Untargeted Urinary 1H NMR-Based Metabolomic Pattern as a Potential Platform in Breast Cancer Detection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Urine Collection
2.3. NMR Measurements
2.4. Statistical Analysis
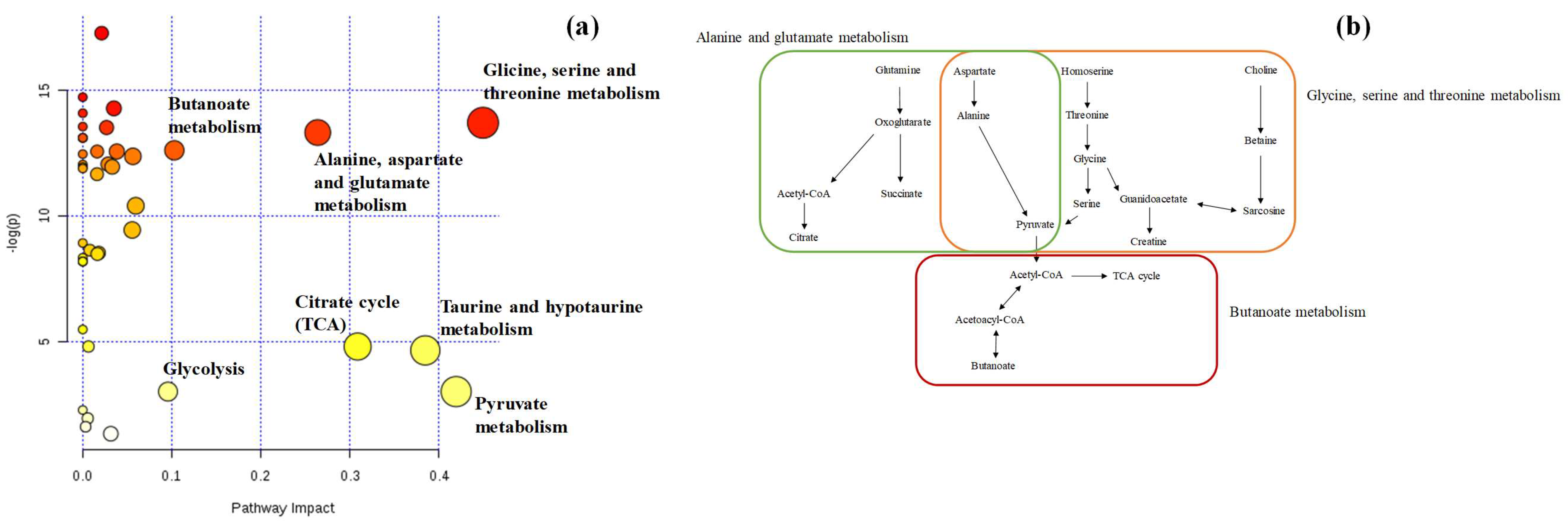
3. Results and Discussion
3.1. Urinary Metabolomic Pattern Based on 1H NMR
3.2. Multivariate Statistical Analysis of Urinary Metabolomic Profile
4. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kretschmer, C.; Sterner-Kock, A.; Siedentopf, F.; Schoenegg, W.; Schlag, P.M.; Kemmner, W. Identification of early molecular markers for breast cancer. Mol. Cancer 2011, 10, 15. [Google Scholar] [CrossRef] [PubMed]
- Slupsky, C.M.; Steed, H.; Wells, T.H.; Dabbs, K.; Schepansky, A.; Capstick, V.; Faught, W.; Sawyer, M.B. Urine metabolite analysis offers potential early diagnosis of ovarian and breast cancers. Clin. Cancer Res. 2010, 16, 5835–5841. [Google Scholar] [CrossRef] [PubMed]
- Duffy, M.J. Serum Tumor Markers in Breast Cancer: Are They of Clinical Value? Clin. Chem. 2006, 52, 345–351. [Google Scholar] [CrossRef] [Green Version]
- Asiago, V.M.; Alvarado, L.Z.; Shanaiah, N.; Gowda, G.A.N.; Owusu-Sarfo, K.; Ballas, R.A.; Raftery, D. Early Detection of Recurrent Breast Cancer Using Metabolite Profiling. Cancer Res. 2010, 70, 8309–8318. [Google Scholar] [CrossRef] [Green Version]
- Brase, J.C.; Wuttig, D.; Kuner, R.; Sültmann, H. Serum microRNAs as non-invasive biomarkers for cancer. Mol. Cancer 2010, 9, 306. [Google Scholar] [CrossRef]
- Kim, Y.; Koo, I.; Jung, B.H.; Chung, B.C.; Lee, D. Multivariate classification of urine metabolome profiles for breast cancer diagnosis. BMC Bioinform. 2010, 11, S4. [Google Scholar] [CrossRef]
- Woo, H.M.; Kim, K.M.; Choi, M.H.; Jung, B.H.; Lee, J.; Kong, G.; Nam, S.J.; Kim, S.; Bai, S.W.; Chung, B.C. Mass spectrometry based metabolomic approaches in urinary biomarker study of women’s cancers. Clin. Chim. Acta 2009, 400, 63–69. [Google Scholar] [CrossRef]
- Nam, H.; Chung, B.C.; Kim, Y.; Lee, K.; Lee, D. Combining tissue transcriptomics and urine metabolomics for breast cancer biomarker identification. Bioinformatics 2009, 25, 3151–3157. [Google Scholar] [CrossRef] [Green Version]
- Henneges, C.; Bullinger, D.; Fux, R.; Friese, N.; Seeger, H.; Neubauer, H.; Laufer, S.; Gleiter, C.H.; Schwab, M.; Zell, A.; et al. Prediction of breast cancer by profiling of urinary RNA metabolites using Support Vector Machine-based feature selection. BMC Cancer 2009, 9, 104. [Google Scholar] [CrossRef]
- Abaffy, T.; Möller, M.; Riemer, D.D.; Milikowski, C. A Case Report-Volatile Metabolomic Signature of Malignant Melanoma using Matching Skin as a Control. J. Cancer Sci. Ther. 2011, 03, 140–144. [Google Scholar] [CrossRef] [PubMed]
