New Antibacterial Peptides from the Freshwater Mollusk Pomacea poeyana (Pilsbry, 1927)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Invertebrate Collection and Sample Preparation
2.2. Microorganism Strains and Growth Conditions
2.3. Cell Culture
2.4. Sample Fractionation and Peptide Sequencing Analysis by Nanolc-ESI-MS-MS
2.5. Antimicrobial Activity Prediction
2.6. Peptide Synthesis
2.7. Structural Prediction
2.8. Circular Dichroism Measurement
2.9. Bioassays Against Bacteria
2.10. 3H-Uracil Proliferation Assay
2.11. Antifungal Bioassays
2.12. Antiviral Bioassays
2.13. Cell Viability Assays
3. Results
3.1. Identification of Antimicrobial Peptides in Pomacea poeyana Extracts and Prediction of Antimicrobial Activity
3.2. Structural Prediction
3.3. Circular Dichroism Spectroscopy
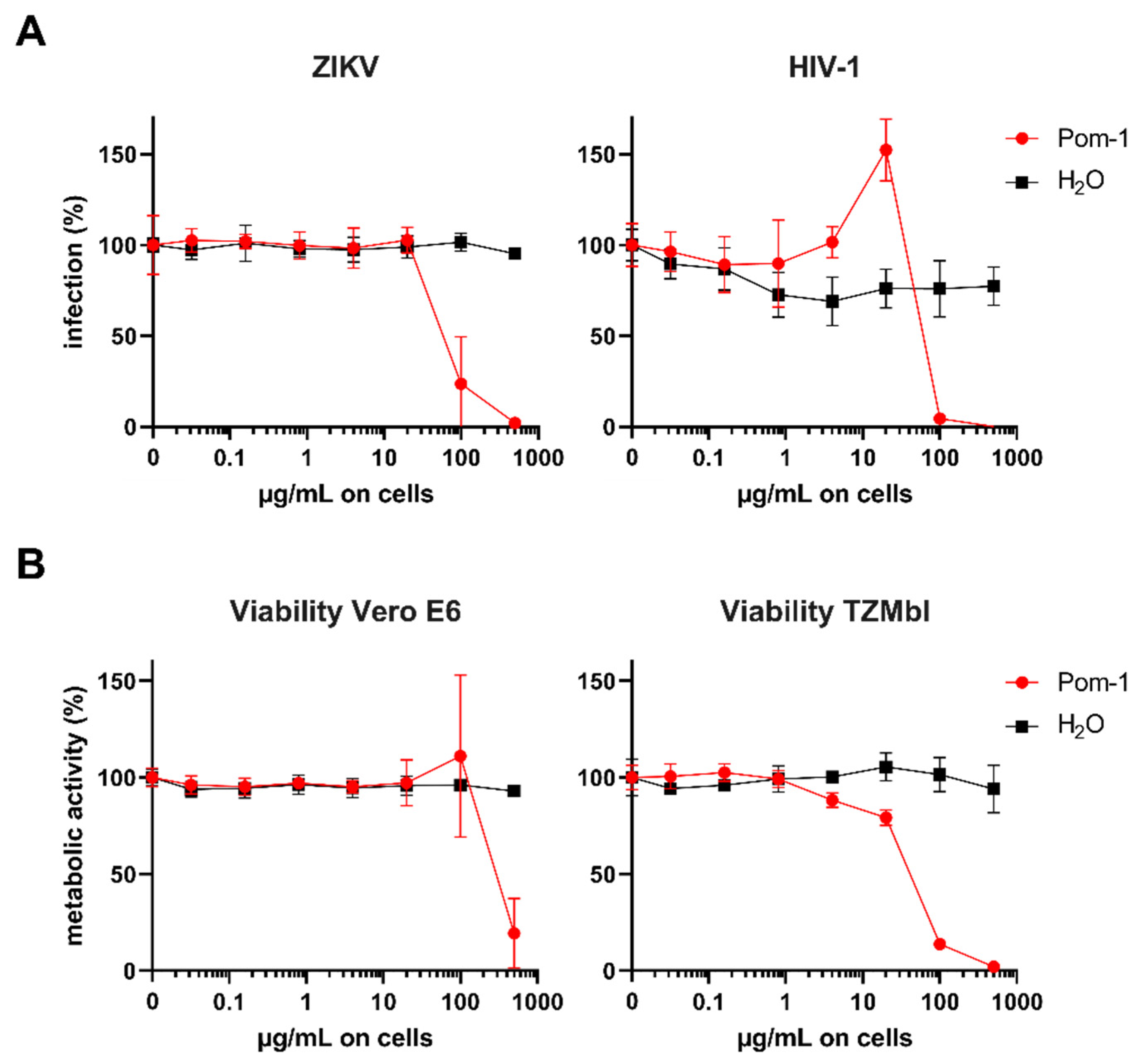
3.4. In Vitro Antimicrobial Activity
3.5. Cytotoxic Effect on Human Macrophages
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Nakatsuji, T.; Gallo, R.L. Antimicrobial Peptides: Old Molecules with New Ideas. J. Investig. Dermatol. 2012, 132, 887–895. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tran, D.; Koon, H.W. Antimicrobial peptides: An emerging approach to treating colitis. J. Colitis Diverculitis 2016, 1, 1000e002. [Google Scholar]
- Kumar, P.; Kizhakkedathu, J.N.; Straus, S.K. Antimicrobial Peptides: Diversity, Mechanism of Action and Strategies to Improve the Activity and Biocompatibility In Vivo. Biomolecules 2018, 8, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hancock, R.E.W.; Haney, E.F.; Gill, E.E. The immunology of host defense peptides: Beyond antimicrobial activity. Nat. Rev. Immunol. 2016, 16, 321–334. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Qiu, L.; Zhou, Z.; Song, L. Research progress on the mollusc immunity in China. Dev. Comp. Immunol. 2013, 39, 2–10. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, M.; Kimura, K.; Kisugi, J.; Muramoto, K.; Kamiya, H. Isolation and characterization of a Novel cytolitic Factor in Purple Fluid of the Sea Hare, Aplysia kuroai. Cancer Res. 1989, 49, 3834–3838. [Google Scholar] [PubMed]
