Extracellular Microbial Metabolomics: The State of the Art
Abstract
:1. Introduction
1.1. Applications and Implications of Metabolic Footprinting
1.2. Dynamic or Time-Resolved Metabolic Footprinting
2. Sample Preparation for the Analysis of Extracellular Metabolites
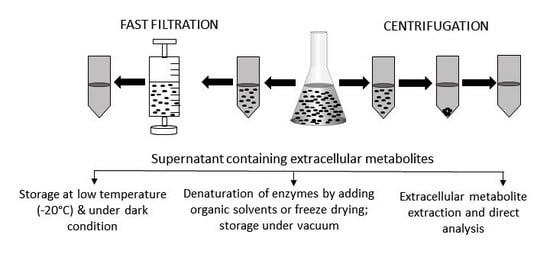
2.1. Metabolites in Solution
- (a)
- liquid-liquid separation where metabolites of interest are separated into an immiscible solvent,
- (b)
- using a column or solid-phase matrix to trap the metabolites, and
- (c)
- selective solubilization, which is the complete evaporation of the solvent to concentrate the sample and the metabolites are then dissolved with suitable solvents.
2.1.1. Solid-Phase Extraction
2.1.2. Solid-Phase Microextraction
2.2. Metabolites in Gas Phase
SPME and Headspace Analysis of Volatile Compounds
- Direct headspace analysis where volatile compounds are collected into a syringe and analyzed by GC.
- Headspace SPME (HS-SPME) which is a coupled technique in which the gas sample in the headspace is trapped on the SPME fiber [6].
3. Concentration of Extracellular Samples to Improve Detection
3.1. Freeze-Drying
3.2. Vacuum-Drying
4. Storage of Extracellular Microbial Samples
5. Integration of Extracellular Metabolomics Data to Genome Scale Metabolic Models
6. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Mashego, M.R.; Van Gulik, W.M.; Heijnen, J.J. Metabolome dynamic responses of Saccharomyces cerevisiae to simultaneous rapid perturbations in external electron acceptor and electron donor. FEMS Yeast Res. 2007, 7, 48–66. [Google Scholar] [CrossRef] [PubMed]
- Tredwell, G.D.; Edwards-Jones, B.; Leak, D.J.; Bundy, J.G. The development of metabolomic sampling procedures for pichia pastoris, and baseline metabolome data. PLoS ONE 2011, 6, e16286. [Google Scholar] [CrossRef] [PubMed]
- Villas-Bôas, S.G.; Bruheim, P. The potential of metabolomics tools in bioremediation studies. OMICS A J. Integr. Biol. 2007, 11, 305–313. [Google Scholar] [CrossRef] [PubMed]
- Kaderbhai, N.N.; Broadhurst, D.I.; Ellis, D.I.; Goodacre, R.; Kell, D.B. Functional genomics via metabolic footprinting: Monitoring metabolite secretion by Escherichia coli tryptophan metabolism mutants using FT-IR and direct injection electrospray mass spectrometry. Comp. Funct. Genom. 2003, 4, 376–391. [Google Scholar] [CrossRef] [PubMed]
- Mapelli, V.; Olsson, L.; Nielsen, J. Metabolic footprinting in microbiology: Methods and applications in functional genomics and biotechnology. Trends Biotechnol. 2008, 26, 490–497. [Google Scholar] [CrossRef] [PubMed]
- Villas-Bôas, S.G.; Roessner, U.; Hansen, M.E.; Smedsgaard, J.; Nielsen, J. Metabolome Analsis An Introduction; Wiley: Hoboken, NJ, USA, 2007. [Google Scholar]
- Oldiges, M.; Lütz, S.; Pflug, S.; Schroer, K.; Stein, N.; Wiendahl, C. Metabolomics: Current state and evolving methodologies and tools. Appl. Microbiol. Biotechnol. 2007, 76, 495–511. [Google Scholar] [CrossRef] [PubMed]