- Carrola, J.; Rocha, C.M.; Barros, A.S.; Gil, A.M.; Goodfellow, B.J.; Carreira, I.M.; Bernardo, J.; Gomes, A.; Sousa, V.; Carvalho, L.; et al. Metabolic signatures of lung cancer in biofluids: NMR-based metabonomics of urine. J. Proteome Res. 2011, 10, 221–230. [Google Scholar] [CrossRef]
- Abaffy, T.; Möller, M.G.; Riemer, D.D.; Milikowski, C.; DeFazio, R.A. Comparative analysis of volatile metabolomics signals from melanoma and benign skin: A pilot study. Metabolomics 2013, 9, 998–1008. [Google Scholar] [CrossRef]
- Filipiak, W.; Filipiak, A.; Sponring, A.; Schmid, T.; Zelger, B.; Ager, C.; Klodzinska, E.; Denz, H.; Pizzini, A.; Lucciarini, P.; et al. Comparative analyses of volatile organic compounds (VOCs) from patients, tumors and transformed cell lines for the validation of lung cancer-derived breath markers. J. Breath Res. 2014, 8, 027111. [Google Scholar] [CrossRef] [PubMed]
- Filipiak, W.; Sponring, A.; Filipiak, A.; Ager, C.; Schubert, J.; Miekisch, W.; Amann, A.; Troppmair, J. TD-GC-MS analysis of volatile metabolites of human lung cancer and normal cells in vitro. Cancer Epidemiol. Biomark. Prev. 2010, 19, 182–195. [Google Scholar] [CrossRef]
- Silva, C.L.; Passos, M.; Câmara, J.S. Solid phase microextraction, mass spectrometry and metabolomic approaches for detection of potential urinary cancer biomarkers—A powerful strategy for breast cancer diagnosis. Talanta 2012, 89, 360–368. [Google Scholar] [CrossRef] [PubMed]
- Matsumura, K.; Opiekun, M.; Oka, H.; Vachani, A.; Albelda, S.M.; Yamazaki, K.; Beauchamp, G.K. Urinary Volatile Compounds as Biomarkers for Lung Cancer: A Proof of Principle Study Using Odor Signatures in Mouse Models of Lung Cancer. PLoS ONE 2010, 5, e8819. [Google Scholar] [CrossRef] [PubMed]
- Silva, C.L.; Passos, M.; Câmara, J.S. Investigation of urinary volatile organic metabolites as potential cancer biomarkers by solid-phase microextraction in combination with gas chromatography-mass spectrometry. Br. J. Cancer 2011, 105, 1894–1904. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Zhang, R.; Song, Y.; He, J.; Sun, J.; Bai, J.; An, Z.; Dong, L.; Zhan, Q.; Abliz, Z. RRLC-MS/MS-based metabonomics combined with in-depth analysis of metabolic correlation network: Finding potential biomarkers for breast cancer. Analyst 2009, 134, 2003. [Google Scholar] [CrossRef]
- Gu, H.; Pan, Z.; Xi, B.; Asiago, V.; Musselman, B.; Raftery, D. Principal component directed partial least squares analysis for combining nuclear magnetic resonance and mass spectrometry data in metabolomics: Application to the detection of breast cancer. Anal. Chim. Acta 2011, 686, 57–63. [Google Scholar] [CrossRef] [Green Version]
