Activation Studies of the β-Carbonic Anhydrase from the Pathogenic Protozoan Entamoeba histolytica with Amino Acids and Amines
Abstract
:1. Introduction
2. Results and Discussion
- (i)
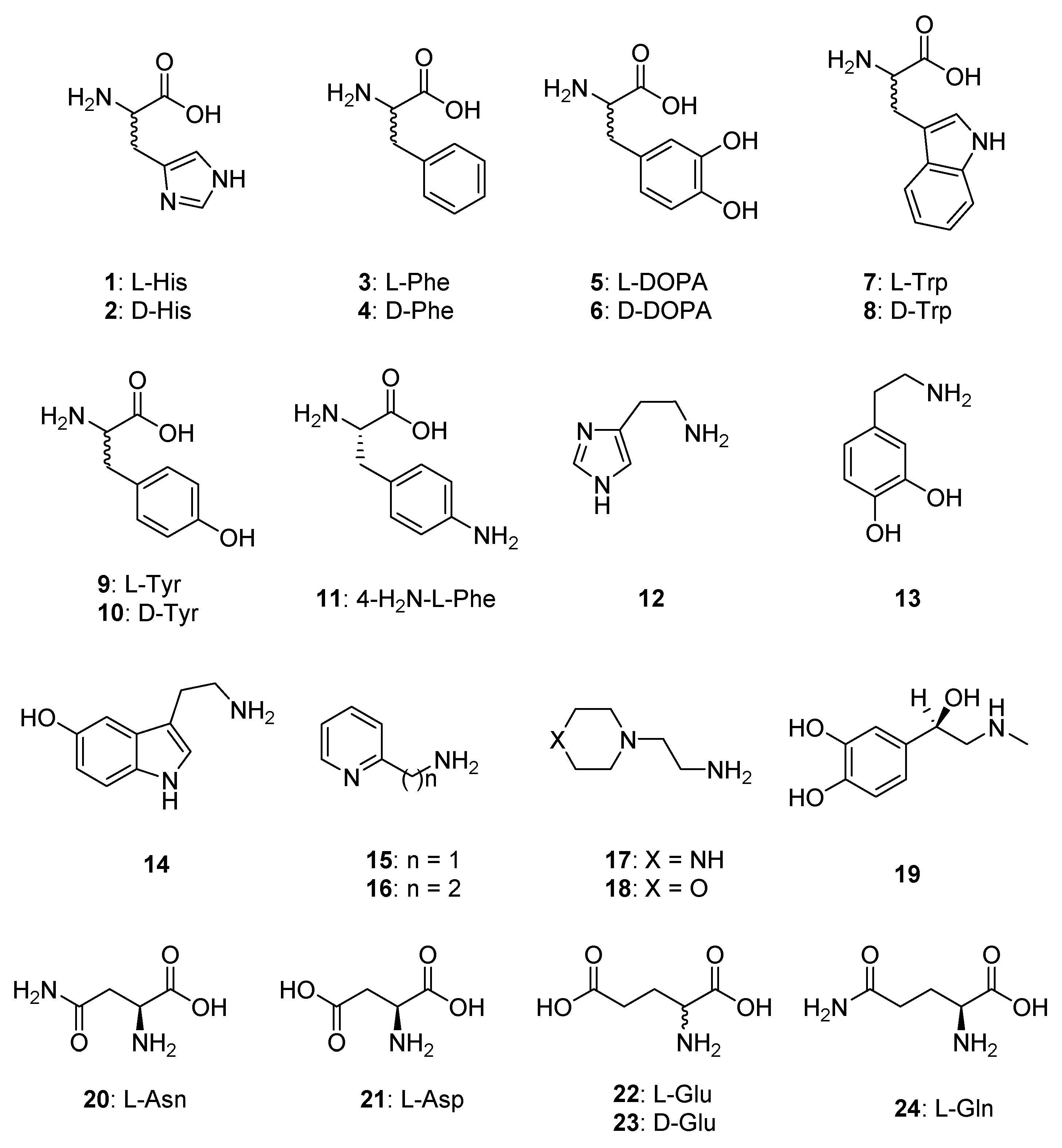
- Some heterocyclic-alkyl amines, such as 2-pyridyl-methyl/ethyl-amine 15, 16 and 4-(2-aminoethyl)-morpholine, were devoid of EhiCA activating properties up to 100 µM concentration of activator in the assay system. All these compounds are structurally related, possessing a heterocyclic ring and aminomethyl/aminoethyl moieties in their molecules.
