fermentations



2.3. Identification of Volatile Metabolites
| Compound | GC-MS | MCC-IMS | peak | reduced inverse ion mobility 1/K0 (Vs cm−2) | MCC-IMS retention time (s) | GC-MS retention time (min) |
|---|---|---|---|---|---|---|
| 2,3-butanediol | x | x | 0.575 | 4.5 | 26.517 | |
| 2,3-hexanedione | x | x | C3-7 | 0.570 | 19.4 | 9.633 |
| 2-pentanone | x | x | C3-5 | 0.554 | 6.4 | 5.029 |
| acetoin | x | x | 0.532 | 8.5 | 16.932 | |
| benzaldehyde | x | x | 0.566 | 37.5 | 25.366 | |
| butyric acid | x | x | 0.630 | 24.4 | 30.127 | |
| ethanol | x | x | C3-15 | 0.516 | 4.0 | 4.193 |
| isobutyric acid | x | x | C3-8 | 0.618 | 10.4 | 27.827 |
| isovaleric acid | x | ND | − | − | 31.594 | |
| 2-phenylethanol | x | x | 0.578 | 4.5 | 39.670 |
3. Experimental Section
3.1. Yeast Strains and Growth Conditions
3.2. Analytics
3.3. SPME GC-MS Measurements for Validation of Volatile Metabolites
3.4. MCC-IMS Measurements
3.5. Volatile Metabolite Identification
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- McGovern, P.E.; Glusker, D.L.; Exner, L.J.; Voigt, M.M. Neolithic resinated wine. Nature 1996, 381, 480–481. [Google Scholar]
- Mortimer, R.K. Evolution and variation of the yeast (Saccharomyces). Genome Res. 2000, 10, 891. [Google Scholar]
- McGovern, P.E.; Zhang, J.H.; Tang, J.G.; Zhang, Z.Q.; Hall, G.R.; Moreau, R.A.; Nunez, A.; Butrym, E.D.; Richards, M.P.; Wang, C.S.; et al. Fermented beverages of pre- and proto-historic china. Proc. Natl. Acad. Sci. 2004, 101, 17593–17598. [Google Scholar] [CrossRef]
- Sicard, D.; Legras, J.L. Bread, beer and wine: Yeast domestication in the Saccharomyces sensu stricto complex. Comptes Rendus Biol. 2011, 334, 229–236. [Google Scholar]
- Paddon, C.J.; Westfall, P.J.; Pitera, D.J.; Benjamin, K.; Fisher, K.; McPhee, D.; Leavell, M.D.; Tai, A.; Main, A.; Eng, D.; et al. High-level semi-synthetic production of the potent antimalarial artemisinin. Nature 2013, 496, 528–532. [Google Scholar]
- Kjeldsen, T. Yeast secretory expression of insulin precursors. Appl. Microbiol. Biotechnol. 2000, 54, 277–286. [Google Scholar] [CrossRef]
- Thim, L.; Hansen, M.T.; Norris, K.; Hoegh, I.; Boel, E.; Forstrom, J.; Ammerer, G.; Fiil, N.P. Secretion and processing of insulin precursors in yeast. Proc. Natl. Acad. Sci. 1986, 83, 6766–6770. [Google Scholar]
- Basso, L.C.; de Amorim, H.V.; de Oliveira, A.J.; Lopes, M.L. Yeast selection for fuel ethanol production in brazil. FEMS Yeast Res. 2008, 8, 1155–1163. [Google Scholar]
- Quantz, M. Versuchsanstalt der Hefeindustrie e.V., Berlin, Germany. Available online: http://www.vh-berlin.org/ (accessed 11 July 2014).
- Goffeau, A.; Barrell, B.G.; Bussey, H.; Davis, R.W.; Dujon, B.; Feldmann, H.; Galibert, F.; Hoheisel, J.D.; Jacq, C.; Johnston, M.; et al. Life with 6000 genes. Science 1996, 274, 546, 563–547. [Google Scholar]
- Saccharomyces cerevisiae Genome Snapshot/Overview. Available online: http://www.yeastgenome.org/cache/genomeSnapshot.html (accessed 2 May 2014).
