Image Segmentation of the Ventricular Septum in Fetal Cardiac Ultrasound Videos Based on Deep Learning Using Time-Series Information
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cropping Module
2.2. Segmentation Module
2.3. Calibration Module
2.4. CSC Network
2.5. Training Procedure
2.6. Data Preparation
2.6.1. Data Acquisition Method
2.6.2. Data Preprocessing
2.7. Metrics
2.8. Experiments and Comparison
3. Results
3.1. Data Characteristics
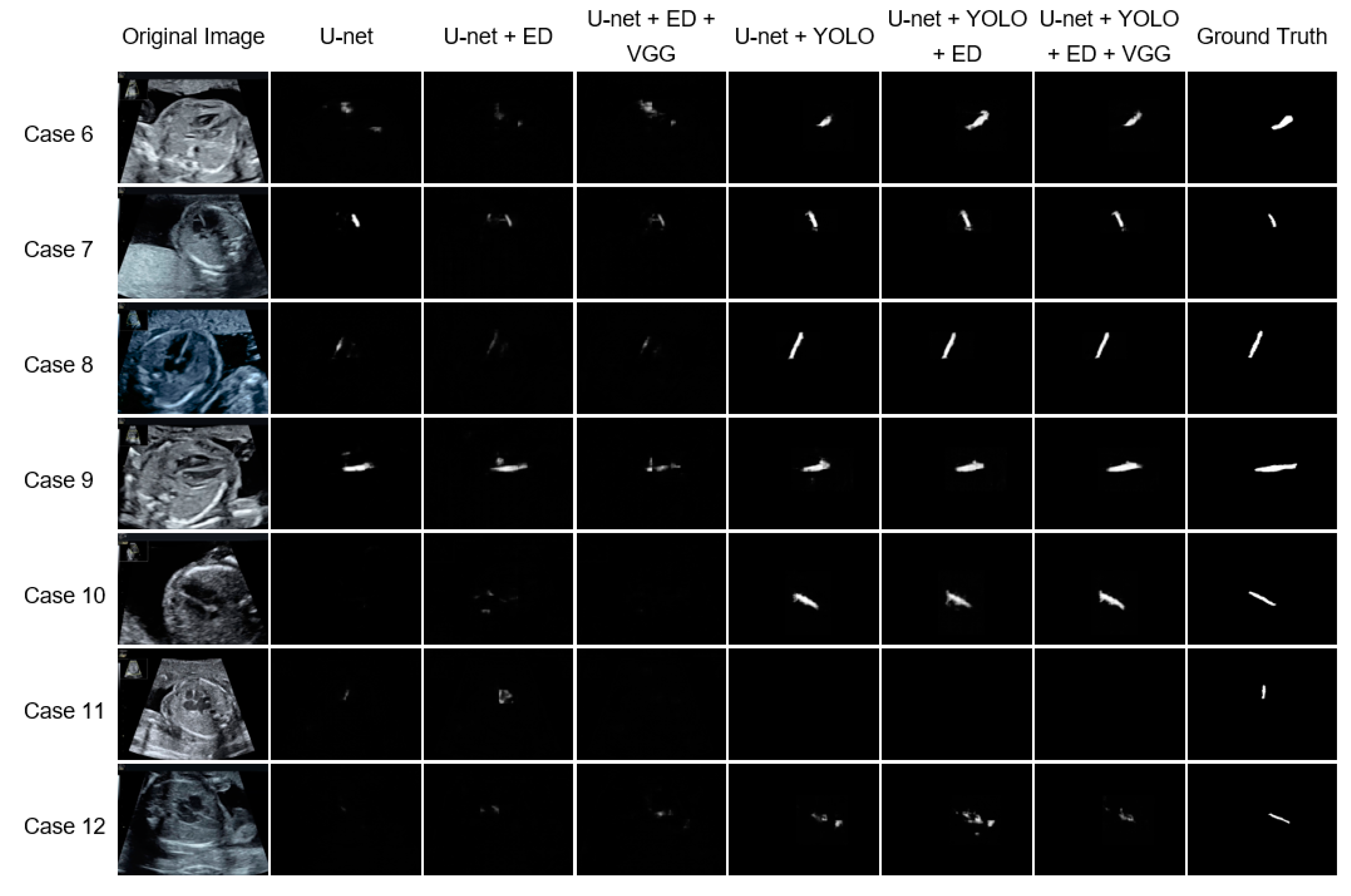
3.2. Comparison with the Existing Methods
3.3. Comparison of Modules
3.4. Effects of Cardiac Axis Orientation and Ventricular Systolic State
4. Discussion
4.1. Combination Analysis of Modules
4.2. Heart Axis and Ventricular Systole
4.3. Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A

Appendix B
References
- Hoffman, J.I.; Kaplan, S. The incidence of congenital heart disease. J. Am. Coll. Cardiol. 2002, 39, 1890–1900. [Google Scholar] [CrossRef] [Green Version]
- Wren, C.; Reinhardt, Z.; Khawaja, K. Twenty-year trends in diagnosis of life-threatening neonatal cardiovascular malformations. Arch. Dis. Child. Fetal Neonatal Ed. 2008, 93, F33–F35. [Google Scholar] [CrossRef] [PubMed]
- Dolk, H.; Loane, M.; Garne, E. A European Surveillance of Congenital Anomalies (EUROCAT) Working Group Congenital Heart Defects in Europe. Circulation 2011, 123, 841–849. