An Insight into the Algal Evolution and Genomics
Abstract
:1. Introduction
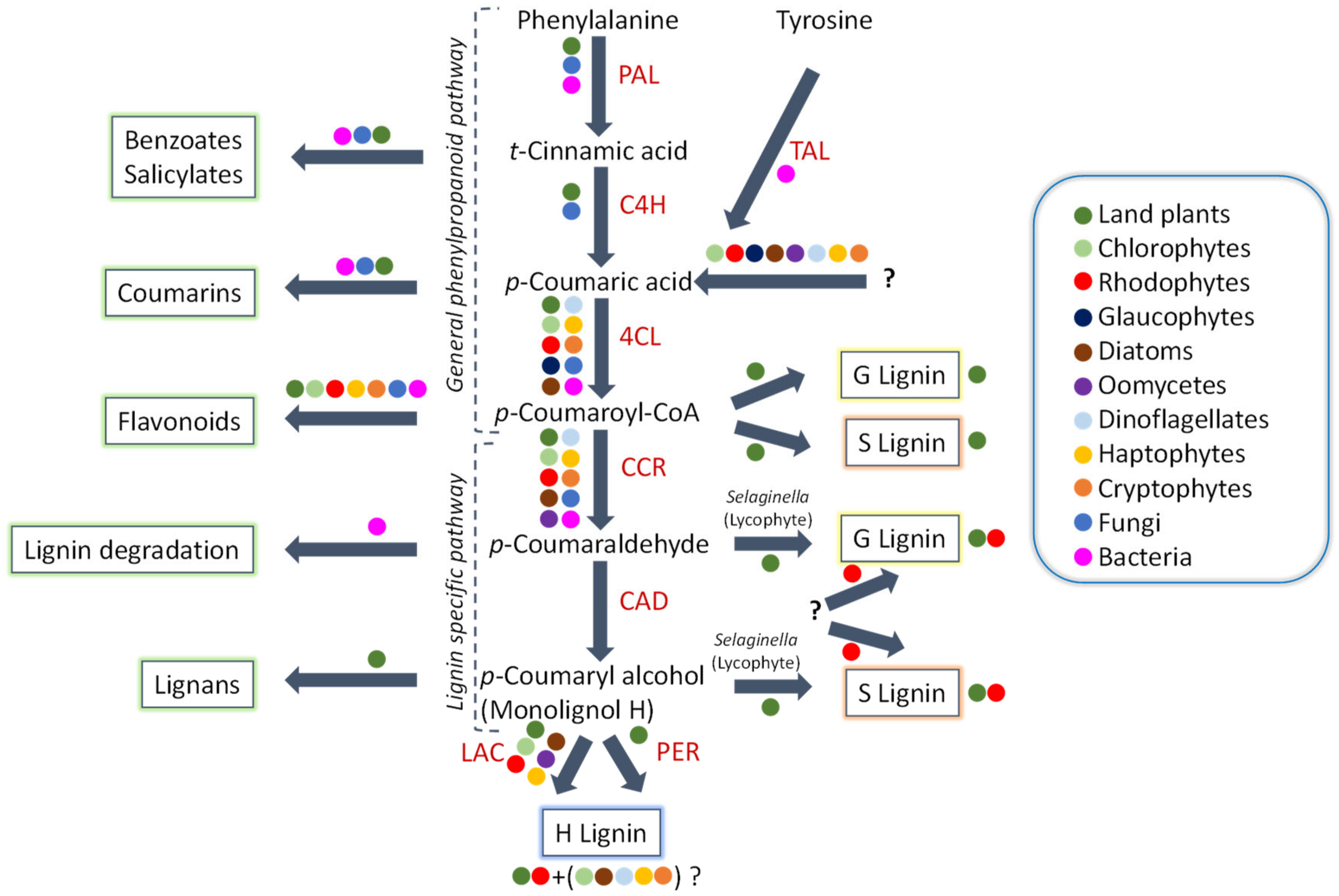
2. Insights to Algal Evolution
3. Algal Genomes
4. Conclusions and Future Prospects
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gilbert, J.A.; Dupont, C.L. Microbial metagenomics: Beyond the genome. Ann. Rev. Mar. Sci. 2011, 3, 347–371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kong, J.; Huh, S.; Won, J.I.; Yoon, J.; Kim, B.; Kim, K. GAAP: A Genome Assembly + Annotation Pipeline. Biomed. Res. Int. 2019, 2019, 4767354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blaby-Haas, C.E.; Merchant, S.S. Comparative and Functional Algal Genomics. Annu. Rev. Plant Biol. 2019, 70, 605–638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tirichine, L.; Bowler, C. Decoding algal genomes: Tracing back the history of photosynthetic life on Earth. Plant J. 2011, 66, 45–57. [Google Scholar] [CrossRef] [PubMed]
- Adelheid, S. The Structure and Reproduction of the Algae Vol. I; Cambridge University Press: London, UK, 1967; Volume 7, pp. 168–169. [Google Scholar]
- Bajpai, P. Characteristics of Algae. In Third Generation Biofuels; Springer: Singapore, 2019; pp. 11–15. [Google Scholar] [CrossRef]
- Andersen, R. Diversity of eukaryotic algae. Biodivers. Conserv. 1992, 1, 267–292. [Google Scholar] [CrossRef]
- Sukenik, A.; Zohary, T.; Padisák, J. Cyanoprokaryota and other prokaryotic algae. In Encyclopedia of Inland Waters; Elsevier Inc.: Amsterdam, The Netherlands, 2009; pp. 138–148. [Google Scholar]
- Reyes-Prieto, A.; Weber, A.P.; Bhattacharya, D. The origin and establishment of the plastid in algae and plants. Annu. Rev. Genet. 2007, 41, 147–168. [Google Scholar] [CrossRef] [Green Version]
- Keeling, P.J.; Burger, G.; Durnford, D.G.; Lang, B.F.; Lee, R.W.; Pearlman, R.E.; Roger, A.J.; Gray, M.W. The tree of eukaryotes. Trends Ecol. Evol. 2005, 20, 670–676. [Google Scholar] [CrossRef]
- Keeling, P.J. Diversity and evolutionary history of plastids and their hosts. Am. J. Bot. 2004, 91, 1481–1493. [Google Scholar] [CrossRef] [Green Version]
- Baweja, P.; Sahoo, D. Classification of algae. In The Algae World; Dinabandhu, S., Seckbach, J., Eds.; Springer: New York, NY, USA, 2015; pp. 31–55. [Google Scholar]
- Stevenson, C.S.; Capper, E.A.; Roshak, A.K.; Marquez, B.; Eichman, C.; Jackson, J.R.; Mattern, M.; Gerwick, W.H.; Jacobs, R.S.; Marshall, L.A.; et al. The identification and characterization of the marine natural product scytonemin as a novel antiproliferative pharmacophore. J. Pharm. Exp. Therap. 2002, 303, 858–866. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.-C.; Emau, P.; Jiang, Y.; Agy, M.B.; Shattock, R.J.; Schmidt, A.; Morton, W.R.; Gustafson, K.R.; Boyd, M.R. Cyanovirin-N inhibits AIDS virus infections in vaginal transmission models. Aids Res. Hum. Retrovir. 2004, 20, 11–18. [Google Scholar] [CrossRef]