- McGovern, A.C.; Broadhurst, D.; Taylor, J.; Kaderbhai, N.; Winson, M.K.; Small, D.A.; Rowland, J.J.; Kell, D.B.; Goodacre, R. Monitoring of complex industrial bioprocesses for metabolite concentrations using modern spectroscopies and machine learning: Application to gibberellic acid production. Biotechnol. Bioeng. 2002, 78, 527–538. [Google Scholar] [CrossRef] [PubMed]
- Carneiro, S.; Villas-Bôas, S.G.; Ferreira, E.C.; Rocha, I. Metabolic footprint analysis of recombinant Escherichia coli strains during fed-batch fermentations. Mol. Biosyst. 2011, 7, 899–910. [Google Scholar] [CrossRef] [PubMed]
- Graf, A.; Dragosits, M.; Gasser, B.; Mattanovich, D. Yeast systems biotechnology for the production of heterologous proteins. FEMS Yeast Res. 2009, 9, 335–348. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.J.; Yang, H.J.; Kim, M.J.; Han, E.S.; Kim, H.J.; Kwon, D.Y. Metabolomic analysis of meju during fermentation by ultra performance liquid chromatography-quadrupole-time of flight mass spectrometry (UPLC-Q-TOF MS). Food Chem. 2011, 127, 1056–1064. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.E.; Hong, Y.S.; Lee, C.H. Characterization of fermentative behaviors of lactic acid bacteria in grape wines through 1H NMR- and GC-based metabolic profiling. J. Agric. Food Chem. 2009, 57, 4810–4817. [Google Scholar] [CrossRef] [PubMed]
- Rossouw, D.; Næs, T.; Bauer, F.F. Linking gene regulation and the exo-metabolome: A comparative transcriptomics approach to identify genes that impact on the production of volatile aroma compounds in yeast. BMC Genom. 2008, 9. [Google Scholar] [CrossRef] [PubMed]
- Howell, K.S.; Cozzolino, D.; Bartowsky, E.J.; Fleet, G.H.; Henschke, P.A. Metabolic profiling as a tool for revealing saccharomyces interactions during wine fermentation. FEMS Yeast Res. 2006, 6, 91–101. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, X.; Han, Z.; Bai, Z.; Zhuang, G.; Shim, H. Progress in decontamination by halophilic microorganisms in saline wastewater and soil. Environ. Pollut. 2010, 158, 1119–1126. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.R.; Luo, Z.H.; Kwok-Kei Chow, R.; Vrijmoed, L.L.P. Purification and characterization of an extracellular laccase from the anthracene-degrading fungus fusarium solani MAS2. Bioresour. Technol. 2010, 101, 9772–9777. [Google Scholar] [CrossRef] [PubMed]
- Parrilli, E.; Papa, R.; Tutino, M.L.; Sannia, G. Engineering of a psychrophilic bacterium for the bioremediation of aromatic compounds. Bioeng. Bugs 2010, 1, 213–216. [Google Scholar] [CrossRef] [PubMed]
- Plácido, J.; Imam, T.; Capareda, S. Evaluation of ligninolytic enzymes, ultrasonication and liquid hot water as pretreatments for bioethanol production from cotton gin trash. Bioresour. Technol. 2013, 139, 203–208. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.P.; Trivedi, R.K. Ethanol, an Economical & Environmentally Feasible Way of Biofuel from Cellulosic Materials: Process and Discussion. Available online: https://www.ripublication.com/Volume/ijaerv7n11_spl..htm (accessed on 22 August 2017).
