Age-Dependent and Tissue-Specific Alterations in the rDNA Clusters of the Panax ginseng C. A. Meyer Cultivated Cell Lines
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Cytological Analysis
2.3. Isolation of Total DNA and Polymerase Chain Reaction Amplification
2.4. Cloning and Sequencing of the 18S rDNA Sequences
2.5. Prediction of 18S rRNA Secondary Structure
2.6. Statistical Analysis and Phylogenetic Reconstructions
3. Results
3.1. Cytogenetic Analysis
3.2. Nucleotide Polymorphisms of the 18S rDNA Sequences in Cultivated Cell Lines
3.3. Distribution of Nucleotide Diversity along the 18S rDNA Sequences
3.4. Secondary Structures of the 18S rRNA ES3 and Helices
3.5. Phylogenetic Reconstructions for the 18S rDNA Gene Sequences
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gantait, S.; Mitra, M.; Chen, J.-T. Biotechnological Interventions for ginsenosides production. Biomolecules 2020, 10, 538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, Y.S.; Hahn, E.J.; Murthy, H.N.; Paek, K.Y. Adventitious root growth and ginsenoside accumulation in Panax ginseng cultures as affected by methyl jasmonate. Biotechnol. Lett. 2004, 26, 1612–1622. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Galera, S.; Pelacho, A.M.; Gené, A.; Capell, T.; Christou, P. The genetic manipulation of medicinal and aromatic plants. Plant Cell Rep. 2007, 26, 1689–1715. [Google Scholar] [CrossRef] [PubMed]
- Verpoorte, R.; van der Heijden, R.; Memelink, J. Engineering the plant cell factory forsecondary metabolite production. Transgenic Res. 2000, 9, 323–343. [Google Scholar] [CrossRef] [PubMed]
- Espinosa-Leal, C.A.; Puente-Garza, C.A.; Garsia-Lara, S. In vitro plant tissue culture: Means for production of biological active compounds. Planta 2018, 248, 1–18. [Google Scholar] [CrossRef]
- Rani, V.; Raina, S.N. Genetic fidelity of organized meristem-derived micropropagated plants: A critical reappraisal. Vitro Cell. Dev. Biol. Plant 2000, 36, 319–330. [Google Scholar] [CrossRef]
- Kaeppler, S.M.; Kaeppler, H.F.; Rhee, Y. Epigenetic aspects of somaclonal variation in plants. Plant Mol. Biol. 2000, 43, 179–188. [Google Scholar] [CrossRef]
- Krishna, H.; Alizadeh, M.; Singh, D.; Singh, U.; Chauhan, N.; Eftekhari, M.; Sadh, R.K. Somaclonal variations and their applications in horticultural crops improvement. Biotech 2016, 6, 54. [Google Scholar] [CrossRef] [Green Version]