- Hannon, M.; Gimpel, J.; Tran, M.; Rasala, B.; Mayfield, S. Biofuels from algae: Challenges and potential. Biofuels 2010, 1, 763–784. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Sharma, N. Industrial and biotechnological applications of algae: A review. J. Adv. Plant Biol. 2017, 1, 1. [Google Scholar] [CrossRef]
- Scieszka, S.; Klewicka, E. Algae in food: A general review. Crit. Rev. Food Sci. Nutr. 2019, 59, 3538–3547. [Google Scholar] [CrossRef] [PubMed]
- Bowler, C.; Allen, A.E. The contribution of genomics to the understanding of algal evolution. In Unravelling the Algae: The Past, Present, Future of Algal Systematics; CRC Press: Boca Raton, FL, USA, 2007; p. 331. [Google Scholar]
- Yoon, H.S.; Hackett, J.D.; Ciniglia, C.; Pinto, G.; Bhattacharya, D. A molecular timeline for the origin of photosynthetic eukaryotes. Mol. Biol. Evol. 2004, 21, 809–818. [Google Scholar] [CrossRef] [Green Version]
- Timmis, J.N.; Ayliffe, M.A.; Huang, C.Y.; Martin, W. Endosymbiotic gene transfer: Organelle genomes forge eukaryotic chromosomes. Nat. Rev. Genet. 2004, 5, 123–135. [Google Scholar] [CrossRef]
- Mackiewicz, P.; Gagat, P. Monophyly of Archaeplastida supergroup and relationships among its lineages in the light of phylogenetic and phylogenomic studies. Are we close to a consensus? Acta Soc. Bot. Pol. 2014, 83, 263–280. [Google Scholar] [CrossRef] [Green Version]
- Yoon, H.S.; Müller, K.; Sheath, R.; Ott, F.; Bhattacharya, D. Defining the major lineages of red algae (Rhodophyta). J. Phycol. 2006, 42, 482–492. [Google Scholar] [CrossRef]
- Glazer, A.N. Structure and molecular organization of the photosynthetic accessory pigments of cyanobacteria and red algae. Mol. Cell. Biochem. 1977, 18, 125–140. [Google Scholar] [CrossRef]
- Yoon, H.S.; Nelson, W.; Lindstrom, S.C.; Boo, S.M.; Pueschel, C.; Qiu, H.; Bhattacharya, D. Rhodophyta. In Handbook of the Protists; Archibald, J.M., Simpson, A.G.B., Slamovits, C.H., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 89–133. [Google Scholar]
- Salomaki, E.D.; Lane, C.E. Red Algal Mitochondrial Genomes Are More Complete than Previously Reported. Genome Biol. Evol. 2017, 9, 48–63. [Google Scholar] [CrossRef] [Green Version]
- Moreira, D.; López-García, P. Evolution: King-Size Plastid Genomes in a New Red Algal Clade. Curr. Biol. 2017, 27, R651–R653. [Google Scholar] [CrossRef]
- Tanaka, A.; Tanaka, R. Chapter Six-The biochemistry, physiology, and evolution of the chlorophyll cycle. In Advances in Botanical Research; Grimm, B., Ed.; Academic Press: Cambridge, MA, USA, 2019; Volume 90, pp. 183–212. [Google Scholar]
- Busi, M.V.; Barchiesi, J.; Martín, M.; Gomez-Casati, D.F. Starch metabolism in green algae. Starch 2014, 66, 28–40. [Google Scholar] [CrossRef]