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosano, A. Infant mortality and congenital anomalies from 1950 to 1994: An international perspective. J. Epidemiology Community Heal. 2000, 54, 660–666. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petrini, J.R.; Broussard, C.S.; Gilboa, S.M.; Lee, K.A.; Oster, M.; Honein, M.A. Racial differences by gestational age in neonatal deaths attributable to congenital heart defects—United States. Morb. Mortal. Wkly. Rep. 2010, 59, 1208–1211. [Google Scholar] [PubMed]
- Giorgione, V.; Fesslova, V.; Boveri, S.; Candiani, M.; Khalil, A.; Cavoretto, P. Adverse perinatal outcome and placental abnormalities in pregnancies with major fetal congenital heart defects: A retrospective case-control study. Prenat. Diagn. 2020, 40, 1390–1397. [Google Scholar] [CrossRef] [PubMed]
- Inversetti, A.; Fesslova, V.; Deprest, J.; Candiani, M.; Giorgione, V.; Cavoretto, P. Prenatal Growth in Fetuses with Isolated Cyanotic and Non-Cyanotic Congenital Heart Defects. Fetal Diagn. Ther. 2018, 47, 411–419. [Google Scholar] [CrossRef]
- Redmon, J.; Divvala, S.; Girshick, R.; Farhadi, A. You Only Look Once: Unified, Real-Time Object Detection. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; Institute of Electrical and Electronics Engineers (IEEE): New York, NY, USA, 2016; pp. 779–788. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. Available online: https://arxiv.org/abs/1409.1556 (accessed on 8 October 2020).