- (ii)
- L-His, dopamine, 1-(2-aminoethyl)-piperazine and D-Glu were poor EhiCA activators, with activation constants ranging between 30.3 and 78.7 µM (Table 2). There is no strong structural correlation between these three compounds.
- (iii)
- Many of the compounds investigated here showed medium potency efficacy as EhiCA activators, with KAs ranging between 16.5 and 25.6 µM. They include L-Phe, L-DOPA, L-adrenaline, L-Asn, L-Asp, L-Glu and L-Gln. It may be observed that there are no remarkable differences of activity between the pairs L-Asp/L-Asn and L-Glu/L-Gln, whereas D-Glu was more ineffective compared to L-Glu. This is in fact the exception, as for other L-/D-enantiomeric amino acids investigated here, the D-enantiomer was the most effective activator (see later in the text).
- (iv)
- Effective EhiCA activating properties were detected for the following amino acids/amines: D-His, D-Phe, D-DOPA, L- and D-Trp, L- and D-Tyr, 4-amino-L-Tyr, histamine and serotonin, which showed KAs ranging between 1.07 and 10.1 µM. The best activator was D-Tyr (KA of 1.07 µM). In fact for all aromatic amino acids investigated here, the D-enantiomer was more effective as EhiCA activator compared to the corresponding L-enantiomer. For the Phe-Tyr-DOPA subseries, the activity increased by hydroxylation of the Phe, achieving a maximum for Tyr and then slightly decreased with the introduction of an additional OH moiety in DOPA, but always the D-enantiomers were better activators compared to the L-ones. The loss of the carboxyl moiety, such as in histamine and serotonin, did not lead to important changes of activity compared to the corresponding D-amino acids, but in the case of dopamine, the activating efficacy was much lower compared to those of both L- and D-DOPA.
- (v)
- The activation profile of EhiCA with amino acid and amine derivatives is rather different from those of other CAs, among which the protozoan β-CA from Leishmania donovani chagasi (LdcCA) or the α-class human CAs, isoforms hCA I and II. For example 17 was a nanomolar activator for LdcCA whereas its affinity for EhiCA was of only 43.8 µM. For the moment, no EhiCA-selective activators were detected.
3. Materials and Methods
3.1. EhiCA Production and Purification
3.2. CA activity and Activation Measurements
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bua, S.; Haapanen, S.; Kuuslahti, M.; Parkkila, S.; Supuran, C.T. Sulfonamide Inhibition Studies of a New β-Carbonic Anhydrase from the Pathogenic Protozoan Entamoeba histolytica. Int. J. Mol. Sci. 2018, 19, E3946. [Google Scholar] [CrossRef] [PubMed]
- Haapanen, S.; Bua, S.; Kuuslahti, M.; Parkkila, S.; Supuran, C.T. Cloning, Characterization and Anion Inhibition Studies of a β-Carbonic Anhydrase from the Pathogenic Protozoan Entamoeba histolytica. Molecules 2018, 23, E3112. [Google Scholar] [CrossRef] [PubMed]