- Lemieux, C.; Vincent, A.T.; Labarre, A.; Otis, C.; Turmel, M. Chloroplast phylogenomic analysis of chlorophyte green algae identifies a novel lineage sister to the Sphaeropleales (Chlorophyceae). BMC Evol. Biol. 2015, 15, 264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Domozych, D.; Ciancia, M.; Fangel, J.; Mikkelsen, M.; Ulvskov, P.; Willats, W. The Cell Walls of Green Algae: A Journey through Evolution and Diversity. Front. Plant Sci. 2012, 3, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Jackson, C.; Clayden, S.; Reyes-Prieto, A. The Glaucophyta: The blue-green plants in a nutshell. Acta Soc. Bot. Pol. 2015, 84, 149–165. [Google Scholar] [CrossRef] [Green Version]
- Figueroa-Martinez, F.; Jackson, C.; Reyes-Prieto, A. Plastid Genomes from Diverse Glaucophyte Genera Reveal a Largely Conserved Gene Content and Limited Architectural Diversity. Genome Biol. Evol. 2018, 11, 174–188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Price, D.C.; Chan, C.X.; Yoon, H.S.; Yang, E.C.; Qiu, H.; Weber, A.P.M.; Schwacke, R.; Gross, J.; Blouin, N.A.; Lane, C.; et al. Cyanophora paradoxa genome elucidates origin of photosynthesis in algae and plants. Science 2012, 335, 843–847. [Google Scholar] [CrossRef] [Green Version]
- Bowler, C.; Allen, A.E.; Badger, J.H.; Grimwood, J.; Jabbari, K.; Kuo, A.; Maheswari, U.; Martens, C.; Maumus, F.; Otillar, R.P. The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 2008, 456, 239. [Google Scholar] [CrossRef] [PubMed]
- Derelle, E.; Ferraz, C.; Rombauts, S.; Rouzé, P.; Worden, A.Z.; Robbens, S.; Partensky, F.; Degroeve, S.; Echeynié, S.; Cooke, R. Genome analysis of the smallest free-living eukaryote Ostreococcus tauri unveils many unique features. Proc. Natl. Acad. Sci. USA 2006, 103, 11647–11652. [Google Scholar] [CrossRef] [Green Version]
- McFadden, G.I. Primary and secondary endosymbiosis and the origin of plastids. J. Phycol. 2001, 37, 951–959. [Google Scholar] [CrossRef]
- Archibald, J. Nucleomorph genomes: Structure, function, origin and evolution. BioEssays 2007, 29, 392–402. [Google Scholar] [CrossRef]
- Burki, F.; Kaplan, M.; Tikhonenkov, D.V.; Zlatogursky, V.; Minh, B.Q.; Radaykina, L.V.; Smirnov, A.; Mylnikov, A.P.; Keeling, P.J. Untangling the early diversification of eukaryotes: A phylogenomic study of the evolutionary origins of Centrohelida, Haptophyta and Cryptista. Proc. R. Soc. B Biol. Sci. 2016, 283, 20152802. [Google Scholar] [CrossRef] [Green Version]
- Yoon, H.S.; Hackett, J.D.; Van Dolah, F.M.; Nosenko, T.; Lidie, K.L.; Bhattacharya, D. Tertiary Endosymbiosis Driven Genome Evolution in Dinoflagellate Algae. Mol. Biol. Evol. 2005, 22, 1299–1308. [Google Scholar] [CrossRef]
- Shrager, J.; Hauser, C.; Chang, C.-W.; Harris, E.H.; Davies, J.; McDermott, J.; Tamse, R.; Zhang, Z.; Grossman, A.R. Chlamydomonas reinhardtii genome project. A guide to the generation and use of the cDNA information. Plant Physiol. 2003, 131, 401–408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, D.R.; Lee, R.W. Nucleotide diversity of the Chlamydomonas reinhardtii plastid genome: Addressing the mutational-hazard hypothesis. BMC Evol. Biol. 2009, 9, 120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merchant, S.S.; Prochnik, S.E.; Vallon, O.; Harris, E.H.; Karpowicz, S.J.; Witman, G.B.; Terry, A.; Salamov, A.; Fritz-Laylin, L.K.; Maréchal-Drouard, L. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 2007, 318, 245–250. [Google Scholar] [CrossRef] [Green Version]