- Cherry, J.M.; Hong, E.L.; Amundsen, C.; Balakrishnan, R.; Binkley, G.; Chan, E.T.; Christie, K.R.; Costanzo, M.C.; Dwight, S.S.; Engel, S.R.; et al. Saccharomyces genome database: The genomics resource of budding yeast. Nucleic Acids Res. 2012, 40, D700–D705. [Google Scholar]
- Mewes, H.W.; Frishman, D.; Guldener, U.; Mannhaupt, G.; Mayer, K.; Mokrejs, M.; Morgenstern, B.; Munsterkotter, M.; Rudd, S.; Weil, B. Mips: A database for genomes and protein sequences. Nucleic Acids Res. 2002, 30, 31–34. [Google Scholar]
- Costanzo, M.C.; Hogan, J.D.; Cusick, M.E.; Davis, B.P.; Fancher, A.M.; Hodges, P.E.; Kondu, P.; Lengieza, C.; Lew-Smith, J.E.; Lingner, C.; et al. The yeast proteome database (ypd) and Caenorhabditis elegans proteome database (wormpd): Comprehensive resources for the organization and comparison of model organism protein information. Nucleic Acids Res. 2000, 28, 73–76. [Google Scholar] [CrossRef]
- Förster, J.; Famili, I.; Fu, P.; Palsson, B.O.; Nielsen, J. Genome-scale reconstruction of the Saccharomyces cerevisiae metabolic network. Genome Res. 2003, 13, 244–253. [Google Scholar]
- Duarte, N.C.; Herrgard, M.J.; Palsson, B.O. Reconstruction and validation of Saccharomyces cerevisiae ind750, a fully compartmentalized genome-scale metabolic model. Genome Res. 2004, 14, 1298–1309. [Google Scholar]
- Nookaew, I.; Jewett, M.C.; Meechai, A.; Thammarongtham, C.; Laoteng, K.; Cheevadhanarak, S.; Nielsen, J.; Bhumiratana, S. The genome-scale metabolic model iin800 of Saccharomyces cerevisiae and its validation: A scaffold to query lipid metabolism. BMC Syst. Biol. 2008. [Google Scholar] [CrossRef]
- Kuepfer, L.; Sauer, U.; Blank, L.M. Metabolic functions of duplicate genes in Saccharomyces cerevisiae. Genome Res. 2005, 15, 1421–1430. [Google Scholar] [CrossRef]
- Dobson, P.D.; Smallbone, K.; Jameson, D.; Simeonidis, E.; Lanthaler, K.; Pir, P.; Lu, C.; Swainston, N.; Dunn, W.B.; Fisher, P.; et al. Further developments towards a genome-scale metabolic model of yeast. BMC Syst. Biol. 2010. [Google Scholar] [CrossRef]
- Herrgard, M.J.; Swainston, N.; Dobson, P.; Dunn, W.B.; Arga, K.Y.; Arvas, M.; Bluthgen, N.; Borger, S.; Costenoble, R.; Heinemann, M.; et al. A consensus yeast metabolic network reconstruction obtained from a community approach to systems biology. Nat. Biotechnol. 2008, 26, 1155–1160. [Google Scholar] [CrossRef]
- Heavner, B.D.; Smallbone, K.; Swainston, N. Yeastnet. Available online: http://sourceforge.net/projects/yeast/files/ (accessed on 2 September 2014).