- Fu, H.; Xu, Y.; Wong, D.W.K.; Liu, J. Retinal vessel segmentation via deep learning network and fully-connected conditional random fields. In Proceedings of the 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI), Prague, Czech Republic, 13–16 April 2016; Institute of Electrical and Electronics Engineers (IEEE): New York, NY, USA, 2016; pp. 698–701. [Google Scholar]
- Pereira, S.; Pinto, A.; Alves, V.; Silva, C.A. Brain Tumor Segmentation Using Convolutional Neural Networks in MRI Images. IEEE Trans. Med. Imaging 2016, 35, 1240–1251. [Google Scholar] [CrossRef]
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; Institute of Electrical and Electronics Engineers (IEEE): New York, NY, USA, 2015; pp. 3431–3440. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Proceedings of the Research in Attacks, Intrusions, and Defenses, Kyoto, Japan, 2–4 November 2015; Springer Science and Business Media LLC: Berlin, Germany, 2015; pp. 234–241. [Google Scholar]
- Badrinarayanan, V.; Kendall, A.; Cipolla, R. SegNet: A Deep Convolutional Encoder-Decoder Architecture for Image Segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 2481–2495. [Google Scholar] [CrossRef]
- Zhao, H.; Shi, J.; Qi, X.; Wang, X.; Jia, J. Pyramid Scene Parsing Network. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; Institute of Electrical and Electronics Engineers (IEEE): New York, NY, USA, 2017; pp. 6230–6239. [Google Scholar]
- Chen, L.-C.; Papandreou, G.; Kokkinos, I.; Murphy, K.; Yuille, A.L. DeepLab: Semantic Image Segmentation with Deep Convolutional Nets, Atrous Convolution, and Fully Connected CRFs. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 40, 834–848. [Google Scholar] [CrossRef]
- Chen, L.-C.; Zhu, Y.; Papandreou, G.; Schroff, F.; Adam, H. Encoder-Decoder with Atrous Separable Convolution for Semantic Image Segmentation. In Proceedings of the Proceedings of the European Conference on Computer Vision, Munich, Germany, 8–14 September 2018; pp. 801–818. Available online: https://arxiv.org/abs/1802.02611 (accessed on 8 October 2020).
- Litjens, G.J.S.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; Van Der Laak, J.A.W.M.; Van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Gajjala, S.; Agrawal, P.; Tison, G.H.; Hallock, L.A.; Beussink-Nelson, L.; Lassen, M.H.; Fan, E.; Aras, M.A.; Jordan, C.; et al. Fully Automated Echocardiogram Interpretation in Clinical Practice. Circulation 2018, 138, 1623–1635. [Google Scholar] [CrossRef]
- Madani, A.; Ong, J.R.; Tibrewal, A.; Mofrad, M.R.K. Deep echocardiography: Data-efficient supervised and semi-supervised deep learning towards automated diagnosis of cardiac disease. npj Digit. Med. 2018, 1, 1–11. [Google Scholar] [CrossRef]
- Kusunose, K.; Abe, T.; Haga, A.; Fukuda, D.; Yamada, H.; Harada, M.; Sata, M. A Deep Learning Approach for Assessment of Regional Wall Motion Abnormality from Echocardiographic Images. JACC Cardiovasc. Imaging 2020, 13, 374–381. [Google Scholar] [CrossRef]
- Ghesu, F.C.; Krubasik, E.; Georgescu, B.; Singh, V.; Zheng, Y.; Hornegger, J.; Comaniciu, D. Marginal Space Deep Learning: Efficient Architecture for Volumetric Image Parsing. IEEE Trans. Med. Imaging 2016, 35, 1217–1228. [Google Scholar] [CrossRef] [PubMed]
- Pereira, F.; Bueno, A.; Rodriguez, A.; Perrin, D.; Marx, G.; Cardinale, M.; Salgo, I.; Del Nido, P. Automated detection of coarctation of aorta in neonates from two-dimensional echocardiograms. J. Med. Imaging 2017, 4, 14502. [Google Scholar] [CrossRef] [Green Version]
- Yasutomi, S.; Arakaki, T.; Hamamoto, R. Shadow Detection for Ultrasound Images Using Unlabeled Data and Synthetic Shadows. arXiv 2019, arXiv:1908.01439. Available online: https://arxiv.org/abs/1908.01439 (accessed on 8 October 2020).