- Shirley, D.T.; Farr, L.; Watanabe, K.; Moonah, S. A Review of the Global Burden, New Diagnostics, and Current Therapeutics for Amebiasis. Open Forum Infect. Dis. 2018, 5, ofy161. [Google Scholar] [CrossRef]
- Hashmey, N.; Genta, N.; White, N., Jr. Parasites and Diarrhea. I: Protozoans and Diarrhea. J. Travel Med. 1997, 4, 17–31. [Google Scholar] [CrossRef] [PubMed]
- Loftus, B.; Anderson, I.; Davies, R.; Alsmark, U.C.; Samuelson, J.; Amedeo, P.; Roncaglia, P.; Berriman, M.; Hirt, R.P.; Mann, B.J.; et al. The genome of the protist parasite Entamoeba histolytica. Nature 2005, 433, 865–868. [Google Scholar] [CrossRef] [PubMed]
- Andrade, R.M.; Reed, S.L. New drug target in protozoan parasites: The role of thioredoxin reductase. Front. Microbiol. 2015, 6, 975. [Google Scholar] [CrossRef]
- Supuran, C.T. Structure and function of carbonic anhydrases. Biochem. J. 2016, 473, 2023–2032. [Google Scholar] [CrossRef]
- Supuran, C.T. Carbonic Anhydrases and Metabolism. Metabolites 2018, 8, E25. [Google Scholar] [CrossRef]
- Capasso, C.; Supuran, C.T. An overview of the alpha-, beta- and gamma-carbonic anhydrases from Bacteria: Can bacterial carbonic anhydrases shed new light on evolution of bacteria? J. Enzyme Inhib. Med. Chem. 2015, 30, 325–332. [Google Scholar] [CrossRef]
- Supuran, C.T.; Capasso, C. An Overview of the Bacterial Carbonic Anhydrases. Metabolites 2017, 7, E56. [Google Scholar] [CrossRef]
- Supuran, C.T. Carbonic anhydrases: Novel therapeutic applications for inhibitors and activators. Nat. Rev. Drug Discov. 2008, 7, 168–181. [Google Scholar] [CrossRef] [PubMed]
- Neri, D.; Supuran, C.T. Interfering with pH regulation in tumours as a therapeutic strategy. Nat. Rev. Drug Discov. 2011, 10, 767–777. [Google Scholar] [CrossRef] [PubMed]
- Supuran, C.T. Carbonic Anhydrase Inhibition and the Management of Hypoxic Tumors. Metabolites 2017, 7, E48. [Google Scholar] [CrossRef] [PubMed]
- Supuran, C.T. Advances in structure-based drug discovery of carbonic anhydrase inhibitors. Expert Opin. Drug Discov. 2017, 12, 61–88. [Google Scholar] [CrossRef] [PubMed]
- Nishimori, I.; Onishi, S.; Takeuchi, H.; Supuran, C.T. The α and β-Classes Carbonic Anhydrases from Helicobacter pylori as Novel Drug Targets. Curr. Pharm. Des. 2008, 14, 622–630. [Google Scholar] [PubMed]
- Supuran, C.T.; Capasso, C. New light on bacterial carbonic anhydrases phylogeny based on the analysis of signal peptide sequences. J. Enzyme Inhib. Med. Chem. 2016, 31, 1254–1260. [Google Scholar] [CrossRef] [PubMed]
- Supuran, C.T.; Capasso, C. Biomedical applications of prokaryotic carbonic anhydrases. Expert Opin. Ther. Pat. 2018, 28, 745–754. [Google Scholar] [CrossRef] [PubMed]
- Zolfaghari Emameh, R.; Barker, H.; Hytönen, V.P.; Tolvanen, M.E.E.; Parkkila, S. Beta carbonic anhydrases: Novel targets for pesticides and anti-parasitic agents in agriculture and livestock husbandry. Parasites Vect. 2014, 7, 403. [Google Scholar] [CrossRef]