- Janech, M.; Krell, A.; Mock, T.; Raymond, J. Ice-binding proteins from sea ice diatoms (Bacillariophyceae). J. Phycol. 2006, 42, 410–416. [Google Scholar] [CrossRef] [Green Version]
- Nakov, T.; Beaulieu, J.M.; Alverson, A.J. Accelerated diversification is related to life history and locomotion in a hyperdiverse lineage of microbial eukaryotes (Diatoms, Bacillariophyta). New Phytol. 2018, 219, 462–473. [Google Scholar] [CrossRef] [Green Version]
- Falciatore, A.; Jaubert, M.; Bouly, J.-P.; Bailleul, B.; Mock, T. Diatom Molecular Research Comes of Age: Model Species for Studying Phytoplankton Biology and Diversity. Plant Cell 2020, 32, 547–572. [Google Scholar] [CrossRef] [Green Version]
- Vancaester, E.; Depuydt, T.; Osuna-Cruz, C.M.; Vandepoele, K. Systematic and functional analysis of horizontal gene transfer events in diatoms. bioRxiv 2020. [Google Scholar] [CrossRef]
- Pierella Karlusich, J.J.; Carrillo, N. Evolution of the acceptor side of photosystem I: Ferredoxin, flavodoxin, and ferredoxin-NADP(+) oxidoreductase. Photosynth. Res. 2017, 134, 235–250. [Google Scholar] [CrossRef] [PubMed]
- Groussman, R.D.; Parker, M.S.; Armbrust, E.V. Diversity and evolutionary history of iron metabolism genes in diatoms. PLoS ONE 2015, 10, e0129081. [Google Scholar] [CrossRef] [Green Version]
- Palenik, B.; Grimwood, J.; Aerts, A.; Rouzé, P.; Salamov, A.; Putnam, N.; Dupont, C.; Jorgensen, R.; Derelle, E.; Rombauts, S.; et al. The tiny eukaryote Ostreococcus provides genomic insights into the paradox of plankton speciation. Proc. Natl. Acad. Sci. USA 2007, 104, 7705–7710. [Google Scholar] [CrossRef] [Green Version]
- Lelandais, G.; Scheiber, I.; Paz-Yepes, J.; Lozano, J.-C.; Botebol, H.; Pilátová, J.; Žárský, V.; Léger, T.; Blaiseau, P.-L.; Bowler, C.; et al. Ostreococcus tauri is a new model green alga for studying iron metabolism in eukaryotic phytoplankton. BMC Genom. 2016, 17, 319. [Google Scholar] [CrossRef]
- Yu, Z.; Zhang, T.; Zhu, Y. Whole-genome re-sequencing and transcriptome reveal cadmium tolerance related genes and pathways in Chlamydomonas reinhardtii. Ecotoxicol. Environ. Saf. 2020, 191, 110231. [Google Scholar] [CrossRef]
- Li, L.; Peng, H.; Tan, S.; Zhou, J.; Fang, Z.; Hu, Z.; Gao, L.; Li, T.; Zhang, W.; Chen, L. Effects of early cold stress on gene expression in Chlamydomonas reinhardtii. Genomics 2020, 112, 1128–1138. [Google Scholar] [CrossRef]
- Hirooka, S.; Hirose, Y.; Kanesaki, Y.; Higuchi, S.; Fujiwara, T.; Onuma, R.; Era, A.; Ohbayashi, R.; Uzuka, A.; Nozaki, H.; et al. Acidophilic green algal genome provides insights into adaptation to an acidic environment. Proc. Natl. Acad. Sci. USA 2017, 114, E8304–E8313. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Qu, C.; Yao, R.; Nie, Y.; Xu, C.; Miao, J.; Zhong, B. The Parallel Molecular Adaptations to the Antarctic Cold Environment in Two Psychrophilic Green Algae. Genome Biol. Evol. 2019, 11, 1897–1908. [Google Scholar] [CrossRef]