- Zain, N.A.M.; Shen, N.S.; Suhaimi, M.S.; Hasan, N.B.; Aziman, S.N. Potential use of liquid pinapple waste for bioethanol production by immobilized bakers‘ yeast. Jurnal Teknologi 2012, 59, 43–47. [Google Scholar]
- Panagiotou, G.; Christakopoulos, P.; Olsson, L. The influence of different cultivation conditions on the metabolome of Fusarium oxysporum. J. Biotechnol. 2005, 118, 304–315. [Google Scholar] [CrossRef] [PubMed]
- Panagiotou, G.; Anderson, M.R.; Grotkjær, T.; Regueira, T.B.; Hofmann, G.; Nielsen, J.; Olsson, L. Systems analysis unfolds the relationship between the phosphoketolase pathway and growth in aspergillus nidulans. PLoS ONE 2008, 3, e3847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Panagiotou, G.; Kouskoumvekaki, I.; Jónsdóttir, S.O.; Olsson, L. Monitoring novel metabolic pathways using metabolomics and machine learning: Induction of the phosphoketolase pathway in aspergillus nidulans cultivations. Metabolomics 2007, 3, 503–516. [Google Scholar] [CrossRef]
- Meijer, S.; Panagiotou, G.; Olsson, L.; Nielsen, J. Physiological characterization of xylose metabolism in aspergillus niger under oxygen-limited conditions. Biotechnol. Bioeng. 2007, 98, 462–475. [Google Scholar] [CrossRef] [PubMed]
- Panagiotou, G.; Pachidou, F.; Petroutsos, D.; Olsson, L.; Christakopoulos, P. Fermentation characteristics of Fusarium oxysporum grown on acetate. Bioresour. Technol. 2008, 99, 7397–7401. [Google Scholar] [CrossRef] [PubMed]
- Villas-Bôas, S.G.; Noel, S.; Lane, G.A.; Attwood, G.; Cookson, A. Extracellular metabolomics: A metabolic footprinting approach to assess fiber degradation in complex media. Anal. Biochem. 2006, 349, 297–305. [Google Scholar] [CrossRef] [PubMed]
- Allen, J.; Davey, H.M.; Broadhurst, D.; Heald, J.K.; Rowland, J.J.; Oliver, S.G.; Kell, D.B. High-throughput classification of yeast mutants for functional genomics using metabolic footprinting. Nat. Biotechnol. 2003, 21, 692–696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mas, S.; Villas-Bôas, S.G.; Hansen, M.E.; Åkesson, M.; Nielsen, J. A comparison of direct infusion ms and GC-MS for metabolic footprinting of yeast mutants. Biotechnol. Bioeng. 2007, 96, 1014–1022. [Google Scholar] [CrossRef] [PubMed]
- Villas-Bôas, S.G.; Moon, C.D.; Noel, S.; Hussein, H.; Kelly, W.J.; Cao, M.; Lane, G.A.; Cookson, A.L.; Attwood, G.T. Phenotypic characterization of transposon-inserted mutants of Clostridium proteoclasticum B316T using extracellular metabolomics. J. Biotechnol. 2008, 134, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, S.; Williams, P. Quorum sensing and social networking in the microbial world. J. R. Soc. Interface 2009, 6, 959–978. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, E.P. Sociomicrobiology: A personal perspective on an emerging research area. Microbe 2010, 5, 206–211. [Google Scholar] [CrossRef]
- Bjarnsholt, T.; Jensen, P.Ø.; Jakobsen, T.H.; Phipps, R.; Nielsen, A.K.; Rybtke, M.T.; Tolker-Nielsen, T.; Givskov, M.; Høiby, N.; Ciofu, O. Quorum sensing and virulence of pseudomonas aeruginosa during lung infection of cystic fibrosis patients. PLoS ONE 2010, 5, e10115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pope, G.A.; MacKenzie, D.A.; Defernez, M.; Aroso, M.A.M.M.; Fuller, L.J.; Mellon, F.A.; Dunn, W.B.; Brown, M.; Goodacre, R.; Kell, D.B.; et al. Metabolic footprinting as a tool for discriminating between brewing yeasts. Yeast 2007, 24, 667–679. [Google Scholar] [CrossRef] [PubMed]