- Scocchi, M.; Mardirossian, M.; Runti, G.; Benincasa, M. Non-Membrane Permeabilizing Modes of Action of Antimicrobial Peptides on Bacteria. Curr. Top. Med. Chem. 2015, 16, 76–88. [Google Scholar] [CrossRef] [PubMed]
- Vázquez, A.; Perera, S. Endemic Freshwater molluscs of Cuba and their conservation status. Trop. Conserv. Sci. 2010, 3, 190–199. [Google Scholar]
- Mohr, K.B.; Zirafi, O.; Hennies, M.; Wiese, S.; Kirchhoff, F.; Münch, J. Sandwich enzyme-linked immunosorbent assay for the quantification of human serum albumin fragment 408–423 in bodily fluids. Anal. Biochem. 2015, 476, 29–35. [Google Scholar] [CrossRef]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef]
- Cox, J.; Neuhauser, N.; Michalski, A.; Scheltema, R.A.; Olsen, J.V.; Mann, M. Andromeda: A Peptide Search Engine Integrated into the MaxQuant Environment. J. Proteome Res. 2011, 10, 1794–1805. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Li, X.; Wang, Z. APD3: The antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 2015, 44, D1087–D1093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waghu, F.H.; Barai, R.S.; Gurung, P.; Idicula-Thomas, S. CAMPR3: A database on sequences, structures and signatures of antimicrobial peptides: Table 1. Nucleic Acids Res. 2015, 44, D1094–D1097. [Google Scholar] [CrossRef] [Green Version]
- Veltri, D.; Kamath, U.; Shehu, A. Deep learning improves antimicrobial peptide recognition. Bioinformatics 2018, 34, 2740–2747. [Google Scholar] [CrossRef] [Green Version]
- Meher, P.K.; Sahu, T.K.; Saini, V.; Rao, A.R. Predicting antimicrobial peptides with improved accuracy by incorporating the compositional, physico-chemical and structural features into Chou’s general PseAAC. Sci. Rep. 2017, 7, srep42362. [Google Scholar] [CrossRef]