- Syrjänen, L.; Parkkila, S.; Scozzafava, A.; Supuran, C.T. Sulfonamide inhibition studies of the β carbonic anhydrase from Drosophila melanogaster. Bioorg. Med. Chem. Lett. 2014, 24, 2797–2801. [Google Scholar] [CrossRef]
- Vermelho, A.B.; Capaci, G.R.; Rodrigues, I.A.; Cardoso, V.S.; Mazotto, A.M.; Supuran, C.T. Carbonic anhydrases from Trypanosoma and Leishmania as anti-protozoan drug targets. Bioorg. Med. Chem. 2017, 25, 1543–1555. [Google Scholar] [CrossRef]
- Da Silva Cardoso, V.; Vermelho, A.B.; Ricci Junior, E.; Almeida Rodrigues, I.; Mazotto, A.M.; Supuran, C.T. Antileishmanial activity of sulphonamide nanoemulsions targeting the β-carbonic anhydrase from Leishmania species. J. Enzyme Inhib. Med. Chem. 2018, 33, 850–857. [Google Scholar] [CrossRef] [PubMed]
- Vermelho, A.B.; da Silva Cardoso, V.; Ricci Junior, E.; Dos Santos, E.P.; Supuran, C.T. Nanoemulsions of sulfonamide carbonic anhydrase inhibitors strongly inhibit the growth of Trypanosoma cruzi. J. Enzyme Inhib. Med. Chem. 2018, 33, 139–146. [Google Scholar] [CrossRef] [PubMed]
- De Menezes Dda, R.; Calvet, C.M.; Rodrigues, G.C.; de Souza Pereira, M.C.; Almeida, I.R.; de Aguiar, A.P.; Supuran, C.T.; Vermelho, A.B. Hydroxamic acid derivatives: A promising scaffold for rational compound optimization in Chagas disease. J. Enzyme Inhib. Med. Chem. 2016, 31, 964–973. [Google Scholar] [CrossRef] [PubMed]
- Rowlett, R.S. Structure and catalytic mechanism of the β-carbonic anhydrases. Biochim. Biophys. Acta Prot. Proteom. 2010, 1804, 362–373. [Google Scholar] [CrossRef] [PubMed]
- Covarrubias, A.S.; Bergfors, T.; Jones, T.A.; Högbom, M. Structural mechanics of the pH-dependent activity of beta-carbonic anhydrase from Mycobacterium tuberculosis. J. Biol. Chem. 2006, 281, 4993–4999. [Google Scholar] [CrossRef] [PubMed]
- Murray, A.B.; Aggarwal, M.; Pinard, M.; Vullo, D.; Patrauchan, M.; Supuran, C.T.; McKenna, R. Structural Mapping of Anion Inhibitors to β-Carbonic Anhydrase psCA3 from Pseudomonas aeruginosa. Chem. Med. Chem. 2018, 13, 2024–2029. [Google Scholar] [CrossRef] [PubMed]
- Zimmerman, S.A.; Ferry, J.G.; Supuran, C.T. Inhibition of the archaeal beta-class (Cab) and gamma-class (Cam) carbonic anhydrases. Curr. Top. Med. Chem. 2007, 7, 901–908. [Google Scholar] [CrossRef]
- De Simone, G.; Supuran, C.T. (In) organic anions as carbonic anhydrase inhibitors. J. Inorg. Biochem. 2012, 111, 117–129. [Google Scholar] [CrossRef]
- Supuran, C.T. Applications of carbonic anhydrases inhibitors in renal and central nervous system diseases. Expert Opin. Ther. Pat. 2018, 28, 713–721. [Google Scholar] [CrossRef]
- Supuran, C.T. Carbonic anhydrase inhibitors and their potential in a range of therapeutic areas. Expert Opin. Ther. Pat. 2018, 28, 709–712. [Google Scholar] [CrossRef]
- Nocentini, A.; Supuran, C.T. Carbonic anhydrase inhibitors as antitumor/antimetastatic agents: A patent review (2008–2018). Expert Opin Ther Pat. 2018, 28, 729–740. [Google Scholar] [CrossRef] [PubMed]