- Qiu, H.; Price, D.C.; Weber, A.P.M.; Reeb, V.; Chan Yang, E.; Lee, J.M.; Kim, S.Y.; Yoon, H.S.; Bhattacharya, D. Adaptation through horizontal gene transfer in the cryptoendolithic red alga Galdieria phlegrea. Curr. Biol. 2013, 23, R865–R866. [Google Scholar] [CrossRef] [Green Version]
- Nozaki, H.; Takano, H.; Misumi, O.; Terasawa, K.; Matsuzaki, M.; Maruyama, S.; Nishida, K.; Yagisawa, F.; Yoshida, Y.; Fujiwara, T.; et al. A 100%-complete sequence reveals unusually simple genomic features in the hot-spring red alga Cyanidioschyzon merolae. BMC Biol. 2007, 5, 28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsuzaki, M.; Misumi, O.; Shin-i, T.; Maruyama, S.; Takahara, M.; Miyagishima, S.-y.; Mori, T.; Nishida, K.; Yagisawa, F.; Nishida, K.; et al. Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature 2004, 428, 653–657. [Google Scholar] [CrossRef]
- Han, W.; Fan, X.; Teng, L.; Kaczurowski, M.J.S.; Zhang, X.; Xu, D.; Yin, Y.; Ye, N. Identification, classification, and evolution of putative xylosyltransferases from algae. Protoplasma 2019, 256, 1119–1132. [Google Scholar] [CrossRef]
- Cheng, S.; Xian, W.; Fu, Y.; Marin, B.; Keller, J.; Wu, T.; Sun, W.; Li, X.; Xu, Y.; Zhang, Y.; et al. Genomes of Subaerial Zygnematophyceae Provide Insights into Land Plant Evolution. Cell 2019, 179, 1057–1067.e14. [Google Scholar] [CrossRef]
- McClelland, H.L.O.; Barbarin, N.; Beaufort, L.; Hermoso, M.; Ferretti, P.; Greaves, M.; Rickaby, R.E.M. Calcification response of a key phytoplankton family to millennial-scale environmental change. Sci. Rep. 2016, 6, 34263. [Google Scholar] [CrossRef] [Green Version]
- Puerta, M.V.S.; Bachvaroff, T.R.; Delwiche, C.F. The Complete Plastid Genome Sequence of the Haptophyte Emiliania huxleyi: A Comparison to Other Plastid Genomes. DNA Res. 2005, 12, 151–156. [Google Scholar] [CrossRef]
- Wang, G.; Liu, N.; Li, Y.; Zhang, L.; Meinita, M.D.N.; Chen, W.; Liu, T.; Chi, S. The complete plastid genome and phylogenetic analysis of Gracilaria chilensis. Mitochondrial DNA Part B 2020, 5, 1282–1283. [Google Scholar] [CrossRef] [Green Version]
- De Koning, A.P.; Keeling, P.J. The complete plastid genome sequence of the parasitic green alga Helicosporidium sp. is highly reduced and structured. BMC Biol. 2006, 4, 12. [Google Scholar] [CrossRef] [Green Version]
- Kenrick, P.; Crane, P. The origin and early evolution of plants on Land. Nature 1997, 389, 33–39. [Google Scholar] [CrossRef]
- Tronchet, M.; Balague, C.; Kroj, T.; Jouanin, L.; Roby, D. Cinnamyl alcohol dehydrogenases-C and D, key enzymes in lignin biosynthesis, play an essential role in disease resistance in Arabidopsis. Mol. Plant Pathol. 2010, 11, 83–92. [Google Scholar] [CrossRef]