- Bairu, M.W.; Aremu, A.O.; Staden, J.V. Somaclonal variation in plants: Causes and detection methods. Plant Growth Regul. 2011, 63, 147–173. [Google Scholar] [CrossRef]
- Sun, Q.; Sun, H.; Bell, R.L.; Li, H.; Xin, L. Variation of phenotype, ploidy level, and organogenic potential of in vitro regenerated polyploids of Pyrus communis. Plant Cell Tissue Organ Cult. 2011, 107, 131–140. [Google Scholar] [CrossRef]
- Winarto, B.; Rachmawati, F.; Pramanik, D.; Teixeira da Silva, J.A. Morphological and cytological diversity of regenerants derived from half-anther cultures of anthurium. Plant Cell Tissue Organ Cult. 2011, 105, 363–374. [Google Scholar] [CrossRef]
- Sivanesan, I.; Jeong, B.R. Identification of somaclonal variants in proliferating shoot cultures of Senecio cruentus cv. Tokyo Daruma. Plant Cell Tissue Organ Cult. 2012, 111, 247–253. [Google Scholar] [CrossRef]
- Orzechowska, M.; Stępień, K.; Kamińska, T.; Siwińska, D. Chromosome variations in regenerants of Arabidopsis thaliana derived from 2- and 6-week-old callus detected using flow cytometry and FISH analyses. Plant Cell Tissue Organ Cult. 2013, 112, 263–273. [Google Scholar] [CrossRef] [Green Version]
- Khrolenko, Y.A.; Burundukova, O.L.; Lauve, L.S.; Muzarok, T.I.; Makhan’kov, V.V.; Zhuravlev, Y.N. Characterization of the variability of nucleoli in the cells of Panax ginseng Meyer in vivo and in vitro. J. Ginseng Res. 2012, 36, 322–326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kiselev, K.V.; Shumakova, O.A.; Tchernoded, G.K. Mutation of Panax ginseng genes during long-term cultivation of ginseng cell cultures. J. Plant Physiol. 2011, 168, 1280–1285. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, S.; Nason, J.D.; Bhattacharya, D. Extensive ribosomal DNA genic variation in the columnar cactus Lophocereus. J. Mol. Evol. 2001, 53, 124–134. [Google Scholar] [CrossRef] [PubMed]
- Alvenson, A.J.; Kolnick, L. Intragenomic nucleotide polymorphism among small subunit (18S) rDNA paralogs in the diatom genus Skeletonema (Bacillariophyta). J. Phycol. 2005, 41, 1248–1257. [Google Scholar] [CrossRef]
- Chelomina, G.N.; Rozhkovan, K.V.; Ivanov, S.A.; Bulgakov, V.P. Multiplicity of alleles of nuclear 18S rRNA gene of Amur sturgeons: Genes and pseudogenes? Dokl. Biochem. Biophys. 2008, 420, 115–118. [Google Scholar] [CrossRef]
- Chelomina, G.N.; Rozhkovan, K.V.; Voronova, A.N.; Burundukova, O.L.; Muzarok, T.I.; Zhuravlev, Y.N. Variation in the number of nucleoli and incomplete homogenization of 18S ribosomal DNA sequences in leaf cells of the cultivated Oriental ginseng (Panax ginseng Meyer). J. Ginseng Res. 2016, 40, 176–184. [Google Scholar] [CrossRef] [Green Version]