- Oakman, C.; Tenori, L.; Biganzoli, L.; Santarpia, L.; Cappadona, S.; Luchinat, C.; Di Leo, A. Uncovering the metabolomic fingerprint of breast cancer. Int. J. Biochem. Cell Biol. 2011, 43, 1010–1020. [Google Scholar] [CrossRef] [PubMed]
- Bu, Q.; Huang, Y.; Yan, G.; Cen, X.; Zhao, Y.L. Metabolomics: A revolution for novel cancer marker identification. Comb. Chem. High Throughput Screen. 2012, 15, 266–275. [Google Scholar] [CrossRef] [PubMed]
- Santini, G.; Mores, N.; Penas, A.; Capuano, R.; Mondino, C.; Trové, A.; Macagno, F.; Zini, G.; Cattani, P.; Martinelli, E.; et al. Electronic Nose and Exhaled Breath NMR-based Metabolomics Applications in Airways Disease. Curr. Top. Med. Chem. 2016, 16, 1610–1630. [Google Scholar] [CrossRef] [PubMed]
- Figueira, J.; Jonsson, P.; Nordin Adolfsson, A.; Adolfsson, R.; Nyberg, L.; Öhman, A. NMR analysis of the human saliva metabolome distinguishes dementia patients from matched controls. Mol. Biosyst. 2016, 12, 2562–2571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frank, A.; Möller, H.M.; Exner, T.E. Toward the Quantum Chemical Calculation of NMR Chemical Shifts of Proteins. 2. Level of Theory, Basis Set, and Solvents Model Dependence. J. Chem. Theory Comput. 2012, 8, 1480–1492. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Sun, H.; Qiu, S.; Wang, X. NMR-based metabolomics coupled with pattern recognition methods in biomarker discovery and disease diagnosis. Magn. Reson. Chem. 2013, 51, 549–556. [Google Scholar] [CrossRef] [PubMed]
- Alonso, A.; Marsal, S.; Julià, A. Analytical methods in untargeted metabolomics: State of the art in 2015. Front. Bioeng. Biotechnol. 2015, 3, 23. [Google Scholar] [CrossRef]
- Zhang, A.; Sun, H.; Wang, P.; Han, Y.; Wang, X. Recent and potential developments of biofluid analyses in metabolomics. J. Proteom. 2012, 75, 1079–1088. [Google Scholar] [CrossRef]
- Bingol, K. Recent Advances in Targeted and Untargeted Metabolomics by NMR and MS/NMR Methods. High-Throughput 2018, 7, 9. [Google Scholar] [CrossRef] [PubMed]
- Bertram, H.C.; Eggers, N.; Eller, N. Potential of Human Saliva for Nuclear Magnetic Resonance-Based Metabolomics and for Health-Related Biomarker Identification. Anal. Chem. 2009, 81, 9188–9193. [Google Scholar] [CrossRef]
- Tiziani, S.; Lopes, V.; Günther, U.L. Early stage diagnosis of oral cancer using 1H NMR-based metabolomics. Neoplasia 2009, 11, 269–276. [Google Scholar] [CrossRef]
- Louis, E.; Bervoets, L.; Reekmans, G.; De Jonge, E.; Mesotten, L.; Thomeer, M.; Adriaensens, P. Phenotyping human blood plasma by 1H-NMR: A robust protocol based on metabolite spiking and its evaluation in breast cancer. Metabolomics 2015, 11, 225–236. [Google Scholar] [CrossRef]