- Snyder, M.; Gallagher, J.E.G. Systems biology from a yeast omics perspective. FEBS Lett. 2009, 583, 3895–3899. [Google Scholar]
- Patel, S.; Shibamoto, T. Effect of 20 different yeast strains on the production of volatile components in symphony wine. J. Food Compos. Anal. 2003, 16, 469–476. [Google Scholar]
- Perez-Coello, M.S.; Perez, A.I.B.; Iranzo, J.F.U.; Alvarez, P.J.M. Characteristics of wines fermented with different Saccharomyces cerevisiae strains isolated from the la mancha region. Food Microbiol. 1999, 16, 563–573. [Google Scholar] [CrossRef]
- Callejon, R.M.; Clavijo, A.; Ortigueira, P.; Troncoso, A.M.; Paneque, P.; Morales, M.L. Volatile and sensory profile of organic red wines produced by different selected autochthonous and commercial Saccharomyces cerevisiae strains. Anal. Chim. Acta 2010, 660, 68–75. [Google Scholar]
- Torija, M.J.; Beltran, G.; Novo, M.; Poblet, M.; Guillamon, J.M.; Mas, A.; Rozes, N. Effects of fermentation temperature and Saccharomyces species on the cell fatty acid composition and presence of volatile compounds in wine. Int. J. Food Microbiol. 2003, 85, 127–136. [Google Scholar]
- Suomalainen, H.; Nykänen, L. The aroma components produced by yeast in nitrogen-free sugar solution. J. Inst. Brew. 1966, 72, 469–474. [Google Scholar] [CrossRef]
- Polaskova, P.; Herszage, J.; Ebeler, S.E. Wine flavor: Chemistry in a glass. Chem. Soc. Rev. 2008, 37, 2478–2489. [Google Scholar]
- Lemfack, M.C.; Nickel, J.; Dunkel, M.; Preissner, R.; Piechulla, B. mVOC: A database of microbial volatiles. Nucleic Acids Res. 2014, 42, D744–D748. [Google Scholar]
- El-Sayaed, A.M. The pherobase: Database of insect pheromones and semiochemicals. Available online: http://www.pherobase.com (accessed on 20 August 2014).
- Dunkel, M.; Schmidt, U.; Struck, S.; Berger, L.; Gruening, B.; Hossbach, J.; Jaeger, I.S.; Effmert, U.; Piechulla, B.; Eriksson, R.; et al. Superscent—A database of flavors and scents. Nucleic Acids Res. 2009, 37, D291–D294. [Google Scholar] [CrossRef]
- Arn, H.; Acree, T.E. Flavornet: A database of aroma compounds based on odor potency in natural products. In Food Flavors: Formation, Analysis, and Packaging Influences; Contis, E.T., Ho, C.T., Mussinan, C.J., Parliment, T.H., Shahidi, F., Spanier, A.M., Eds.; Elsevier Science Bv: Amsterdam, The Netherland, 1998; Volume 40, p. 27. [Google Scholar]
- De Lacy Costello, B.; Amann, A.; Al-Kateb, H.; Flynn, C.; Filipiak, W.; Khalid, T.; Osborne, D.; Ratcliffe, N.M. A review of the volatiles from the healthy human body. J. Breath Res. 2014. [Google Scholar] [CrossRef]
- Aung, H.W.; Henry, S.A.; Walker, L.P. Revising the representation of fatty acid, glycerolipid, and glycerophospholipid metabolism in the consensus model of yeast metabolism. Ind. Biotechnol. 2013, 9, 215–228. [Google Scholar]
- Hazelwood, L.A.; Daran, J.M.; van Maris, A.J.; Pronk, J.T.; Dickinson, J.R. The Ehrlich pathway for fusel alcohol production: A century of research on Saccharomyces cerevisiae metabolism. Appl. Environ. Microbiol. 2008, 74, 2259–2266. [Google Scholar] [CrossRef]
- Bruce, A.; Verrall, S.; Hackett, C.A.; Wheatley, R.E. Identification of volatile organic compounds (vocs) from bacteria and yeast causing growth inhibition of sapstain fungi. Holzforschung 2004, 58, 193–198. [Google Scholar]