- Villas-Bôas, S.G.; Moxley, J.F.; Åkesson, M.; Stephanopoulos, G.; Nielsen, J. High-throughput metabolic state analysis: The missing link in integrated functional genomics of yeasts. Biochem. J. 2005, 388, 669–677. [Google Scholar] [CrossRef] [PubMed]
- Fan, T.W.M.; Lorkiewicz, P.K.; Sellers, K.; Moseley, H.N.B.; Higashi, R.M.; Lane, A.N. Stable isotope-resolved metabolomics and applications for drug development. Pharm. Ther. 2012, 133, 366–391. [Google Scholar] [CrossRef] [PubMed]
- Pinu, F.; Edwards, P.B.; Jouanneau, S.; Kilmartin, P.; Gardner, R.; Villas-Boas, S. Sauvignon blanc metabolomics: Grape juice metabolites affecting the development of varietal thiols and other aroma compounds in wines. Metabolomics 2013, 556–573. [Google Scholar] [CrossRef]
- Granucci, N.; Pinu, F.R.; Han, T.L.; Villas-Boas, S.G. Can we predict the intracellular metabolic state of a cell based on extracellular metabolite data? Mol. Biosyst. 2015, 11, 3297–3304. [Google Scholar] [CrossRef] [PubMed]
- Aurich, M.K.; Paglia, G.; Rolfsson, O.; Hrafnsdottir, S.; Magnusdottir, M.; Stefaniak, M.M.; Palsson, B.O.; Fleming, R.M.T.; Thiele, I. Prediction of intracellular metabolic states from extracellular metabolomic data. Metabolomics 2015, 11, 603–619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aurich, M.K.; Fleming, R.M.T.; Thiele, I. Metabotools: A comprehensive toolbox for analysis of genome-scale metabolic models. Front. Physiol. 2016, 7, 24. [Google Scholar] [CrossRef] [PubMed]
- Behrends, V.; Ebbels, T.M.D.; Williams, H.D.; Bundy, J.G. Time-resolved metabolic footprinting for nonlinear modeling of bacterial substrate utilization. Appl. Environ. Microbiol. 2009, 75, 2453–2463. [Google Scholar] [CrossRef] [PubMed]
- Chumnanpuen, P.; Hansen, M.A.E.; Smedsgaard, J.; Nielsen, J. Dynamic metabolic footprinting reveals the key components of metabolic network in yeast saccharomyces cerevisiae. Int. J. Genom. 2014. Available online: https://www.hindawi.com/journals/ijg/2014/894296/abs/ (accessed on 19 August 2017).
- Sue, T.; Obolonkin, V.; Griffiths, H.; Villas-Bôas, S.G. An exometabolomics approach to monitoring microbial contamination in microalgal fermentation processes by using metabolic footprint analysis. Appl. Environ. Microbiol. 2011, 77, 7605–7610. [Google Scholar] [CrossRef] [PubMed]
- Behrends, V.; Ryall, B.; Wang, X.; Bundy, J.G.; Williams, H.D. Metabolic profiling of pseudomonas aeruginosa demonstrates that the anti-sigma factor muca modulates osmotic stress tolerance. Mol. Biosyst. 2010, 6, 562–569. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Zeng, J.; Zhou, L.N.; Hu, C.X.; Yin, P.Y.; Lin, X.H. A new strategy for analyzing time-series data using dynamic networks: Identifying prospective biomarkers of hepatocellular carcinoma. Sci. Rep. 2016, 6, 32448. [Google Scholar] [CrossRef] [PubMed]
- Smilde, A.K.; Westerhuis, J.A.; Hoefsloot, H.C.J.; Bijlsma, S.; Rubingh, C.M.; Vis, D.J.; Jellema, R.H.; Pijl, H.; Roelfsema, F.; van der Greef, J. Dynamic metabolomic data analysis: A tutorial review. Metabolomics 2010, 6, 3–17. [Google Scholar] [CrossRef] [PubMed]
- Villas-Bôas, S.G.; Koulman, A.; Lane, G.A. Analytical methods from the perspective of method standardization. In Metabolomics; Springer: Berlin/Heidelberg, Germany, 2007; Volume 18, pp. 11–52. [Google Scholar]
- Smart, K.F.; Aggio, R.B.M.; Van Houtte, J.R.; Villas-Boas, S.G. Analytical platform for metabolome analysis of microbial cells using methyl chloroformate derivatization followed by gas chromatography-mass spectrometry. Nat. Protoc. 2010, 5, 1709–1729. [Google Scholar] [CrossRef] [PubMed]