- Bulgakov, V.P.; Khodakovskaya, M.V.; Labetskaya, N.V.; Chernoded, G.K.; Zhuravlev, Y.N. The impact of plant rolC oncogene on ginsenoside production by ginseng hairy root culture. Phytochemistry 1998, 49, 1929–1934. [Google Scholar] [CrossRef]
- Muratova, E.N. Nucleolus staining methods for karyotype analysis of conifers. Bot. J. 1995, 80, 82–86. (In Russian) [Google Scholar]
- Kiselev, K.V.; Bulgakov, V.P. Stability of the rolC gene and its expression in 15-year old cell cultures of Panax ginseng. Appl. Biochem. Microbiol. 2009, 45, 252–258. [Google Scholar] [CrossRef]
- Krieger, J.; Hett, A.K.; Fuerst, P.A.; Birstein, V.J.; Ludwig, A. Unusual intraindividual variation of the nuclear 18S rRNA gene is widespread within the Acipenseridae. J. Hered. 2006, 97, 218–225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Armache, J.P.; Jarasch, A.; Anger, A.M.; Villa, E.; Becker, T.; Bhushan, S.; Jossinet, F.; Habeck, M.; Dindar, G.; Franckenberg, S.; et al. Cryo-EM structure and rRNA model of a translating eukaryotic 80S ribosome at 5.5-A resolution. Proc. Natl. Acad. Sci. USA 2010, 107, 19748–19753. [Google Scholar] [CrossRef] [Green Version]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucl. Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [Green Version]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [Green Version]
- Bandelt, H.J.; Forster, P.; Röhl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef]
- Excoffier, L.; Laval, G.; Schneider, S. Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evol. Bioinform. Online 2005, 1, 47–50. [Google Scholar]
- Limoli, C.L.; Kaplan, M.I.; Corcoran, J.; Meyers, M.; Boothman, D.A.; Morgan, W.F. Chromosomal instability and its relationship to other end points of genomic instability. Cancer Res. 1997, 57, 5557–5563. [Google Scholar]
- Kobayashi, T. A new role of the rDNA and nucleolus in the nucleus rDNA instability maintains genome integrity. Bioessays 2008, 30, 267–272. [Google Scholar] [CrossRef] [PubMed]
- Schöfer, C.; Weipoltshammer, K. Nucleolus and chromatin. Histochem. Cell Biol. 2018, 150, 209–225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kunakh, V.A. Evolution of cell populations in vitro: Peculiarities, driving forces, mechanisms and consequences. Biopolym. Cell 2013, 29, 295–310. [Google Scholar] [CrossRef]