- Xu, D.; Zhang, Y. Toward optimal fragment generations for ab initio protein structure assembly. Proteins Struct. Funct. Bioinform. 2012, 81, 229–239. [Google Scholar] [CrossRef] [Green Version]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; Beer, T.A.P.D.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.-K.; Lee, E.; Shin, S.; Jeong, K.-W.; Lee, J.-Y.; Bae, S.-Y.; Kim, S.-H.; Lee, J.; Kim, S.R.; Lee, D.G.; et al. Structure and Function of Papiliocin with Antimicrobial and Anti-inflammatory Activities Isolated from the Swallowtail Butterfly, Papilio xuthus. J. Biol. Chem. 2011, 286, 41296–41311. [Google Scholar] [CrossRef] [Green Version]
- Williams, C.J.; Headd, J.J.; Moriarty, N.W.; Prisant, M.G.; Videau, L.L.; Deis, L.N.; Verma, V.; Keedy, D.A.; Hintze, B.J.; Chen, V.B.; et al. MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 2017, 27, 293–315. [Google Scholar] [CrossRef] [PubMed]
- Snider, C.; Jayasinghe, S.; Hristova, K.; White, S.H. MPEx: A tool for exploring membrane proteins. Protein Sci. 2009, 18, 2624–2628. [Google Scholar] [CrossRef] [Green Version]
- Turner, J.; Cho, Y.; Dinh, N.-N.; Waring, A.J.; Lehrer, R.I. Activities of LL-37, a Cathelin-Associated Antimicrobial Peptide of Human Neutrophils. Antimicrob. Agents Chemother. 1998, 42, 2206–2214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clinical and Laboratory Standards Institute. Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts; Approved Standard, 3rd ed. In CLSI Document M27-A3; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2008; pp. 1–25. [Google Scholar]
- Lambert, R.J.W.; Pearson, J. Susceptibility testing: Accurate and reproducible minimum inhibitory concentration (MIC) and non-inhibitory concentration (NIC) values. J. Appl. Microbiol. 2000, 88, 784–790. [Google Scholar] [CrossRef] [PubMed]