- Japelt, K.B.; Christensen, J.H.; Villas-Boas, S.G. Metabolic fingerprinting of lactobacillus paracasei: The optimal quenching strategy. Microb. Cell Fact. 2015, 14, 10. [Google Scholar] [CrossRef] [PubMed]
- Duportet, X.; Aggio, R.B.M.; Carneiro, S.; Villas-Bôas, S.G. The biological interpretation of metabolomic data can be misled by the extraction method used. Metabolomics 2012, 8, 410–421. [Google Scholar] [CrossRef]
- Álvarez-Sánchez, B.; Priego-Capote, F.; Castro, M.D.L.D. Metabolomics analysis II. Preparation of biological samples prior to detection. Trends Anal. Chem. 2010, 29, 120–127. [Google Scholar] [CrossRef]
- Dettmer, K.; Aronov, P.A.; Hammock, B.D. Mass spectrometry-based metabolomics. Mass Spectrom. Rev. 2007, 26, 51–78. [Google Scholar] [CrossRef] [PubMed]
- Kraly, J.R.; Holcomb, R.E.; Guan, Q.; Henry, C.S. Review: Microfluidic applications in metabolomics and metabolic profiling. Anal. Chim. Acta 2009, 653, 23–35. [Google Scholar] [CrossRef] [PubMed]
- Mitra, S. Sample Preparation Techniques in Analytical Chemsitry; Whiley: Hoboken, NJ, USA, 2003. [Google Scholar]
- Theodoridis, G.; Gika, H.; Franceschi, P.; Caputi, L.; Arapitsas, P.; Scholz, M.; Masuero, D.; Wehrens, R.; Vrhovsek, U.; Mattivi, F. LC-MS based global metabolite profiling of grapes: Solvent extraction protocol optimisation. Metabolomics 2012, 8, 175–185. [Google Scholar] [CrossRef]
- Ström, K.; Sjögren, J.; Broberg, A.; Schnürer, J. Lactobacillus plantarum milab 393 produces the antifungal cyclic dipeptides Cyclo(L-Phe-L-Pro) and Cyclo(L-Phe-Trans-4-OH-L-Pro) and 3-phenyllactic acid. Appl. Environ. Microbiol. 2002, 68, 4322–4327. [Google Scholar] [CrossRef] [PubMed]
- Yang, E.J.; Chang, H.C. Purification of a new antifungal compound produced by lactobacillus plantarum af1 isolated from kimchi. Int. J. Food Microbiol. 2010, 139, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Pawliszyn, J. New directions in sample preparation for analysis of organic compounds. Trends Anal. Chem. 1995, 14, 113–122. [Google Scholar] [CrossRef]
- Pawliszyn, J. Chapter 13 Solid Phase Microextraction; Wiley: Hoboken, NJ, USA, 2002; Volume 37, pp. 389–477. [Google Scholar]
- Risticevic, S.; Chen, Y.; Kudlejova, L.; Vatinno, R.; Baltensperger, B.; Stuff, J.R.; Hein, D.; Pawliszyn, J. Protocol for the development of automated high-throughput SPME-GC methods for the analysis of volatile and semivolatile constituents in wine samples. Nat. Protoc. 2010, 5, 162–176. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, G.; Zhao, W.; Pawliszyn, J. Kinetic calibration for automated headspace liquid-phase microextraction. Anal. Chem. 2005, 77, 8122–8128. [Google Scholar] [CrossRef] [PubMed]
- Vuckovic, D.; Zhang, X.; Cudjoe, E.; Pawliszyn, J. Solid-phase microextraction in bioanalysis: New devices and directions. J. Chromatogr. A 2010, 1217, 4041–4060. [Google Scholar] [CrossRef] [PubMed]
- Lord, H.; Pawliszyn, J. Evolution of solid-phase microextraction technology. J. Chromatogr. A 2000, 885, 153–193. [Google Scholar] [CrossRef]
- Pawliszyn, J. New developments and applications of solvent-free sampling and sample preparation technologies for the investigation of living systems. Aust. J. Chem. 2003, 56, 155–158. [Google Scholar] [CrossRef]
- Ouyang, G.; Pawliszyn, J. Recent developments in SPME for on-site analysis and monitoring. Trends Anal. Chem. 2006, 25, 692–703. [Google Scholar] [CrossRef]
- Jeleń, H.H. Use of solid phase microextraction (SPME) for profiling fungal volatile metabolites. Lett. Appl. Microbiol. 2003, 36, 263–267. [Google Scholar] [CrossRef] [PubMed]