- Hazelwood, L. Cornucopia-Non-Conventional Yeasts for Food Fermentation. In Proceedings of the 25th VH Yeast Conference, Hamburg, Germany, 23–24 April 2012; Research Institute for Baker’s Yeast: Berlin, Germany, 2012; pp. 35–47. [Google Scholar]
- Mateos, J.A.R.; Perez-Nevado, F.; Fernandez, M.R. Influence of Saccharomyces cerevisiae yeast strain on the major volatile compounds of wine. Enzyme Microb. Technol. 2006, 40, 151–157. [Google Scholar]
- Ames, J.M.; Leod, G.M. Volatile components of a yeast extract composition. J. Food Sci. 1985, 50, 125–131. [Google Scholar]
- Baumbach, J.I.; Eiceman, G.A. Ion mobility spectrometry: Arriving on site and moving beyond a low profile. Appl. Spectrosc. 1999, 53, 338–355. [Google Scholar]
- Zhang, Y.; Fraatz, M.A.; Horlamus, F.; Quitmann, H.; Zorn, H. Identification of potent odorants in a novel nonalcoholic beverage produced by fermentation of wort with shiitake (Lentinula edodes). J. Agric. Food Chem. 2014, 62, 4195–4203. [Google Scholar]
- Fenske, J.D.; Paulson, S.E. Human breath emissions of vocs. J. Air Waste Manag. Assoc. 1999, 49, 594–598. [Google Scholar]
- Yinon, J. Field detection and monitoring of explosives. TrAC Trends Anal. Chem. 2002, 21, 292–301. [Google Scholar] [CrossRef]
- Baumbach, J.I. Process analysis using ion mobility spectrometry. Anal. Bioanal. Chem. 2006, 384, 1059–1070. [Google Scholar]
- Maddula, S.; Blank, L.M.; Schmid, A.; Baumbach, J.I. Detection of volatile metabolites of Escherichia coli by multi capillary column coupled ion mobility spectrometry. Anal. Bioanal. Chem. 2009, 394, 791–800. [Google Scholar]
- Xie, Z.; Sielemann, S.; Schmidt, H.; Baumbach, J.I. A novel method for the detection of mtbe: Ion mobility spectrometry coupled to multi capillary column. Int. J. Ion Mobil. Spec. 2000, 4, 77–83. [Google Scholar]
- Kunze, N.; Gopel, J.; Kuhns, M.; Junger, M.; Quintel, M.; Perl, T. Detection and validation of volatile metabolic patterns over different strains of two human pathogenic bacteria during their growth in a complex medium using multi-capillary column-ion mobility spectrometry (MCC-IMS). Appl. Microbiol. Biotechnol. 2013, 97, 3665–3676. [Google Scholar]
- Kotiaho, T.; Lauritsen, F.R.; Degn, H.; Paakkanen, H. Membrane inlet ion mobility spectrometry for online measurement of ethanol in beer and in yeast fermentation. Anal. Chim. Acta 1995, 309, 317–325. [Google Scholar] [CrossRef]
- Kolehmainen, M.; Ronkko, P.; Raatikainen, A. Monitoring of yeast fermentation by ion mobility spectrometry measurement and data visualisation with self-organizing maps. Anal. Chim. Acta 2003, 484, 93–100. [Google Scholar] [CrossRef]
- Vautz, W.; Baumbach, J.I.; Jung, J. Beer fermentation control using ion mobility spectrometry—Results of a pilot study. J. Inst. Brew. 2006, 112, 157–164. [Google Scholar] [CrossRef]
- Stach, J.; Baumbach, J.I. Ion mobility spectrometry-basic elements and applications. Int. J. Ion Mobil. Spec. 2002, 5, 1–21. [Google Scholar]
- Westhoff, M.; Litterst, P.; Freitag, L.; Urfer, W.; Bader, S.; Baumbach, J.I. Ion mobility spectrometry for the detection of volatile organic compounds in exhaled breath of patients with lung cancer: Results of a pilot study. Thorax 2009, 64, 744–748. [Google Scholar] [CrossRef]