- Louis, E.; Adriaensens, P.; Guedens, W.; Bigirumurame, T.; Baeten, K.; Vanhove, K.; Vandeurzen, K.; Darquennes, K.; Vansteenkiste, J.; Dooms, C.; et al. Detection of Lung Cancer through Metabolic Changes Measured in Blood Plasma. J. Thorac. Oncol. 2016, 11, 516–523. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Wang, L.; Zhang, H.; Deng, P.; Chen, J.; Zhou, B.; Hu, J.; Zou, J.; Lu, W.; Xiang, P.; et al. 1H NMR-based metabolic profiling of human rectal cancer tissue. Mol. Cancer 2013, 12, 121. [Google Scholar] [CrossRef]
- Lindon, J.C.; Holmes, E.; Lindon, J.C.; Holmes, E. NMR Spectroscopy of Biofluids, Tissues, and Tissue Extracts. In eMagRes; John Wiley & Sons, Ltd.: Chichester, UK, 2008. [Google Scholar]
- Beckonert, O.; Monnerjahn, J.; Bonk, U.; Leibfritz, D. Visualizing metabolic changes in breast-cancer tissue using1H-NMR spectroscopy and self-organizing maps. NMR Biomed. 2003, 16, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Sitter, B.; Bathen, T.F.; Singstad, T.E.; Fjøsne, H.E.; Lundgren, S.; Halgunset, J.; Gribbestad, I.S. Quantification of metabolites in breast cancer patients with different clinical prognosis using HR MAS MR spectroscopy. NMR Biomed. 2010, 23, 424–431. [Google Scholar] [CrossRef] [PubMed]
- Madhu, B.; Dadulescu, M.; Griffiths, J. Artefacts in 1H NMR-based metabolomic studies on cell cultures. Magn. Reson. Mater. Phys. Biol. Med. 2015, 28, 161–171. [Google Scholar] [CrossRef] [PubMed]
- Aranibar, N.; Borys, M.; Mackin, N.A.; Ly, V.; Abu-Absi, N.; Abu-Absi, S.; Niemitz, M.; Schilling, B.; Li, Z.J.; Brock, B.; et al. NMR-based metabolomics of mammalian cell and tissue cultures. J. Biomol. NMR 2011, 49, 195–206. [Google Scholar] [CrossRef] [PubMed]
- Bradley, S.A.; Ouyang, A.; Purdie, J.; Smitka, T.A.; Wang, T.; Kaerner, A. Fermentanomics: Monitoring Mammalian Cell Cultures with NMR Spectroscopy. J. Am. Chem. Soc. 2010, 132, 9531–9533. [Google Scholar] [CrossRef] [PubMed]
- Kostidis, S.; Addie, R.D.; Morreau, H.; Mayboroda, O.A.; Giera, M. Quantitative NMR analysis of intra- and extracellular metabolism of mammalian cells: A tutorial. Anal. Chim. Acta 2017, 980, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Jewison, T.; Guo, A.C.; Wilson, M.; Knox, C.; Liu, Y.; Djoumbou, Y.; Mandal, R.; Aziat, F.; Dong, E.; et al. HMDB 3.0—The Human Metabolome Database in 2013. Nucleic Acids Res. 2012, 41, D801–D807. [Google Scholar] [CrossRef]
- Bouatra, S.; Aziat, F.; Mandal, R.; Guo, A.C.; Wilson, M.R.; Knox, C.; Bjorndahl, T.C.; Krishnamurthy, R.; Saleem, F.; Liu, P.; et al. The Human Urine Metabolome. PLoS ONE 2013, 8, e73076. [Google Scholar]
- Xia, J.; Wishart, D.S.; Xia, J.; Wishart, D.S. Using MetaboAnalyst 3.0 for Comprehensive Metabolomics Data Analysis. In Current Protocols in Bioinformatics; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2016; pp. 14.10.1–14.10.91. ISBN 9780471250951. [Google Scholar]