- Müller, J.A.; Harms, M.; Krüger, F.; Groß, R.; Joas, S.; Hayn, M.; Dietz, A.N.; Lippold, S.; Von Einem, J.; Schubert, A.; et al. Semen inhibits Zika virus infection of cells and tissues from the anogenital region. Nat. Commun. 2018, 9, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Kemperman, R.; Kuipers, A.; Karsens, H.; Nauta, A.; Kuipers, O.; Kok, J. Identification and Characterization of Two Novel Clostridial Bacteriocins, Circularin A and Closticin 574. Appl. Environ. Microbiol. 2003, 69, 1589–1597. [Google Scholar] [CrossRef] [Green Version]
- Mak, P.; Chmiel, D.; Gacek, G.J. Antibacterial peptides of the moth Galleria mellonella. Acta Biochim. Pol. 2001, 48, 1191–1195. [Google Scholar] [CrossRef] [Green Version]
- Perera, G.; Walls, J.G. Apple Sanils in the Aquarium; T.F.H. Publications Inc: Neptune City, NJ, USA, 1996. [Google Scholar]
- Vieira, C.S.; Waniek, P.J.; Mattos, D.P.; Castro, D.P.; Mello, C.B.; Ratcliffe, N.A.; Garcia, E.S.; Azambuja, P. Humoral responses in Rhodnius prolixus: Bacterial feeding induces differential patterns of antibacterial activity and enhances mRNA levels of antimicrobial peptides in the midgut. Parasites Vectors 2014, 7, 232. [Google Scholar] [CrossRef] [Green Version]
- Mitta, G.; Vandenbulcke, F.; Roch, P. Original involvement of antimicrobial peptides in mussel innate immunity. FEBS Lett. 2000, 486, 185–190. [Google Scholar] [CrossRef] [Green Version]
- Dolashka, P.; Dolashki, A.; Voelter, W.; Van Beeumen, J.; Stefanovic, S. Antimicrobial activity of peptides from the hemolymph of Helix lucorum snails. Int.J.Curr. Microbiol. App. Sci. 2015, 4, 1061–1071. [Google Scholar]
- Zannella, C.; Mosca, F.; Mariani, F.; Franci, G.; Folliero, V.; Galdiero, M.; Tiscar, P.G.; Galdiero, M. Microbial Diseases of Bivalve Mollusks: Infections, Immunology and Antimicrobial Defense. Mar. Drugs 2017, 15, 182. [Google Scholar] [CrossRef]
- Seo, J.-K.; Lee, M.J.; Nam, B.-H.; Park, N.G. cgMolluscidin, a novel dibasic residue repeat rich antimicrobial peptide, purified from the gill of the Pacific oyster, Crassostrea gigas Fish Shellfish. Immunology 2013, 35, 480–488. [Google Scholar] [CrossRef]