- Arnaout, R.; Curran, L.; Zhao, Y.; Levine, J.; Chinn, E.; Moon-Grady, A. Expert-level prenatal detection of complex congenital heart disease from screening ultrasound using deep learning. medRxiv 2020. [Google Scholar] [CrossRef]
- Redmon, J.; Farhadi, A. YOLO9000: Better, Faster, Stronger. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; Institute of Electrical and Electronics Engineers (IEEE): New York, NY, USA, 2017; pp. 6517–6525. [Google Scholar]
- Bhatkalkar, B.J.; Reddy, D.R.; Prabhu, S.; Bhandary, S.V. Improving the Performance of Convolutional Neural Network for the Segmentation of Optic Disc in Fundus Images Using Attention Gates and Conditional Random Fields. IEEE Access 2020, 8, 29299–29310. [Google Scholar] [CrossRef]
- Yang, T.; Yoshimura, Y.; Morita, A.; Namiki, T.; Nakaguchi, T. Pyramid Predictive Attention Network for Medical Image Segmentation. IEICE Trans. Fundam. Electron. Commun. Comput. Sci. 2019, E102, 1225–1234. [Google Scholar] [CrossRef] [Green Version]
- Donofrio, M.T.; Moon-Grady, A.J.; Hornberger, L.K.; Copel, J.A.; Sklansky, M.S.; Abuhamad, A.; Cuneo, B.F.; Huhta, J.C.; Jonas, R.A.; Krishnan, A.; et al. Diagnosis and Treatment of Fetal Cardiac Disease. Circulation 2014, 129, 2183–2242. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.-T.; Huang, J.-B.; Schwing, A.G. MaskRNN: Instance Level Video Object Segmentation. In Proceedings of the Proceedings of the International Conference on Advances in Neural Information Processing Systems, Montreal, QC, Canada, 3–8 December 2018; pp. 325–334. [Google Scholar]
- Xu, Y.-S.; Fu, T.-J.; Yang, H.-K.; Lee, C.-Y. Dynamic Video Segmentation Network. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; Institute of Electrical and Electronics Engineers (IEEE): New York, NY, USA, 2018; pp. 6556–6565. [Google Scholar]
- Yu, L.; Guo, Y.; Wang, Y.; Yu, J.-H.; Chen, P. Segmentation of Fetal Left Ventricle in Echocardiographic Sequences Based on Dynamic Convolutional Neural Networks. IEEE Trans. Biomed. Eng. 2017, 64, 1886–1895. [Google Scholar] [CrossRef] [PubMed]




| Ventricular Systolic State | Apical | Non-Apical | Total |
|---|---|---|---|
| Systole | 183 | 118 | 301 |
| Diastole | 114 | 200 | 314 |
| Total | 297 | 318 | 615 |
| Method | mIoU | mDice | ||
|---|---|---|---|---|
| Original Image | Cropped Image | Original Image | Cropped Image | |
| DeepLab v3+ | 0.0224 ± 0.0085 | 0.0382 ± 0.0140 | ||
| U-net | 0.1519 ± 0.0596 | 0.2238 ± 0.0777 | ||
| CSC (Ours) | 0.5543 ± 0.0081 | 0.5598 ± 0.0067 | 0.6891 ± 0.0104 | 0.6950 ± 0.0074 |
| U-Net | YOLO | ED | VGG | mIoU | mDice | ||
|---|---|---|---|---|---|---|---|
| Original Image | Cropped Image | Original Image | Cropped Image | ||||
| ✓ | 0.1519 ± 0.0596 | 0.2238 ± 0.0777 | |||||