- Boulon, S.; Westman, B.J.; Hutten, S.; Boisvert, F.M.; Lamond, A.I. The nucleolus under stress. Mol. Cell 2010, 40, 216–227. [Google Scholar] [CrossRef]
- Show, P.; Brown, J. Nucleoli: Composition, Function, and Dynamics. Plant Physiol. 2012, 158, 44–51. [Google Scholar] [CrossRef] [Green Version]
- Neelakandan, A.K.; Wang, K. Recent progress in the understanding of tissue culture-induced genome level changes in plants and potential applications. Plant Cell Rep. 2012, 31, 597–620. [Google Scholar] [CrossRef]
- Sahijram, L.; Soneji, J.R.; Bollama, K.T. Analyzing somaclonal variation in micropropagated bananes (Musa spp.). Vitro Cell Dev. Biol. Plant 2003, 39, 551–556. [Google Scholar] [CrossRef]
- Yi, T.; Lowry, P.P.; Plunkett, G.M. Chromosomal evolution in Araliaceae and close relatives. Taxon 2004, 53, 987–1005. [Google Scholar] [CrossRef]
- Choi, H.I.; Waminal, N.E.; Park, H.M.; Kim, N.H.; Choi, B.S.; Park, M.; Choi, D.; Lim, Y.P.; Kwon, S.J.; Park, B.S.; et al. Major repeat components covering one-third of the ginseng (Panax ginseng C.A. Meyer) genome and evidence for allotetraploidy. Plant J. 2014, 77, 906–916. [Google Scholar] [CrossRef]
- Rodionov, A.V.; Kotsinyan, A.R.; Gnutikov, A.A.; Dobroradova, M.A.; Machs, E.M. Variability of the ITS15.8S rDNAITS2 Sequence During the Divergence of Sweet Grass Species (Glyceria R. Br.). Russ. J. Genet. Appl. Res. 2013, 3, 83–90. [Google Scholar]
- Waminal, N.E.; Park, H.M.; Ryu, K.B.; Kim, J.H.; Yang, T.J.; Kim, H.H. Karyotype analysis of Panax ginseng C.A. Meyer, 1843 (Araliaceae) based on rDNA loci and DAPI band distribution. Comp. Cytogen. 2012, 6, 425–441. [Google Scholar]
- Mentewan, A.B.; Jacobsen, M.J.; Flowers, R.A. Incomplete homogenization of 18S ribosomal DNA coding regions in Arabidopsis thaliana. BMC Res. Notes 2011, 4, 93. [Google Scholar]
- Wendel, J.F. Genome evolution in polyploids. Plant Mol. Biol. 2000, 42, 225–249. [Google Scholar] [CrossRef]
- Marquez, L.M.; Miller, D.J.; MacKenzie, J.B.; van Oppen, M.J.H. Pseudogenes contribute to the extreme diversity of nuclear ribosomal DNA in the hard coral Acropora. Mol. Biol. Evol. 2003, 20, 1077–1086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crease, T.J.; Lynch, M. Ribosomal DNA variation in Dafnia pulex. Genetics 1991, 141, 1327–1337. [Google Scholar]
- Dover, G.A. Concerted evolution, molecular drive and natural selection. Curr. Biol. 1994, 4, 1165–1166. [Google Scholar] [CrossRef]