- Peng, K.-C.; Lee, S.-H.; Hour, A.-L.; Pan, C.-Y.; Lee, L.-H.; Chen, J.-Y. Five Different Piscidins from Nile Tilapia, Oreochromis niloticus: Analysis of Their Expressions and Biological Functions. PLoS ONE 2012, 7, e50263. [Google Scholar] [CrossRef] [PubMed]
- Baker, D.; Sali, A. Protein Structure Prediction and Structural Genomics. Science 2001, 294, 93–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bradley, P.; Misura, K.M.S.; Baker, D. Toward High-Resolution de Novo Structure Prediction for Small Proteins. Science 2005, 309, 1868–1871. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moult, J.; Fidelis, K.; Kryshtafovych, A.; Rost, B.; Tramontano, A. Critical assessment of methods of protein structure prediction-Round VIII. Proteins Struct. Funct. Bioinform. 2009, 77, 1–4. [Google Scholar] [CrossRef]
- Iwai, H.; Nakajima, Y.; Natori, S.; Arata, Y.; Shimada, I. Solution conformation of an antibacterial peptide, sarcotoxin IA, as determined by 1H-NMR. Eur. J. Biochem. 1993, 217, 639–644. [Google Scholar] [CrossRef]
- Terwilligert, T.C.; Eisenbergg, D. The Structure of Melittin. J. Biol. Chem. 1982, 257, 6016–6022. [Google Scholar]
- Oñate-Garzón, J.F.; Manrique-Moreno, M.; González, E.P. Actividad antimicrobiana de péptidos catiónicos diseñados a partir de un péptido neutro. Acta Biológica Colombiana 2017, 22, 35. [Google Scholar] [CrossRef]
- Lee, E.; Jeong, K.-W.; Lee, J.; Shin, A.; Kim, J.-K.; Lee, J.; Lee, D.G.; Kim, Y. Structure-activity relationships of cecropin-like peptides and their interactions with phospholipid membrane. BMB Rep. 2013, 46, 282–287. [Google Scholar] [CrossRef] [PubMed]
- Silvestro, L.; Weiser, J.N.; Axelsen, P.H. Antibacterial and Antimembrane Activities of Cecropin A in Escherichia coli. Antimicrob. Agents Chemother. 2000, 44, 602–607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bechinger, B.; Zasloff, M.; Opella, S.J. Structure and orientation of the antibiotic peptide magainin in membranes by solid-state nuclear magnetic resonance spectroscopy. Protein Sci. 1993, 2, 2077–2084. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chatterjee, M.; Anju, C.; Biswas, L.; Kumar, V.A.; Mohan, C.G.; Biswas, R. Antibiotic resistance in Pseudomonas aeruginosa and alternative therapeutic options. Int. J. Med Microbiol. 2016, 306, 48–58. [Google Scholar] [CrossRef] [PubMed]
- Paczosa, M.K.; Mecsas, J. Klebsiella pneumoniae: Going on the Offense with a Strong Defense. Microbiol. Mol. Biol. Rev. 2016, 80, 629–661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gahan, C.G.M.; Ehill, C. Listeria monocytogenes: Survival and adaptation in the gastrointestinal tract. Front. Cell. Infect. Microbiol. 2014, 4, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lomonaco, S.; Nucera, D.; Filipello, V. The evolution and epidemiology of Listeria monocytogenes in Europe and the United States. Infect. Genet. Evol. 2015, 35, 172–183. [Google Scholar] [CrossRef]
- Borrero, R.; Álvarez, N.; Reyes, F.; Sarmiento, M.E.; Acosta, A. Mycobacterium tuberculosis: Factores de virulencia. VacciMonitor 2011, 20, 34–38. [Google Scholar]
- Kościuczuk, E.M.; Lisowski, P.; Jarczak, J.; Strzałkowska, N.; Jóźwik, A.; Horbańczuk, J.; Krzyżewski, J.; Zwierzchowski, L.; Bagnicka, E. Cathelicidins: Family of antimicrobial peptides. A review. Mol. Biol. Rep. 2012, 39, 10957–10970. [Google Scholar] [CrossRef] [Green Version]
- Hancock, R.E.W.; Sahl, H.-G. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotechnol. 2006, 24, 1551–1557. [Google Scholar] [CrossRef]
- Strandberg, E.; Tiltak, D.; Ehni, S.; Wadhwani, P.; Ulrich, A.S. Lipid shapeis a key factorfor membrane interactionof amphipatic helical peptides. Biochim. Biophys. Acta BBA Biomembr. 2012, 1818, 1764–1776. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.; Jacob, B.; Jang, M.; Kwak, C.; Lee, Y.; Son, K.; Lee, S.; Jung, I.D.; Jeong, M.S.; Kwon, S.