- Ruzsanyi, V.; Baumbach, J.I.; Eiceman, G.A. Detection of the mold markers using ion mobility spectrometry. Int. J. Ion Mobil. Spec. 2003, 6, 53–57. [Google Scholar]
- Crabtree, H.G. Observations on the carbohydrate metabolism of tumours. Biochem. J. 1929, 23, 536–545. [Google Scholar]
- De Deken, R.H. The crabtree effect: A regulatory system in yeast. J. Gen. Microbiol. 1966, 44, 149–156. [Google Scholar]
- Randez-Gil, F.; Corcoles-Saez, I.; Prieto, J.A. Genetic and phenotypic characteristics of baker’s yeast: Relevance to baking. Annu. Rev. Food Sci. Technol. 2013, 4, 191–214. [Google Scholar]
- Ejiofor, A.O.; Okafor, N.; Ugwueze, E.N. Development of baking yeast from nigerian palm-wine yeasts. World J. Microbiol. Biotechnol. 1994, 10, 199–202. [Google Scholar] [CrossRef]
- Fischer, K. Straightforward Prognostication of Durability of Baker’s Yeast. In Proceedings of the 18th VH Yeast Conference, Berlin, Germany, 25–26, April, 2005; Research Institute for Baker’s Yeast: Berlin, Germany, 2005; pp. 7–18. [Google Scholar]
- Porro, D.; Sauer, M.; Branduardi, P.; Mattanovich, D. Recombinant protein production in yeasts. Mol. Biotechnol. 2005, 31, 245–259. [Google Scholar] [CrossRef]
- Frick, O.; Wittmann, C. Characterization of the metabolic shift between oxidative and fermentative growth in Saccharomyces cerevisiae by comparative 13C flux analysis. Microb. Cell Fact. 2005. [Google Scholar] [CrossRef]
- Jouhten, P.; Rintala, E.; Huuskonen, A.; Tamminen, A.; Toivari, M.; Wiebe, M.; Ruohonen, L.; Penttila, M.; Maaheimo, H. Oxygen dependence of metabolic fluxes and energy generation of Saccharomyces cerevisiae cen.Pk113–1a. BMC Syst. Biol. 2008. [Google Scholar] [CrossRef]
- Chen, G.C.; Jordan, F. Brewers-yeast pyruvate decarboxylase produces acetoin from acetaldehyde—A novel tool to study the mechanism of steps subsequent to carbon-dioxide loss. Biochemistry 1984, 23, 3576–3582. [Google Scholar] [CrossRef]
- Heidlas, J.; Tressl, R. Purification and properties of two oxidoreductases catalyzing the enantioselective reduction of diacetyl and other diketones from baker’s yeast. Eur. J. Biochem. 1990, 188, 165–174. [Google Scholar]
- Romano, P.; Suzzi, G. Origin and production of acetoin during wine yeast fermentation. Appl. Environ. Microbiol. 1996, 62, 309–315. [Google Scholar]
- Bartowsky, E.J.; Henschke, P.A. The “buttery” attribute of wine-diacetyl-desirability, spoilage and beyond. Int. J. Food Microbiol. 2004, 96, 235–252. [Google Scholar]
- Gonzalez, E.; Fernandez, M.R.; Larroy, C.; Pares, X.; Biosca, J.A. Characterization and functional role of Saccharomyces cerevisiae 2,3-butanediol dehydrogenase. Chem.-Biol. Interact. 2001, 130–132, 425–434. [Google Scholar] [CrossRef]
- Pronk, J.T.; Steensma, H.Y.; van Dijken, J.P. Pyruvate metabolism in Saccharomyces cerevisiae. Yeast 1996, 12, 1607–1633. [Google Scholar]
- Ng, C.Y.; Jung, M.Y.; Lee, J.; Oh, M.K. Production of 2,3-butanediol in Saccharomyces cerevisiaeby in silico aided metabolic engineering. Microb. Cell Fact. 2012. [Google Scholar] [CrossRef]
- Walker, V.; Mills, G.A. 2-pentanone production from hexanoic acid by Penicillium roqueforti from blue cheese: Is this the pathway used in humans? Sci. World J. 2014. Available online: http://www.hindawi.com/journals/tswj/2014/215783/ (accessed on 20 August 2014).