- Nickrent, D.L.; Starr, E.M. High rates of nucleotide substitution in nuclear small-subunit (18S) rDNA from holoparasitic flowering plants. J. Mol. Evol. 1994, 39, 62–70. [Google Scholar] [CrossRef]
- Rodin, S.N.; Riggs, A.D. Epigenetic silencing may aid evolution by gene duplication. J. Mol. Evol. 2003, 56, 718–729. [Google Scholar] [CrossRef]
- Zhao, Y.E.; Wang, Z.H.; Xu, Y.; Wu, L.P.; Hu, L. Secondary structure prediction for complete rDNA sequences (18S, 5.8S, and 28S rDNA) of Demodex folliculorum, and comparison of divergent domains structures across Acari. Exp. Parasitol. 2013, 135, 370–381. [Google Scholar] [CrossRef]
- Voronova, A.N.; Chelomina, G.N. The SSU rRNA secondary structures of the Plagiorchiida species (Digenea), its applications in systematics and evolutionary inferences. Infect. Genet. Evol. 2020, 78, 104042. [Google Scholar] [CrossRef]
- Aleshin, V.V.; Kedrova, O.S.; Milyutina, I.A.; Vladychenskay, N.S.; Petrov, N.B. Secondary structure of some elements of 18S rRNA suggests that strongylid and a part of rhabditid nematodes are monophyletic. FEBS Lett. 1998, 429, 4–8. [Google Scholar] [CrossRef] [Green Version]
- Hoerter, J.A.H.; Lambert, M.N.; Pereira, M.J.; Walter, N.G. Dynamics inherent in helix 27 from Escherichia coli 16S ribosomal RNA. Biochemistry 2004, 43, 14624–14636. [Google Scholar] [CrossRef] [PubMed]
- Velichutina, I.V.; Dresios, J.; Hong, J.Y.; Li, C.; Mankin, A.; Synetos, D.; Liebman, S.W. Mutations in helix 27 of the yeast Saccharomyces cerevisiae 18S rRNA affect the function of the decoding center of the ribosome. RNA 2000, 6, 1174–1184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alkemar, G.; Nygard, O.D.D. A possible tertiary rRNA interaction between expansion segments ES3 and ES6 in eukaryotic 40S ribosomal subunits. RNA 2003, 9, 20–24. [Google Scholar] [CrossRef] [Green Version]
- Holzer, A.S.; Wootten, R.; Sommerville, C. The secondary structure of the unusually long 18S rbosomal RNA of the myxozoan Sphaerospora truttae and structural evolutionary trends in the Myxozoa. Int. J. Parasitol. 2007, 37, 1281–1295. [Google Scholar] [CrossRef]
- Osabe, K.; Clement, J.D.; Bedon, F.; Pettolino, F.A.; Ziolkowski, L.; Llewellyn, D.S.; Finnegan, E.J.; Wilson, I.W. Genetic and DNA methylation changes in cotton (Gossypium) genotypes and tissues. PLoS ONE 2014, 9, e86049. [Google Scholar] [CrossRef] [Green Version]