- Monteiro, M.S.; Barros, A.S.; Pinto, J.; Carvalho, M.; Pires-Luís, A.S.; Henrique, R.; Jerónimo, C.; Bastos, M.; de Lourdes Bastos, M.; Gil, A.M.; et al. Nuclear Magnetic Resonance metabolomics reveals an excretory metabolic signature of renal cell carcinoma. Sci. Rep. 2016, 6, 37275. [Google Scholar] [CrossRef]
- Cross, A.J.; Pollock, J.R.A.; Bingham, S.A. Haem, not protein or inorganic iron, is responsible for endogenous intestinal N-nitrosation arising from red meat. Cancer Res. 2003, 63, 2358–2360. [Google Scholar]
- Ohshima, H.; Kawabata, T. Mechanism of N-nitrosodimethylamine formation from trimethylamine and trimethylaminoxide. IARC Sci. Publ. 1978, 19, 143–153. [Google Scholar] [CrossRef]
- Awwad, H.M.; Geisel, J.; Obeid, R. Determination of trimethylamine, trimethylamine N-oxide, and taurine in human plasma and urine by UHPLC–MS/MS technique. J. Chromatogr. B 2016, 1038, 12–18. [Google Scholar] [CrossRef]
- Chalmers, R.A.; Bain, M.D.; Michelakakis, H.; Zschocke, J.; Iles, R.A. Diagnosis and management of trimethylaminuria (FMO3 deficiency) in children. J. Inherit. Metab. Dis. 2006, 29, 162–172. [Google Scholar] [CrossRef]
- Mackay, R.J.; McEntyre, C.J.; Henderson, C.; Lever, M.; George, P.M. Trimethylaminuria: Causes and diagnosis of a socially distressing condition. Clin. Biochem. Rev. 2011, 32, 33–43. [Google Scholar] [PubMed]
- Zhao, L.; Dong, M.; Liao, S.; Du, Y.; Zhou, Q.; Zheng, H.; Chen, M.; Ji, J.; Gao, H. Identification of key metabolic changes in renal interstitial fibrosis rats using metabonomics and pharmacology. Sci. Rep. 2016, 6, 27194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barr, D.B.; Wilder, L.C.; Caudill, S.P.; Gonzalez, A.J.; Needham, L.L.; Pirkle, J.L. Urinary Creatinine Concentrations in the U.S. Population: Implications for Urinary Biologic Monitoring Measurements. Environ. Health Perspect. 2004, 113, 192–200. [Google Scholar] [CrossRef] [PubMed]
- Nankivell, B.J. Abnormal Laboratory Results: Creatinine clearance and the assessment of renal function. Aust. Prescr. 2001, 24, 15–17. [Google Scholar] [CrossRef] [Green Version]
- Gowda, S.; Desai, P.B.; Kulkarni, S.S.; Hull, V.V.; Math, A.A.K.; Vernekar, S.N. Markers of renal function tests. N. Am. J. Med. Sci. 2010, 2, 170–173. [Google Scholar]
- Wang, L.; Tang, Y.; Liu, S.; Mao, S.; Ling, Y.; Liu, D.; He, X.; Wang, X. Metabonomic profiling of serum and urine by (1)H NMR-based spectroscopy discriminates patients with chronic obstructive pulmonary disease and healthy individuals. PLoS ONE 2013, 8, e65675. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, S.; Roy, R.; Singh, S.; Kumar, P.; Dalela, D.; Sankhwar, S.N.; Goel, A.; Sonkar, A.A. Taurine–A possible fingerprint biomarker in non-muscle invasive bladder cancer: A pilot study by 1H NMR spectroscopy. Cancer Biomark. 