-H.; et al. Development of a novel short 12-meric papiliocin-derived peptide that is effective against Gram-negative sepsis. Sci. Rep. 2019, 9, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Guilhelmelli, F.; Vilela, N.; Albuquerque, P.; Derengowski, L.D.S.; Silva-Pereira, I.; Kyaw, C.M. Antibiotic development challenges: The various mechanisms of action of antimicrobial peptides and of bacterial resistance. Front. Microbiol. 2013, 4, 353. [Google Scholar] [CrossRef] [Green Version]
- Matsuzaki, K. Control of cell selectivity of antimicrobial peptides. Biochim. Biophys. Acta BBA Biomembr. 2009, 1788, 1687–1692. [Google Scholar] [CrossRef] [Green Version]
- Lai, Z.; Tan, P.; Zhu, Y.; Shao, C.; Shan, A.; Li, L. Highly Stabilized α-Helical Coiled Coils Kill Gram-Negative Bacteria by Multicomplementary Mechanisms under Acidic Condition. ACS Appl. Mater. Interfaces 2019, 11, 22113–22128. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.-H.; Hall, K.N.; Aguilar, M.-I. Antimicrobial Peptide Structure and Mechanism of Action: A Focus on the Role of Membrane Structure. Curr. Top. Med. Chem. 2015, 16, 25–39. [Google Scholar] [CrossRef] [PubMed]
- Bobone, S.; Stella, L. Selectivity of Antimicrobial Peptides: A Complex Interplay of Multiple Equilibria. In Antimicrobial Peptides; Matsuzaki, K., Ed.; Springer: Singapore, 2019. [Google Scholar] [CrossRef]
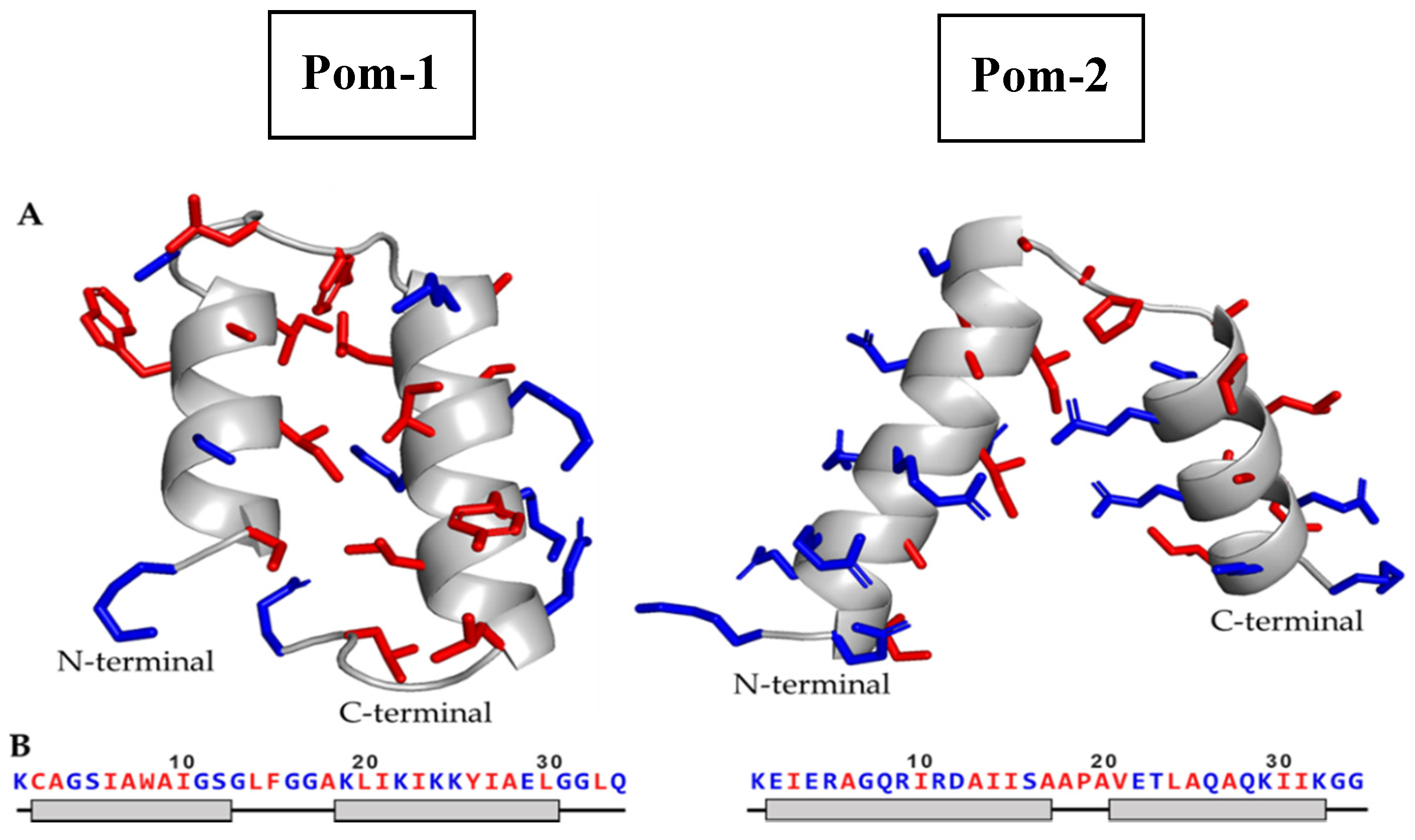
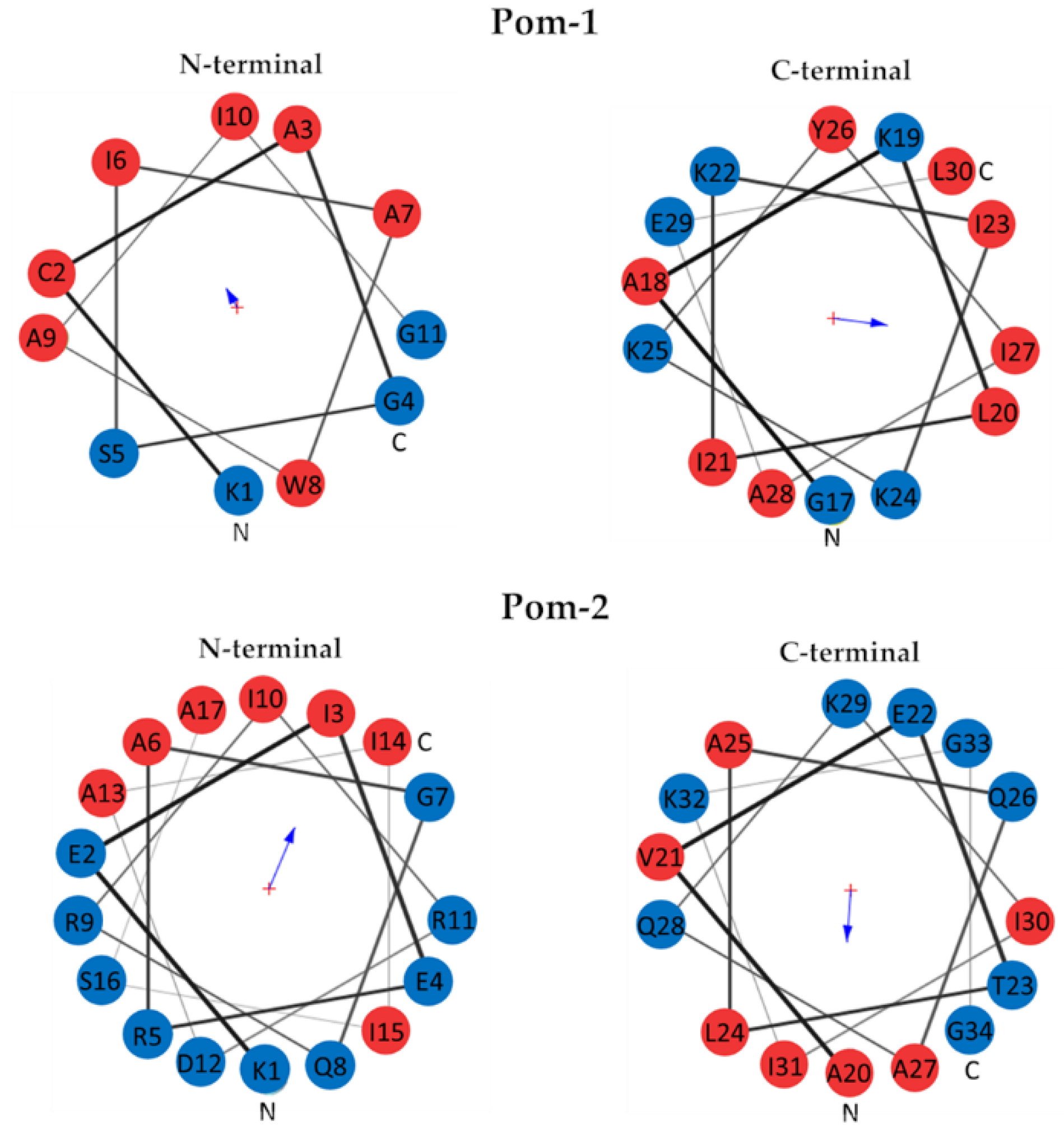


| Model | MolProbity Score | Clash Score | Poor Rotamers | Favored Rotamers | Ramachandran Outliers | Ramachandran Favored | Cβ Deviations | Bad Bonds | Bad Angles |
|---|---|---|---|---|---|---|---|---|---|
| Pom-1_1 | 2.72 | 5.9 | 2/22 | 17/22 | 2/32 | 27/32 | 3/27 | 0/246 | 2/326 |
| Pom-1_2 | 3.1 | 7.8 | 7/22 | 14/22 | 2/32 | 29/32 | 1/27 | 0/246 | 0/326 |
| Pom-1_3 | 2.98 | 3.9 | 3/22 | 15/22 | 3/32 | 18/32 | 0/27 | 0/246 | 1/326 |
| Pom-1_4 | 2.86 | 2.0 | 7/22 | 11/22 | 2/32 | 25/32 | 1/27 | 0/246 | 3/326 |
| Pom-1_5 | 1.99 | 0.0 | 4/22 | 16/22 | 0/32 | 29/32 | 2/27 | 0/246 | 0/326 |