- Bitonti, M.B.; Cozza, R.; Chiappetta, A.; Giannino, D.; Castiglione, M.R.; Dewitte, W.; Mariotti, D.; Van Onckelen, H.; Innocenti, A.M. Distinct nuclear organization, DNA methylation pattern and cytokinin distribution mark juvenile, juvenile-like and adult vegetative apical meristems in peach (Prunus persica (L.) Batsch). J. Exp. Bot. 2002, 53, 1047–1054. [Google Scholar] [CrossRef]
- Li, M.R.; Shi, F.X.; Zhou, Y.X.; Li, Y.L.; Wang, X.F.; Zhang, C.; Wang, X.T.; Liu, B.; Xiao, H.X.; Li, L.F. Genetic and epigenetic diversities shed light on domestication of cultivated Ginseng (Panax ginseng). Mol. Plant 2015, 8, 1612–1622. [Google Scholar] [CrossRef] [Green Version]
- Parks, M.M.; Kurylo, C.M.; Dass, R.A.; Bojmar, L.; Lyden, D.; Vincent, C.T.; Blanchard, S.C. Variant ribosomal RNA alleles are conserved and exhibit tissue-specific expression. Sci. Adv. 2018, 4, eaao0665. [Google Scholar] [CrossRef] [Green Version]
- Parks, M.M.; Kurylo, C.M.; Batchelder, J.E.; Vincent, C.T.; Blanchard, S.C. Implications of sequence variation on the evolution of rRNA. Chromosome Res. 2019, 27, 89–93. [Google Scholar] [CrossRef] [Green Version]
- Symonová, R. Integrative rDNAomics—Importance of the oldest repetitive fraction of the eukaryote genome. Genes 2019, 10, 345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Genuth, N.R.; Barna, M. The discovery of ribosome heterogeneity and its implications for gene regulation and organismal life. Mol. Cell 2018, 71, 364–374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mills, E.W.; Rachel Green, R. Ribosomopathies: There’s strength in numbers. Science 2017, 358, eaan2755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, W.; Joo, M.; Yeom, J.H.; Shin, E.; Lee, M.; Choi, H.K.; Hwang, J.; Kim, Y.I.; Seo, R.; Lee, J.E.; et al. Divergent rRNAs as regulators of gene expression at the ribosome level. Nat. Microbiol. 2019, 4, 515–526. [Google Scholar] [CrossRef] [PubMed]
- Battle, D.J.; Doudna, J.A. Specificity of RNA-RNA helix recognition. Proc. Natl. Acad. Sci. USA 2002, 99, 11676–11681. [Google Scholar] [CrossRef] [Green Version]
- Zluvova, J.; Janousek, B.; Vyskot, B. Immunohistochemical study of DNA methylation dynamics during plant development. J. Exp. Bot. 2001, 52, 2265–2273. [Google Scholar] [CrossRef]
- Alvarez, M.E.; Nota, F.; Cambiagno, D.A. Epigenetic control of plant immunity. Mol. Plant Pathol. 2010, 11, 563–576. [Google Scholar] [CrossRef]