- Martone, P.T.; Estevez, J.M.; Lu, F.; Ruel, K.; Denny, M.W.; Somerville, C.; Ralph, J. Discovery of lignin in seaweed reveals convergent evolution of cell-wall architecture. Curr. Biol. 2009, 19, 169–175. [Google Scholar] [CrossRef] [Green Version]
- Labeeuw, L.; Martone, P.T.; Boucher, Y.; Case, R.J. Ancient origin of the biosynthesis of lignin precursors. Biol. Direct. 2015, 10, 23. [Google Scholar] [CrossRef] [Green Version]
- Dahlin, L.; Guarnieri, M. Recent advances in algal genetic tool development. Curr. Biotechnol. 2016, 5, 192–197. [Google Scholar] [CrossRef]
- Scranton, M.A.; Ostrand, J.T.; Fields, F.J.; Mayfield, S.P. Chlamydomonas as a model for biofuels and bio-products production. Plant J. 2015, 82, 523–531. [Google Scholar] [CrossRef] [Green Version]
- Lauersen, K.J.; Wichmann, J.; Baier, T.; Kampranis, S.C.; Pateraki, I.; Møller, B.L.; Kruse, O. Phototrophic production of heterologous diterpenoids and a hydroxy-functionalized derivative from Chlamydomonas reinhardtii. Metab. Eng. 2018, 49, 116–127. [Google Scholar] [CrossRef]
- Radakovits, R.; Jinkerson, R.E.; Darzins, A.; Posewitz, M.C. Genetic engineering of algae for enhanced biofuel production. Eukaryot. Cell 2010, 9, 486–501. [Google Scholar] [CrossRef] [Green Version]
- Manuell, A.L.; Beligni, M.V.; Elder, J.H.; Siefker, D.T.; Tran, M.; Weber, A.; McDonald, T.L.; Mayfield, S.P. Robust expression of a bioactive mammalian protein in Chlamydomonas chloroplast. Plant Biotechnol. J. 2007, 5, 402–412. [Google Scholar] [CrossRef] [PubMed]
- Eichler-Stahlberg, A.; Weisheit, W.; Ruecker, O.; Heitzer, M. Strategies to facilitate transgene expression in Chlamydomonas reinhardtii. Planta 2009, 229, 873–883. [Google Scholar] [CrossRef]
- Walsh, G. Biopharmaceutical benchmarks. Nat. Biotechnol. 2000, 18, 831–833. [Google Scholar] [CrossRef]
- Yang, Z.; Chen, F.; Li, D.; Zhang, Z.; Liu, Y.; Zheng, D.; Wang, Y.; Shen, G. Expression of human soluble TRAIL in Chlamydomonas reinhardtii chloroplast. Chin. Sci. Bull. 2006, 51, 1703–1709. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khan, A.K.; Kausar, H.; Jaferi, S.S.; Drouet, S.; Hano, C.; Abbasi, B.H.; Anjum, S. An Insight into the Algal Evolution and Genomics. Biomolecules 2020, 10, 1524. https://doi.org/10.3390/biom10111524
Khan AK, Kausar H, Jaferi SS, Drouet S, Hano C, Abbasi BH, Anjum S. An Insight into the Algal Evolution and Genomics. Biomolecules. 2020; 10(11):1524. https://doi.org/10.3390/biom10111524
Chicago/Turabian StyleKhan, Amna Komal, Humera Kausar, Syyada Samra Jaferi, Samantha Drouet, Christophe Hano, Bilal Haider Abbasi, and Sumaira Anjum. 2020. "An Insight into the Algal Evolution and Genomics" Biomolecules 10, no. 11: 1524. https://doi.org/10.3390/biom10111524
APA StyleKhan, A. K., Kausar, H., Jaferi, S. S., Drouet, S., Hano, C., Abbasi, B. H., & Anjum, S. (2020). An Insight into the Algal Evolution and Genomics. Biomolecules, 10(11), 1524. https://doi.org/10.3390/biom10111524