| Pom-2 | 1.41 | 0.0 | 2/24 | 21/24 | 1/30 | 29/30 | 1/31 | 0/245 | 0/328 |
| Peptide | Helix (%) | Sheets (%) | Turns (%) | Random (%) |
|---|---|---|---|---|
| Pom-1 | 22.2 | 8.9 | 20.8 | 48.1 |
| Pom-2 | 21.0 | 2.8 | 21.7 | 54.5 |
| Bacterial Species | Pom-1 | Pom-2 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Concentration (µg/mL) | 5 | 10 | 20 | 30 | 40 | 50 | 5 | 10 | 20 | 30 | 40 | 50 |
| Pseudomonas aeruginosa ATCC27853 | 0.4 | 0.55 | 0.65 | 0.7 | 0.8 | 0.85 | - | - | - | 0.2 | 0.2 | 0.45 |
| Lysteria monocytogenes ATCC BAA-679/EGD-e | 0.25 | 0.35 | 0.35 | 0.35 | 0.4 | 0.45 | - | - | - | 0.15 | 0.3 | 0.4 |
| Klebsiella pneumoniae ATCC 70063 | - | - | 0.3 | 0.3 | 0.4 | 0.43 | - | - | - | - | - | - |
| Virus | ZIKV (Vero E6) | HIV-1 (TZMb1) |
|---|---|---|
| IC50 | 88.05 | 80.58 |
| CC50 | 438.00 | 40.16 |
| SI | 4.97 | 0.50 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
González García, M.; Rodríguez, A.; Alba, A.; Vázquez, A.A.; Morales Vicente, F.E.; Pérez-Erviti, J.; Spellerberg, B.; Stenger, S.; Grieshober, M.; Conzelmann, C.; et al. New Antibacterial Peptides from the Freshwater Mollusk Pomacea poeyana (Pilsbry, 1927). Biomolecules 2020, 10, 1473. https://doi.org/10.3390/biom10111473
González García M, Rodríguez A, Alba A, Vázquez AA, Morales Vicente FE, Pérez-Erviti J, Spellerberg B, Stenger S, Grieshober M, Conzelmann C, et al. New Antibacterial Peptides from the Freshwater Mollusk Pomacea poeyana (Pilsbry, 1927). Biomolecules. 2020; 10(11):1473. https://doi.org/10.3390/biom10111473
Chicago/Turabian StyleGonzález García, Melaine, Armando Rodríguez, Annia Alba, Antonio A. Vázquez, Fidel E. Morales Vicente, Julio Pérez-Erviti, Barbara Spellerberg, Steffen Stenger, Mark Grieshober, Carina Conzelmann, and et al. 2020. "New Antibacterial Peptides from the Freshwater Mollusk Pomacea poeyana (Pilsbry, 1927)" Biomolecules 10, no. 11: 1473. https://doi.org/10.3390/biom10111473