- Supuran, C.T. Carbonic anhydrase activators. Future Med. Chem. 2018, 10, 561–573. [Google Scholar] [CrossRef] [PubMed]
- Canto de Souza, L.; Provensi, G.; Vullo, D.; Carta, F.; Scozzafava, A.; Costa, A.; Schmidt, S.D.; Passani, M.B.; Supuran, C.T.; Blandina, P. Carbonic anhydrase activation enhances object recognition memory in mice through phosphorylation of the extracellular signal-regulated kinase in the cortex and the hippocampus. Neuropharmacology 2017, 118, 148–156. [Google Scholar] [CrossRef] [PubMed]
- Angeli, A.; Kuuslahti, M.; Parkkila, S.; Supuran, C.T. Activation studies with amines and amino acids of the α-carbonic anhydrase from the pathogenic protozoan Trypanosoma cruzi. Bioorg. Med. Chem. 2018, 26, 4187–4190. [Google Scholar] [CrossRef] [PubMed]
- Angeli, A.; Del Prete, S.; Alasmary, F.A.S.; Alqahtani, L.S.; AlOthman, Z.; Donald, W.A.; Capasso, C.; Supuran, C.T. The first activation studies of the η-carbonic anhydrase from the malaria parasite Plasmodium falciparum with amines and amino acids. Bioorg. Chem. 2018, 80, 94–98. [Google Scholar] [CrossRef] [PubMed]
- Stefanucci, A.; Angeli, A.; Dimmito, M.P.; Luisi, G.; Del Prete, S.; Capasso, C.; Donald, W.A.; Mollica, A.; Supuran, C.T. Activation of β- and γ-carbonic anhydrases from pathogenic bacteria with tripeptides. J. Enzyme Inhib. Med. Chem. 2018, 33, 945–950. [Google Scholar] [CrossRef] [PubMed]
- Angeli, A.; Alasmary, F.A.S.; Del Prete, S.; Osman, S.M.; AlOthman, Z.; Donald, W.A.; Capasso, C.; Supuran, C.T. The first activation study of a δ-carbonic anhydrase: TweCAδ from the diatom Thalassiosira weissflogii is effectively activated by amines and amino acids. J. Enzyme Inhib. Med. Chem. 2018, 33, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Khalifah, R.G. The carbon dioxide hydration activity of carbonic anhydrase. I. Stop-flow kinetic studies on the native human isoenzymes B and C. J. Biol. Chem. 1971, 246, 2561–2573. [Google Scholar] [PubMed]
- Briganti, F.; Mangani, S.; Orioli, P.; Scozzafava, A.; Vernaglione, G.; Supuran, C.T. Carbonic anhydrase activators: X-ray crystallographic and spectroscopic investigations for the interaction of isozymes I and II with histamine. Biochemistry 1997, 36, 10384–10392. [Google Scholar] [CrossRef] [PubMed]
- Clare, B.W.; Supuran, C.T. Carbonic anhydrase activators. 3: Structure-activity correlations for a series of isozyme II activators. J. Pharm. Sci. 1994, 83, 768–773. [Google Scholar] [CrossRef] [PubMed]
- Temperini, C.; Scozzafava, A.; Vullo, D.; Supuran, C.T. Carbonic anhydrase activators. Activation of isoforms I, II, IV, VA, VII, and XIV with L- and D-phenylalanine and crystallographic analysis of their adducts with isozyme II: Stereospecific recognition within the active site of an enzyme and its consequences for the drug design. J. Med. Chem. 2006, 49, 3019–3027. [Google Scholar] [PubMed]