| ✓ | ✓ | 0.0633 ± 0.0372 | 0.0996 ± 0.0538 | ||||
| ✓ | ✓ | ✓ | 0.0902 ± 0.0304 | 0.1400 ± 0.0442 | |||
| ✓ | ✓ | 0.5373 ± 0.0134 | 0.5424 ± 0.0107 | 0.6724 ± 0.0188 | 0.6782 ± 0.0153 | ||
| ✓ | ✓ | ✓ | 0.5533 ± 0.0139 | 0.5587 ± 0.0138 | 0.6885 ± 0.0141 | 0.6944 ± 0.0123 | |
| ✓ | ✓ | ✓ | ✓ | 0.5543 ± 0.0081 | 0.5598 ± 0.0067 | 0.6891 ± 0.0104 | 0.6950 ± 0.0074 |
| U-Net | YOLO | ED | VGG | mIoU | mDice | ||
|---|---|---|---|---|---|---|---|
| Apical | Non-Apical | Apical | Non-Apical | ||||
| ✓ | 0.1878 ±0.1097 | 0.1213 ± 0.0186 | 0.2697 ± 0.1410 | 0.1845 ± 0.0261 | |||
| ✓ | ✓ | 0.5793 ± 0.0315 | 0.4990 ± 0.0058 | 0.7064 ± 0.0405 | 0.6417 ± 0.0086 | ||
| ✓ | ✓ | ✓ | 0.5889 ± 0.0265 | 0.5210 ± 0.0160 | 0.7146 ± 0.0351 | 0.6653 ± 0.0140 | |
| ✓ | ✓ | ✓ | ✓ | 0.5855 ± 0.0167 | 0.5255 ± 0.0016 | 0.7114 ± 0.0264 | 0.6688 ± 0.0026 |
| U-Net | YOLO | ED | VGG | mIoU | mDice | ||
|---|---|---|---|---|---|---|---|
| Systole | Diastole | Systole | Diastole | ||||
| ✓ | 0.1397 ± 0.0686 | 0.1631 ± 0.0528 | 0.2072 ± 0.0914 | 0.2388 ± 0.0677 | |||
| ✓ | ✓ | 0.5255 ± 0.0158 | 0.5491 ± 0.0114 | 0.6567 ± 0.0235 | 0.6882 ± 0.0146 | ||
| ✓ | ✓ | ✓ | 0.5413 ± 0.0196 | 0.5655 ± 0.0065 | 0.6733 ± 0.0186 | 0.7037 ± 0.0067 | |
| ✓ | ✓ | ✓ | ✓ | 0.5435 ± 0.0102 | 0.5648 ± 0.0073 | 0.6755 ± 0.0127 | 0.7026 ± 0.0073 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dozen, A.; Komatsu, M.; Sakai, A.; Komatsu, R.; Shozu, K.; Machino, H.; Yasutomi, S.; Arakaki, T.; Asada, K.; Kaneko, S.; et al. Image Segmentation of the Ventricular Septum in Fetal Cardiac Ultrasound Videos Based on Deep Learning Using Time-Series Information. Biomolecules 2020, 10, 1526. https://doi.org/10.3390/biom10111526
Dozen A, Komatsu M, Sakai A, Komatsu R, Shozu K, Machino H, Yasutomi S, Arakaki T, Asada K, Kaneko S, et al. Image Segmentation of the Ventricular Septum in Fetal Cardiac Ultrasound Videos Based on Deep Learning Using Time-Series Information. Biomolecules. 2020; 10(11):1526. https://doi.org/10.3390/biom10111526
Chicago/Turabian StyleDozen, Ai, Masaaki Komatsu, Akira Sakai, Reina Komatsu, Kanto Shozu, Hidenori Machino, Suguru Yasutomi, Tatsuya Arakaki, Ken Asada, Syuzo Kaneko, and et al. 2020. "Image Segmentation of the Ventricular Septum in Fetal Cardiac Ultrasound Videos Based on Deep Learning Using Time-Series Information" Biomolecules 10, no. 11: 1526. https://doi.org/10.3390/biom10111526
APA StyleDozen, A., Komatsu, M., Sakai, A., Komatsu, R., Shozu, K., Machino, H., Yasutomi, S., Arakaki, T., Asada, K., Kaneko, S., Matsuoka, R., Aoki, D., Sekizawa, A., & Hamamoto, R. (2020). Image Segmentation of the Ventricular Septum in Fetal Cardiac Ultrasound Videos Based on Deep Learning Using Time-Series Information. Biomolecules, 10(11), 1526. https://doi.org/10.3390/biom10111526