- Baranek, M.; Krizan, B.; Ondrusikova, E.; Pidra, M. DNA methylation changes in grapevine somaclones following in vitro culture and thermotherapy. Plant Cell Tissue Organ Cult. 2010, 101, 11–22. [Google Scholar] [CrossRef]
- Finnegan, E.J.; Genger, R.K.; Kovac, K.; Peacock, W.J.; Dennis, E.S. DNA methylation and the promotion of flowering by vernalization. Proc. Nat. Acad. Sci. USA 1998, 95, 5824–5829. [Google Scholar] [CrossRef] [Green Version]
- Ngezahayo, F.; Wang, X.L.; Yu, X.M.; Jiang, L.L.; Chu, Y.J.; Shen, B.H.; Yan, Z.K.; Liu, B. Habitat-induced reciprocal transformation in the root phenotype of Oriental ginseng is associated with alteration in DNA methylation. Chin. Sci. Bull. 2011, 56, 1685–1690. [Google Scholar] [CrossRef] [Green Version]
- Lopez, C.M.R.; Wetten, A.C.; Wilkinson, M.J. Progressive erosion of genetic and epigenetic variation in callus-derived cocoa (Theobroma cacao) plants. New Phytol. 2010, 186, 856–868. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Active DNA demethylation mediated by DNA glycosylases. Annu. Rev. Genet. 2009, 43, 143–166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jack, K.; Bellodi, C.; Landry, D.M.; Niederer, R.O.; Meskauskas, A.; Musalgaonkar, S.; Kopmar, N.; Krasnykh, O.; Dean, A.M.; Thompson, S.R.; et al. rRNA pseudouridylation defects affect ribosomal ligand binding and translational fidelity from yeast to human cells. Mol. Cell 2011, 44, 660–666. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nachmani, D.; Bothmer, A.H.; Grisendi, S.; Mele, A.; Bothmer, D.; Lee, J.D.; Monteleone, E.; Cheng, K.; Zhang, Y.; Bester, A.C.; et al. Germline NPM1 mutations lead to altered rRNA 2′-O-methylation and cause dyskeratosis congenita. Nat. Genet. 2019, 51, 1518–1529. [Google Scholar] [CrossRef]
- Shen, H.; Stoute, J.; Liu, K.F. Structural and catalytic roles of the human 18 S rRNA methyltransferases DIMT1 in ribosome assembly and translation. J. Biol. Chem. 2020, 295, 12058–12070. [Google Scholar] [CrossRef]
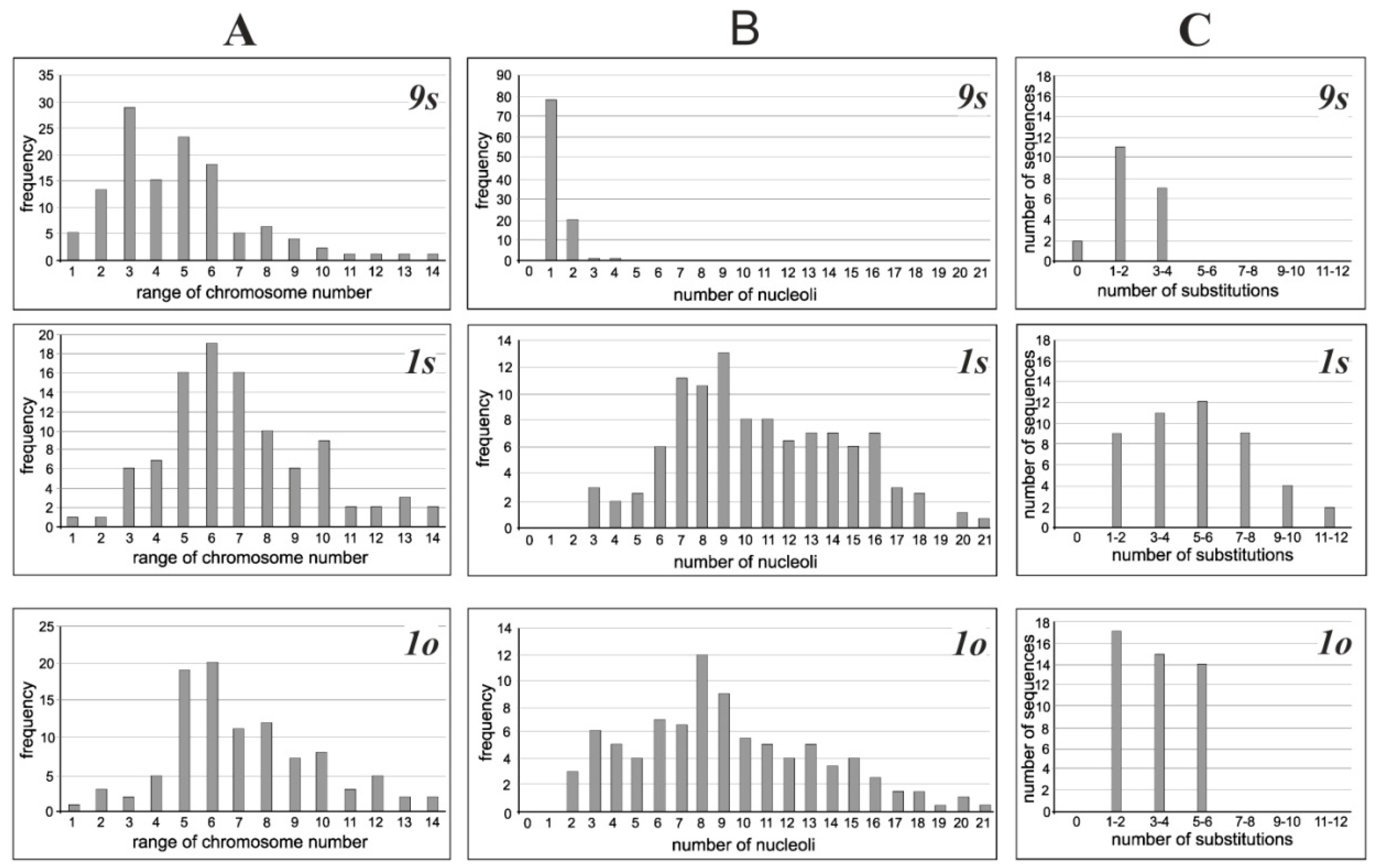


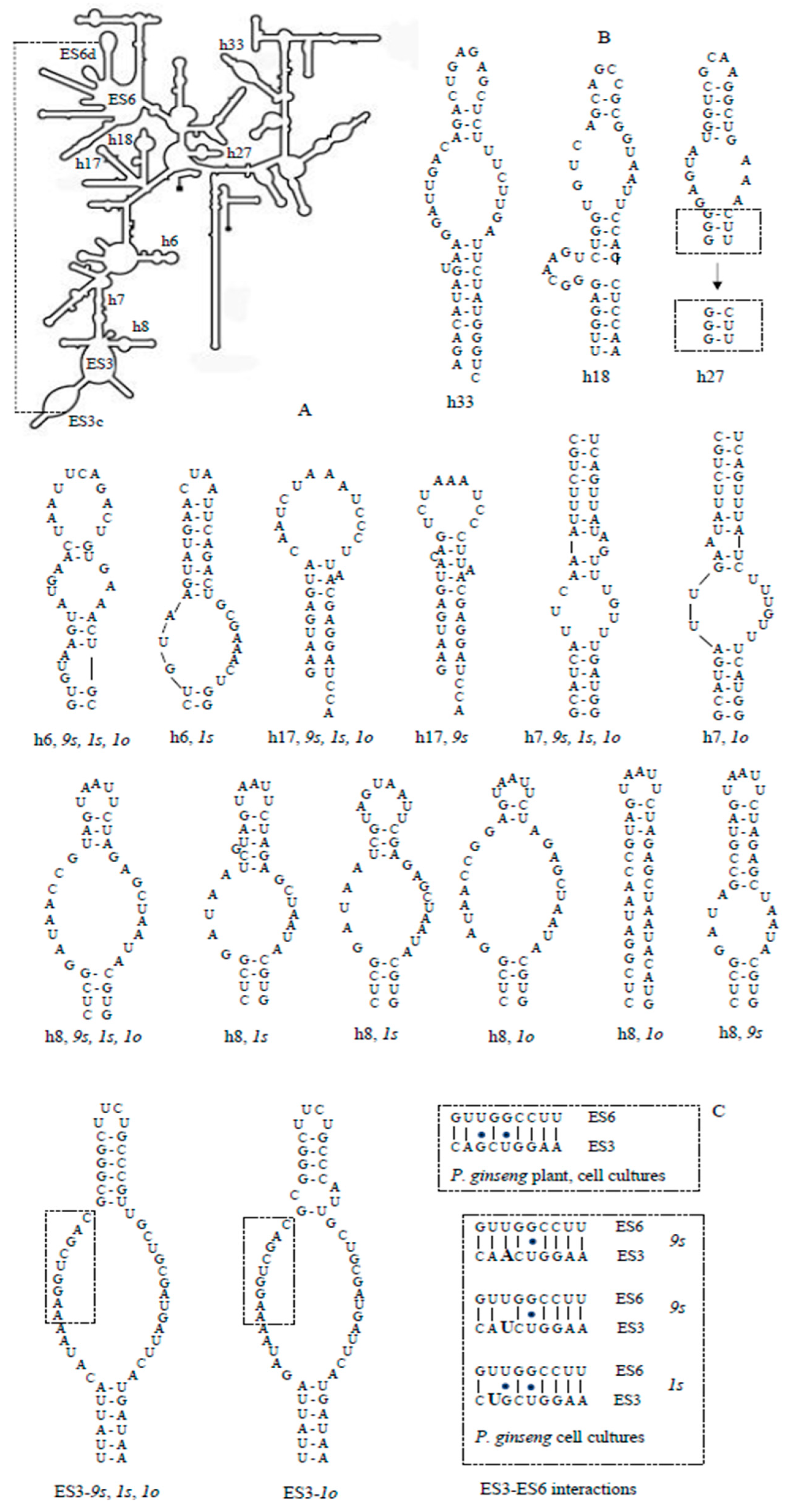

| Cell Lines | The Number of Nucleoli in Interphase Nuclei | The Range of Chromosome Number Variation | The Average Number of Chromosomes | |
|---|---|---|---|---|
| Macro Nucleoli (≥2 μm) | Micro Nucleoli (<2 μm) | |||
| 9s | 1.2 ± 0.1 | - | 6–130 | 42.3 ± 2.4 |
| 1s | 5.6 ± 0.1 | 4.3± 0.2 | 6–150 | 63.8 ± 2.8 |
| 1o | 6.3 ± 0.1 | 5.5 ± 0.2 | 6–150 | 65.5 ± 2.5 |