2010, 6, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.; Jung, Y.; Bang, E.J.; Cho, S.; Jang, Y.J.; Kwak, J.M.; Ryu, D.H.; Park, S.; Hwang, G.S. Noninvasive Diagnosis and Evaluation of Curative Surgery for Gastric Cancer by Using NMR-based Metabolomic Profiling. Ann. Surg. Oncol. 2014, 21, 736–742. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Wang, Y.; Zhang, X.; Zhou, J.; Wang, Y.; Zhang, X.; Zhou, J.; Wang, Y.; Zhang, X. Metabonomics studies on serum and urine of patients with breast cancer using 1H-NMR spectroscopy. Oncotarget 2017, 5–14. [Google Scholar]
- Cala, M.; Aldana, J.; Sánchez, J.; Guio, J.; Meesters, R.J.W. Urinary metabolite and lipid alterations in Colombian Hispanic women with breast cancer: A pilot study. J. Pharm. Biomed. Anal. 2018, 152, 234–241. [Google Scholar] [CrossRef] [PubMed]
- Cha, Y.J.; Kim, E.S.; Koo, J.S. Amino acid transporters and glutamine metabolism in breast cancer. Int. J. Mol. Sci. 2018, 19, 907. [Google Scholar] [CrossRef]
- Xia, J.; Broadhurst, D.I.; Wilson, M.; Wishart, D.S. Translational biomarker discovery in clinical metabolomics: An introductory tutorial. Metabolomics 2013, 9, 280–299. [Google Scholar] [CrossRef] [PubMed]
- Mishra, P.; Ambs, S. Metabolic signatures of human breast cancer. Mol. Cell. Oncol. 2015, 2, e992217. [Google Scholar] [CrossRef] [PubMed] [Green Version]






| Sample Group | N° Urine Samples | Age Range/Years | Mean Age ± SD 1 |
|---|---|---|---|
| Breast Cancer (BC) | n = 40 | 40–74 | 59 ± 10 |
| Control (CTL) | n = 38 | 40–72 | 53 ± 8 |
| Peak n° | Metabolite | δ (ppm) | Relative Concentrations (mM) | Variation | K-S 3 (Normality) | FO (%) 6 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTL | BC | CTL | BC | Mean Comparison 5 | CTL | BC | ||||||||||
| Multiplicity | Min 1 | Max 2 | Average | Min | Max | Average | Statistic 4 | p-Value | Statistic | p-Value | ||||||
| 6 | α-hydroxyisobutyrate | 1.35 (s) 7 | 1.21 | 6.64 | 3.94 | 0.15 | 0.89 | 0.55 | ↓ | 0.118 | 0.200 | 0.138 | 0.200 | 1.10 × 10−12 | 78 | 100 |
| 18 | creatinine | 3.03 (s), 4.05 (s) | 116.84 | 381.60 | 200.56 | 10.49 | 216.36 | 83.87 | ↓ | 0.231 | 0.200 | 0.265 | 0.103 | 9.23 × 10−11 | 100 | 100 |
| 13 | pyruvate | 2.36 (s) | 1.90 | 4.35 | 3.34 | 0.47 | 3.71 | 1.86 | ↓ | 0.317 | 0.032 | 0.114 | 0.200 | 3.86 × 10−10 | 100 | 100 |
| 5 | threonine | 1.32 (d) 8, 3.58 (d), 4.25 (m) 9 | 2.15 | 6.81 | 4.33 | 0.74 | 4.09 | 2.36 | ↓ | 0.203 | 0.200 | 0.197 | 0.200 | 5.12 × 10−10 | 98 | 93 |
| 17 | α-oxoglutarate | 2.43 (t) 10, 2.99 (t) | 3.00 | 10.54 | 6.33 | 0.82 | 6.13 | 3.07 | ↓ | 0.245 | 0.200 | 0.207 | 0.200 | 8.78 × 10−10 | 95 | 92 |
| 3 | β-hydroxyisovalerate | 1.26 (s), 2.35 (s) | 0.71 | 1.64 | 1.19 | 0.15 | 1.16 | 0.57 | ↓ | 0.166 | 0.200 | 0.298 | 0.035 | 7.15 × 10−9 | 98 | 100 |