- Pawliszyn, J. Water analysis by solid phase microextraction based on physical chemical properties of the coating. Anal. Chem. 1997, 69, 1992–1998. [Google Scholar]
- Stansbridge, E.M.; Mills, G.A.; Walker, V. Automated headspace gas chromatographic analysis of faecal short-chain fatty acids. J. Chromatogr.-Biomed. Appl. 1993, 621, 7–13. [Google Scholar] [CrossRef]
- Demyttenaere, J.C.R.; Moriña, R.M.; Sandra, P. Monitoring and fast detection of mycotoxin-producing fungi based on headspace solid-phase microextraction and headspace sorptive extraction of the volatile metabolites. J. Chromatogr. A 2003, 985, 127–135. [Google Scholar] [CrossRef]
- Siripatrawan, U.; Linz, J.E.; Harte, B.R. Detection of Escherichia coli in packaged alfalfa sprouts with an electronic nose and an artificial neural network. J. Food Prot. 2006, 69, 1844–1850. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.A.; Yu, S.J.; Zhang, L.; Chen, X.D. The effects of ac electric field on wine maturation. Innov. Food Sci. Emerg. Technol. 2008, 9, 463–468. [Google Scholar] [CrossRef]
- Villas-Bôas, S.G.; Højer-Pedersen, J.; Åkesson, M.; Smedsgaard, J.; Nielsen, J. Global metabolite analysis of yeast: Evaluation of sample preparation methods. Yeast 2005, 22, 1155–1169. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Nail, S.L.; Pikal, M.J. Freeze-drying process design by manometric temperature measurement: Design of a smart freeze-dryer. Pharm. Res. 2005, 22, 685–700. [Google Scholar] [CrossRef] [PubMed]
- Fischer, T. Lyophilizer qualification: Some practical advice. Drugs Pharm. Sci. 2004, 135, 517–534. [Google Scholar]
- Oikawa, A.; Otsuka, T.; Jikumaru, Y.; Yamaguchi, S.; Matsuda, F.; Nakabayashi, R.; Takashina, T.; Isuzugawa, K.; Saito, K.; Shiratake, K. Effects of freeze-drying of samples on metabolite levels in metabolome analyses. J. Sep. Sci. 2011, 34, 3561–3567. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.H.; Yu, H.L.; Ba, Z.Y.; Chen, J.Y.; Sun, H.G.; Han, B.Z. Sampling methods for NMR-based metabolomics of staphylococcus aureus. Biotechnol. J. 2010, 5, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Pinu, F.R.; De Carvalho-Silva, S.; Uetanabaro, A.P.T.; Villas-Boas, S.G. Vinegar metabolomics: An explorative study of commercial balsamic vinegars using gas chromatography-mass spectrometry. Metabolites 2016, 6, 22. [Google Scholar] [CrossRef] [PubMed]
- Forster, J.; Famili, I.; Fu, P.; Palsson, B.O.; Nielsen, J. Genome-scale reconstruction of the Saccharomyces cerevisiae metabolic network. Genom. Res. 2003, 13, 244–253. [Google Scholar] [CrossRef] [PubMed]
- Heavner, B.D.; Price, N.D. Comparative analysis of yeast metabolic network models highlights progress, opportunities for metabolic reconstruction. PLoS Comput. Biol. 2015, 11, e1004530. [Google Scholar] [CrossRef] [PubMed]
- Otero, J.M.; Cimini, D.; Patil, K.R.; Poulsen, S.G.; Olsson, L.; Nielsen, J. Industrial systems biology of Saccharomyces cerevisiae enables novel succinic acid cell factory. PLoS ONE 2013, 8, e54144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arvas, M. Genome scale metabolic model reconstruction for the needs of industrial biotechnology. New Biotechnol. 2016, 33. [Google Scholar] [CrossRef]
- Becker, S.A.; Feist, A.M.; Mo, M.L.; Hannum, G.; Palsson, B.O.; Herrgard, M.J. Quantitative prediction of cellular metabolism with constraint-based models: The cobra toolbox. Nat. Protoc. 2007, 2, 727–738. [Google Scholar] [CrossRef] [PubMed]