| Cell Lines | Ts/Tv | Transitions | Transversions | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A → G | T → C | G → A * | C → T * | A → T | A → C | T → A | T → G | G → T | C → G | G → C | C → A | ||
| 9s (n = 20) | 9.25 | 10 (24.4) | 19 (46.3) | 7 (17.1) | 2 (4.8) | 0 | 0 | 1 (2.4) | 1 (2.4) | 1 (2.4) | 0 | 0 | 0 |
| 1s (n = 46) | 2.60 | 49 (20.4) | 39 (16.3) | 23 (9.6) | 55 (22.9) | 43 (17.9) | 7 (2.9) | 6 (2.5) | 1 (0.4) | 5 (2.1) | 1 (0.4) | 4 (1.6) | 7 (2.9) |
| 1o (n = 46) | 4.30 | 38 (25.0) | 34 (22.4) | 28 (18.4) | 24 (15.8) | 10 (6.6) | 4 (2.6) | 4 (2.6) | 2 (1.3) | 3 (2.0) | 1 (0.7) | 1 (0.7) | 1 (0.7) |
| Plant (n = 30) | 7.40 | 10 (23.8) | 12 (28.6) | 11 (26.2) | 4 (9.5) | 0 | 0 | 0 | 1 (2.3) | 2 (4.7) | 1 (2.3) | 0 | 1 (2.3) |
| Sample | G + C, % | Met, % | Am, % | Met/Am | Substitutions per Gene | ||
|---|---|---|---|---|---|---|---|
| All Types | Methylation | Amination | |||||
| 9s (n = 20) | 49.36 | 21.95 | 51.2 | 0.43 | 2.05 | 0.45 | 1.05 |
| 1s (n = 46) | 49.31 | 32.50 | 19.1 | 1.70 | 5.20 | 1.70 | 1.00 |
| 1o (n = 46) | 49.34 | 34.20 | 26.3 | 1.30 | 3.30 | 1.13 | 0.87 |
| Plant (n = 30) | 49.33 | 35.71 | 30.95 | 1.15 | 1.40 | 0.20 | 0.43 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chelomina, G.N.; Rozhkovan, K.V.; Burundukova, O.L.; Gorpenchenko, T.Y.; Khrolenko, Y.A.; Zhuravlev, Y.N. Age-Dependent and Tissue-Specific Alterations in the rDNA Clusters of the Panax ginseng C. A. Meyer Cultivated Cell Lines. Biomolecules 2020, 10, 1410. https://doi.org/10.3390/biom10101410
Chelomina GN, Rozhkovan KV, Burundukova OL, Gorpenchenko TY, Khrolenko YA, Zhuravlev YN. Age-Dependent and Tissue-Specific Alterations in the rDNA Clusters of the Panax ginseng C. A. Meyer Cultivated Cell Lines. Biomolecules. 2020; 10(10):1410. https://doi.org/10.3390/biom10101410
Chicago/Turabian StyleChelomina, Galina N., Konstantin V. Rozhkovan, Olga L. Burundukova, Tatiana Y. Gorpenchenko, Yulia A. Khrolenko, and Yuri N. Zhuravlev. 2020. "Age-Dependent and Tissue-Specific Alterations in the rDNA Clusters of the Panax ginseng C. A. Meyer Cultivated Cell Lines" Biomolecules 10, no. 10: 1410. https://doi.org/10.3390/biom10101410
APA StyleChelomina, G. N., Rozhkovan, K. V., Burundukova, O. L., Gorpenchenko, T. Y., Khrolenko, Y. A., & Zhuravlev, Y. N. (2020). Age-Dependent and Tissue-Specific Alterations in the rDNA Clusters of the Panax ginseng C. A. Meyer Cultivated Cell Lines. Biomolecules, 10(10), 1410. https://doi.org/10.3390/biom10101410