| 16 | dimethylamine | 2.72 (s) | 3.43 | 9.71 | 7.00 | 0.31 | 8.07 | 3.61 | ↓ | 0.232 | 0.200 | 0.267 | 0.097 | 1.00 × 10−8 | 95 | 100 |
| 8 | acetate | 1.91 (s) | 1.30 | 3.30 | 2.03 | 0.56 | 2.44 | 1.36 | ↓ | 0.202 | 0.200 | 0.206 | 0.200 | 1.00 × 10−7 | 95 | 96 |
| 4 | lactate | 1.32 (d), 4.11 (m) | 2.14 | 6.41 | 3.99 | 0.75 | 4.71 | 2.88 | ↓ | 0.174 | 0.200 | 0.153 | 0.200 | 1.10 × 10-7 | 98 | 100 |
| 20 | choline | 3.19 (s), 3.51 (m), 4.06 (m) | 0.93 | 2.45 | 1.61 | 0.25 | 2.07 | 1.08 | ↓ | 0.221 | 0.200 | 0.223 | 0.200 | 1.20 × 10−7 | 100 | 96 |
| 28 | serine | 3.84 (q) 11, 3.94 (q), 3.98 (q) | 15.00 | 60.92 | 29.59 | 5.03 | 30.12 | 15.32 | ↓ | 0.231 | 0.200 | 0.220 | 0.200 | 3.59 × 10−7 | 95 | 92 |
| 11 | carnitine | 2.40 (s), 2.45 (s), 3.21 (s) | 0.65 | 2.95 | 1.70 | 0.15 | 1.97 | 0.86 | ↓ | 0.218 | 0.200 | 0.200 | 0.200 | 1.46 × 10−6 | 100 | 80 |
| 14 | succinate | 2.39 (s) | 0.70 | 1.61 | 1.04 | 0.18 | 1.25 | 0.68 | ↓ | 0.192 | 0.200 | 0.146 | 0.200 | 1.88 × 10−6 | 100 | 96 |
| 9 | glutamine | 2.10 (t), 2.42 (m), 3.77 (t) | 1.47 | 11.19 | 7.13 | 1.65 | 10.81 | 4.78 | ↓ | 0.219 | 0.200 | 0.242 | 0.186 | 2.87 × 10−6 | 93 | 67 |
| 12 | 4-cresol sulphate | 2.24 (s), 6.82 (m), 7.23 (m) | 1.22 | 11.91 | 5.51 | 0.79 | 4.92 | 2.09 | ↓ | 0.216 | 0.200 | 0.191 | 0.200 | 1.38 × 10−5 | 100 | 84 |
| 36 | formate | 8.44 (s) | 0.18 | 1.58 | 0.70 | 1.44 | 6.66 | 3.75 | ↑ | 0.256 | 0.184 | 0.349 | 0.005 | 1.54 × 10−5 | 77 | 79 |
| 29 | creatine | 3.03 (s), 3.92 (s) | 6.19 | 40.74 | 20.77 | 1.50 | 17.00 | 7.46 | ↓ | 0.194 | 0.200 | 0.225 | 0.200 | 3.29 × 10−5 | 98 | 92 |
| 27 | guanidoacetate | 3.79 (s) | 7.98 | 21.04 | 12.41 | 1.71 | 14.89 | 8.72 | ↓ | 0.206 | 0.200 | 0.171 | 0.200 | 5.38 × 10−5 | 100 | 92 |
| 19 | cis-aconitate | 3.11 (d), 5.72 (m) | 4.08 | 12.31 | 7.51 | 0.59 | 12.20 | 5.14 | ↓ | 0.167 | 0.200 | 0.207 | 0.200 | 8.28 × 10−5 | 95 | 100 |
| 7 | alanine | 1.47 (d), 3.78 (m) | 1.59 | 4.48 | 2.90 | 0.57 | 3.51 | 2.29 | ↓ | 0.202 | 0.200 | 0.255 | 0.136 | 8.42 × 10−5 | 100 | 100 |
| 1 | valine | 1.03 (d), 2.26 (m), 3.60 (d)4 | 0.32 | 1.14 | 0.60 | 0.27 | 1.24 | 0.50 | ↓ | 0.270 | 0.131 | 0.310 | 0.023 | 1.35 × 10−4 | 95 | 68 |
| 10 | acetone | 2.22 (s) | 0.78 | 2.69 | 1.60 | 0.33 | 2.31 | 1.28 | ↓ | 0.167 | 0.200 | 0.171 | 0.200 | 1.64 × 10−4 | 100 | 92 |
| 22 | trimethylamine N-oxide | 3.25 (s) | 3.37 | 15.65 | 8.96 | 1.31 | 18.04 | 5.63 | ↓ | 0.173 | 0.200 | 0.300 | 0.033 | 2.82 × 10−4 | 85 | 92 |
| 26 | mannitol | 3.67 (m), 3.75 (m), 3.79 (d) | 14.24 | 49.04 | 28.85 | 3.67 | 24.04 | 16.10 | ↓ | 0.230 | 0.200 | 0.167 | 0.200 | 6.31 × 10−4 | 100 | 92 |
| 25 | glycine | 3.55 (s) | 1.93 | 43.52 | 18.57 | 1.88 | 22.13 | 10.20 | ↓ | 0.166 | 0.200 | 0.166 | 0.200 | 9.26 × 10−4 | 88 | 100 |