- Angeli, A.; Donald, W.A.; Parkkila, S.; Supuran, C.T. Activation studies with amines and amino acids of the β-carbonic anhydrase from the pathogenic protozoan. Leishmania donovani chagasi. Bioorg. Chem. 2018, 78, 406–410. [Google Scholar] [CrossRef] [PubMed]
| Isozyme | kcat * | KM * | (kcat)L-Trp ** | KA *** (µM) |
|---|---|---|---|---|
| (s−1) | (mM) | (s−1) | L-Trp | |
| hCA I a | 2.0 × 105 | 4.0 | 3.4 × 105 | 44.0 |
| hCA II a | 1.4 × 106 | 9.3 | 4.9 × 106 | 27.0 |
| LdCA | 9.35 × 105 | 15.8 | 1.9 × 106 | 4.02 |
| EhiCA b | 6.7 × 105 | 7.5 | 1.9 × 106 | 5.24 |
| No. | Compound | KA (mM) * | |||
|---|---|---|---|---|---|
| hCA I a | hCA II a | LdcCA b | EhiCA c | ||
| 1 | L-His | 0.03 | 10.9 | 8.21 | 78.7 |
| 2 | D-His | 0.09 | 43 | 4.13 | 9.83 |
| 3 | L-Phe | 0.07 | 0.013 | 9.16 | 16.5 |
| 4 | D-Phe | 86 | 0.035 | 3.95 | 10.1 |
| 5 | L-DOPA | 3.1 | 11.4 | 1.64 | 16.6 |
| 6 | D-DOPA | 4.9 | 7.8 | 5.47 | 4.05 |
| 7 | L-Trp | 44 | 27 | 4.02 | 5.24 |
| 8 | D-Trp | 41 | 12 | 6.18 | 4.95 |
| 9 | L-Tyr | 0.02 | 0.011 | 8.05 | 4.52 |
| 10 | D-Tyr | 0.04 | 0.013 | 1.27 | 1.07 |
| 11 | 4-H2N-L-Phe | 0.24 | 0.15 | 15.9 | 8.12 |
| 12 | Histamine | 2.1 | 125 | 0.74 | 7.38 |
| 13 | Dopamine | 13.5 | 9.2 | 0.81 | 30.8 |
| 14 | Serotonin | 45 | 50 | 0.62 | 4.94 |
| 15 | 2-Pyridyl-methylamine | 26 | 34 | 0.23 | >100 |
| 16 | 2-(2-Aminoethyl)pyridine | 13 | 15 | 0.012 | >100 |
| 17 | 1-(2-Aminoethyl)-piperazine | 7.4 | 2.3 | 0.009 | 43.8 |
| 18 | 4-(2-Aminoethyl)-morpholine | 0.14 | 0.19 | 0.94 | >100 |
| 19 | L-Adrenaline | 0.09 | 96 | 4.89 | 25.6 |
| 20 | L-Asn | 11.3 | >100 | 4.76 | 23.8 |
| 21 | L-Asp | 5.2 | >100 | 0.3 | 23.9 |
| 22 | L-Glu | 6.43 | >100 | 12.9 | 25.5 |
| 23 | D-Glu | 10.7 | >100 | 0.082 | 30.3 |
| 24 | L-Gln | >100 | >50 | 2.51 | 20.1 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bua, S.; Haapanen, S.; Kuuslahti, M.; Parkkila, S.; Supuran, C.T. Activation Studies of the β-Carbonic Anhydrase from the Pathogenic Protozoan Entamoeba histolytica with Amino Acids and Amines. Metabolites 2019, 9, 26. https://doi.org/10.3390/metabo9020026
Bua S, Haapanen S, Kuuslahti M, Parkkila S, Supuran CT. Activation Studies of the β-Carbonic Anhydrase from the Pathogenic Protozoan Entamoeba histolytica with Amino Acids and Amines. Metabolites. 2019; 9(2):26. https://doi.org/10.3390/metabo9020026
Chicago/Turabian StyleBua, Silvia, Susanna Haapanen, Marianne Kuuslahti, Seppo Parkkila, and Claudiu T. Supuran. 2019. "Activation Studies of the β-Carbonic Anhydrase from the Pathogenic Protozoan Entamoeba histolytica with Amino Acids and Amines" Metabolites 9, no. 2: 26. https://doi.org/10.3390/metabo9020026
APA StyleBua, S., Haapanen, S., Kuuslahti, M., Parkkila, S., & Supuran, C. T. (2019). Activation Studies of the β-Carbonic Anhydrase from the Pathogenic Protozoan Entamoeba histolytica with Amino Acids and Amines. Metabolites, 9(2), 26. https://doi.org/10.3390/metabo9020026