- Jewison, T.; Knox, C.; Neveu, V.; Djoumbou, Y.; Guo, A.C.; Lee, J.; Liu, P.; Mandal, R.; Krishnamurthy, R.; Sinelnikov, I.; et al. Ymdb: The yeast metabolome database. Nucleic Acids Res. 2012, 40, D815–D820. [Google Scholar]
- Dickschat, J.S.; Wickel, S.; Bolten, C.J.; Nawrath, T.; Schulz, S.; Wittmann, C. Pyrazine biosynthesis in Corynebacterium glutamicum. Eur. J. Org. Chem. 2010. [Google Scholar] [CrossRef]
- Van Dijken, J.P.; Bauer, J.; Brambilla, L.; Duboc, P.; Francois, J.M.; Gancedo, C.; Giuseppin, M.L.F.; Heijnen, J.J.; Hoare, M.; Lange, H.C.; et al. An interlaboratory comparison of physiological and genetic properties of four Saccharomyces cerevisiae strains. Enzyme Microb. Technol. 2000, 26, 706–714. [Google Scholar]
- Blank, L.M.; Kuepfer, L.; Sauer, U. Large-scale 13C-flux analysis reveals mechanistic principles of metabolic network robustness to null mutations in yeast. Genome Biol. 2005. [Google Scholar] [CrossRef] [Green Version]
- Verduyn, C.; Postma, E.; Scheffers, W.A.; Vandijken, J.P. Effect of benzoic acid on metabolic fluxes in yeasts—A continuous culture study on the regulation of respiration and alcoholic fermentation. Yeast 1992, 8, 501–517. [Google Scholar] [CrossRef]
- Eiceman, G.H.; Karpas, Z.; Hill, H.H. Ion Mobility Spectrometry, 3rd ed.; CRC Press: Boca Raton, FL, USA, 2014. [Google Scholar]
- Bödeker, B.; Vautz, W.; Baumbach, J. Peak finding and referencing in mcc/ims data. Int. J. Ion Mobil. Spec. 2008, 11, 83–87. [Google Scholar]
- Paczia, N.; Nilgen, A.; Lehmann, T.; Gatgens, J.; Wiechert, W.; Noack, S. Extensive exometabolome analysis reveals extended overflow metabolism in various microorganisms. Microb. Cell Fact. 2012. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Halbfeld, C.; Ebert, B.E.; Blank, L.M. Multi-Capillary Column-Ion Mobility Spectrometry of Volatile Metabolites Emitted by Saccharomyces Cerevisiae. Metabolites 2014, 4, 751-774. https://doi.org/10.3390/metabo4030751
Halbfeld C, Ebert BE, Blank LM. Multi-Capillary Column-Ion Mobility Spectrometry of Volatile Metabolites Emitted by Saccharomyces Cerevisiae. Metabolites. 2014; 4(3):751-774. https://doi.org/10.3390/metabo4030751
Chicago/Turabian StyleHalbfeld, Christoph, Birgitta E. Ebert, and Lars M. Blank. 2014. "Multi-Capillary Column-Ion Mobility Spectrometry of Volatile Metabolites Emitted by Saccharomyces Cerevisiae" Metabolites 4, no. 3: 751-774. https://doi.org/10.3390/metabo4030751
APA StyleHalbfeld, C., Ebert, B. E., & Blank, L. M. (2014). Multi-Capillary Column-Ion Mobility Spectrometry of Volatile Metabolites Emitted by Saccharomyces Cerevisiae. Metabolites, 4(3), 751-774. https://doi.org/10.3390/metabo4030751