| 31 | trigonelline | 4.43 (s), 8.07 (t), 8.83 (m), 8.78 (m) | 1.36 | 4.21 | 2.79 | 0.39 | 6.13 | 2.68 | ↓ | 0.168 | 0.200 | 0.264 | 0.106 | 1.35 × 10−3 | 100 | 88 |
| 15 | citrate | 2.53 (d), 2.69 (d) | 9.14 | 85.10 | 33.04 | 10.58 | 55.47 | 29.53 | ↓ | 0.327 | 0.023 | 0.231 | 0.200 | 2.10 × 10−3 | 100 | 100 |
| 23 | taurine | 3.25 (t), 3.42 (t) | 6.66 | 12.95 | 10.18 | 1.93 | 14.46 | 6.80 | ↓ | 0.164 | 0.200 | 0.307 | 0.026 | 2.58 × 10−3 | 98 | 92 |
| 2 | α-hydroxybutyrate | 1.19 (d), 2.27 (m), 2.39 (m) | 0.47 | 1.30 | 0.77 | 0.39 | 3.38 | 1.47 | ↑ | 0.208 | 0.200 | 0.318 | 0.017 | 2.71 × 10−3 | 100 | 95 |
| 21 | betaine | 3.25 (s), 3.89 (s) | 0.52 | 2.62 | 1.37 | 0.22 | 1.96 | 1.05 | ↓ | 0.261 | 0.163 | 0.144 | 0.200 | 1.71 × 10−2 | 93 | 100 |
| 35 | hypoxanthine | 8.18 (s), 8.20 (s) | 0.45 | 2.77 | 1.57 | 0.31 | 2.14 | 0.78 | ↓ | 0.214 | 0.200 | 0.316 | 0.018 | 3.42 × 10−2 | 80 | 64 |
| 34 | 3-methylhistidine | 3.22 (m), 3.31 (m), 3.73 (s), 3.96 (q), 8.08 (s) | 0.56 | 11.63 | 4.33 | 0.54 | 10.87 | 2.44 | ↓ | 0.224 | 0.200 | 0.419 | 0.000 | 1.74 × 10−1 | 93 | 100 |
| 24 | 4-hydroxyphenylacetate | 3.44 (s), 6.85 (d), 7.15 (d) | 0.90 | 4.14 | 1.89 | 0.59 | 5.02 | 1.57 | ↓ | 0.287 | 0.084 | 0.282 | 0.061 | 3.65 × 10−1 | 90 | 88 |
| 32 | histidine | 7.08 (m), 7.87 (d) | 0.12 | 4.06 | 0.95 | 0.33 | 1.09 | 0.57 | ↓ | 0.406 | 0.001 | 0.206 | 0.200 | 3.88 × 10−1 | 88 | 89 |
| 33 | phenylalanine | 7.31 (m), 7.37 (m), 7.43 (m) | 0.97 | 4.84 | 1.98 | 0.91 | 5.99 | 2.66 | ↑ | 0.252 | 0.200 | 0.182 | 0.200 | 6.93 × 10−1 | 76 | 88 |
| 30 | hippurate | 3.96 (d), 7.54 (t), 7.63 (t), 7.82 (d) | 5.56 | 70.01 | 24.73 | 2.27 | 58.30 | 33.76 | ↑ | 0.258 | 0.174 | 0.313 | 0.021 | 7.48 × 10−1 | 93 | 96 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Silva, C.L.; Olival, A.; Perestrelo, R.; Silva, P.; Tomás, H.; Câmara, J.S. Untargeted Urinary 1H NMR-Based Metabolomic Pattern as a Potential Platform in Breast Cancer Detection. Metabolites 2019, 9, 269. https://doi.org/10.3390/metabo9110269
Silva CL, Olival A, Perestrelo R, Silva P, Tomás H, Câmara JS. Untargeted Urinary 1H NMR-Based Metabolomic Pattern as a Potential Platform in Breast Cancer Detection. Metabolites. 2019; 9(11):269. https://doi.org/10.3390/metabo9110269
Chicago/Turabian StyleSilva, Catarina L., Ana Olival, Rosa Perestrelo, Pedro Silva, Helena Tomás, and José S. Câmara. 2019. "Untargeted Urinary 1H NMR-Based Metabolomic Pattern as a Potential Platform in Breast Cancer Detection" Metabolites 9, no. 11: 269. https://doi.org/10.3390/metabo9110269
APA StyleSilva, C. L., Olival, A., Perestrelo, R., Silva, P., Tomás, H., & Câmara, J. S. (2019). Untargeted Urinary 1H NMR-Based Metabolomic Pattern as a Potential Platform in Breast Cancer Detection. Metabolites, 9(11), 269. https://doi.org/10.3390/metabo9110269