- King, Z.A.; Lloyd, C.J.; Feist, A.M.; Palsson, B.O. Next-generation genome-scale models for metabolic engineering. Curr. Opin. Biotechnol. 2015, 35, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Famili, I.; Forster, J.; Nielson, J.; Palsson, B.O. Saccharomyces cerevisiae phenotypes can be predicted by using constraint-based analysis of a genome-scale reconstructed metabolic network. Proc. Natl. Acad. Sci. USA 2003, 100, 13134–13139. [Google Scholar] [CrossRef] [PubMed]
- Lewis, N.E.; Nagarajan, H.; Palsson, B.O. Constraining the metabolic genotype-phenotype relationship using a phylogeny of in silico methods. Nat. Rev. Microbiol. 2012, 10, 291–305. [Google Scholar] [CrossRef] [PubMed]
- Thiele, I.; Palsson, B.O. A protocol for generating a high-quality genome-scale metabolic reconstruction. Nat. Protoc. 2010, 5, 93–121. [Google Scholar] [CrossRef] [PubMed]
- Mo, M.L.; Palsson, B.O.; Herrgard, M.J. Connecting extracellular metabolomic measurements to intracellular flux states in yeast. BMC Syst. Biol. 2009, 3, 17. [Google Scholar] [CrossRef] [PubMed]
- Cakir, T.; Efe, C.; Dikicioglu, D.; Hortacsu, A.; Kirdar, B.; Oliver, S.G. Flux balance analysis of a genome-scale yeast model constrained by exometabolomic data allows metabolic system identification of genetically different strains. Biotechnol. Prog. 2007, 23, 320–326. [Google Scholar] [CrossRef] [PubMed]
- Beale, D.J.; Barratt, R.; Marlow, D.R.; Dunn, M.S.; Palombo, E.A.; Morrison, P.D.; Key, C. Application of metabolomics to understanding biofilms in water distribution systems: A pilot study. Biofouling 2013, 29, 283–294. [Google Scholar] [CrossRef] [PubMed]
- Creek, D.J.; Nijagal, B.; Kim, D.H.; Rojas, F.; Matthews, K.R.; Barrett, M.P. Metabolomics guides rational development of a simplified cell culture medium for drug screening against trypanosoma brucei. Antimicrob. Agent. Chemother. 2013, 57, 2768–2779. [Google Scholar] [CrossRef] [PubMed]
- Courant, F.; Martzolff, A.; Rabin, G.; Antignac, J.P.; Le Bizec, B.; Giraudeau, P.; Tea, I.; Akoka, S.; Couzinet, A.; Cogne, G.; et al. How metabolomics can contribute to bio-processes: A proof of concept study for biomarkers discovery in the context of nitrogen-starved microalgae grown in photobioreactors. Metabolomics 2013, 9, 1286–1300. [Google Scholar] [CrossRef]
- Mohmad-Saberi, S.E.; Hashim, Y.Z.H.Y.; Mel, M.; Amid, A.; Ahmad-Raus, R.; Packeer-Mohamed, V. Metabolomics profiling of extracellular metabolites in CHO-K1 cells cultured in different types of growth media. Cytotechnology 2012, 65, 577–586. [Google Scholar] [CrossRef] [PubMed]



| Phase | |||
|---|---|---|---|
| Parameters | Normal | Reversed | Ion-Exchange |
| Solvent polarity | High | Low | High |
| Range of solvent polarity | Low to medium | High to medium | High |
| Solvents for elution | Acetone, ethyl acetate | Water/methanol/acetonitrile solution | Salts and buffers |
| Loading solvents | Toluene, hexane | Water and buffers | Water and buffers |
| Eluted sample | Less polar | Most polar | Weakly ionized |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pinu, F.R.; Villas-Boas, S.G. Extracellular Microbial Metabolomics: The State of the Art. Metabolites 2017, 7, 43. https://doi.org/10.3390/metabo7030043
Pinu FR, Villas-Boas SG. Extracellular Microbial Metabolomics: The State of the Art. Metabolites. 2017; 7(3):43. https://doi.org/10.3390/metabo7030043
Chicago/Turabian StylePinu, Farhana R., and Silas G. Villas-Boas. 2017. "Extracellular Microbial Metabolomics: The State of the Art" Metabolites 7, no. 3: 43. https://doi.org/10.3390/metabo7030043
APA StylePinu, F. R., & Villas-Boas, S. G. (2017). Extracellular Microbial Metabolomics: The State of the Art. Metabolites, 7(3), 43. https://doi.org/10.3390/metabo7030043