Mapping the Inorganic and Proteomic Differences among Different Types of Human Teeth: A Preliminary Compositional Insight
Abstract
:1. Introduction
2. Materials and Methods
2.1. Collection of Human Teeth and Isolation of Tooth Proteins
2.2. Inductively Coupled Plasma Resonance Mass Spectroscopy (ICP-MS)
2.3. Label-Free Quantification of Tooth Proteins through Mass Spectrometry
2.4. Protein Interaction Studies
3. Results and Discussion
3.1. The Inorganic Analysis Revealed Differences in Teeth Mineral Composition
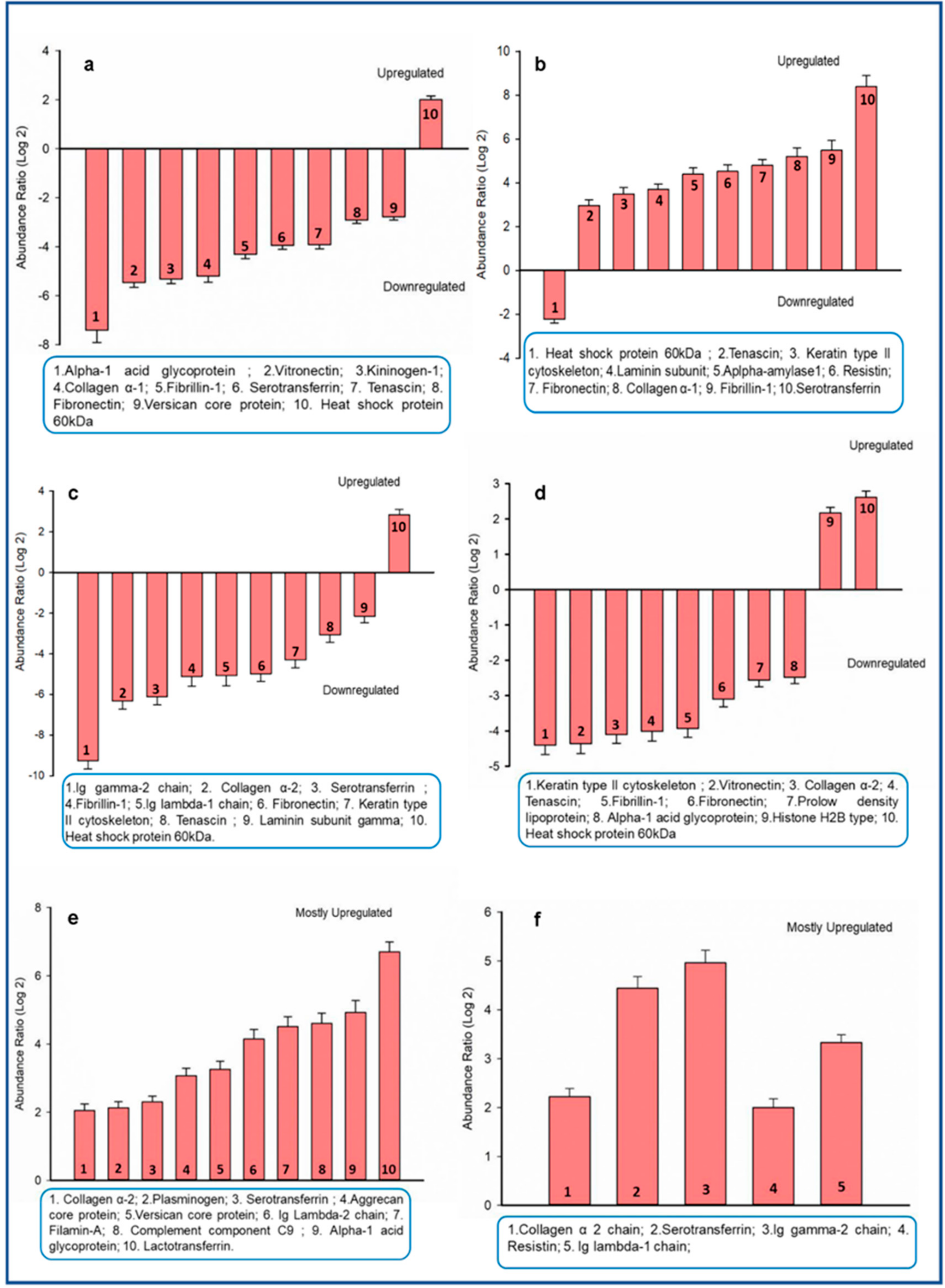
3.2. Proteins in Four Major Teeth Types Differ Qualitatively and Quantitatively
3.3. Protein Interaction Network Studies
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sharma, V.; Srinivasan, A.; Roychoudhury, A.; Rani, K.; Tyagi, M.; Dev, K.; Nikolajeff, F.; Kumar, S. Characterization of protein extracts from different types of human teeth and insight in biomineralization. Sci. Rep. 2019, 9, 9314. [Google Scholar] [CrossRef] [PubMed]
- Sharma, V.; Rani, K.; Roychoudhury, A.; Chawla, A.; Nikolajeff, F.; Kumar, S. Novel insights into regulation of human teeth biomineralization: Deciphering the role of post-translational modifications in a tooth protein extract. Int. J. Mol. Sci. 2019, 20, 4035. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jágr, M.; Eckhardt, A.; Pataridis, S.; Broukal, Z.; Dušková, J.; Mikšík, I. Proteomics of human teeth and saliva. Physiol. Res. 2014, 63, S141–S154. [Google Scholar] [CrossRef]
- Boskey, A.L.; Villarreal-Ramirez, E. Intrinsically disordered proteins and biomineralization. Matrix Biol. 2016, 52, 43–59. [Google Scholar] [CrossRef] [Green Version]
- Sharma, V.; Srinivasan, A.; Nikolajeff, F.; Kumar, S. Biomineralization process in hard tissues: The interaction complexity within protein and inorganic counterparts. Acta Biomater. 2020. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Gasga, J.; Martínez-Piñeiro, E.L.; Rodríguez-Álvarez, G.; Tiznado-Orozco, G.E.; García-García, R.; Brès, E.F. XRD and FTIR crystallinity indices in sound human tooth enamel and synthetic hydroxyapatite. Mater. Sci. Eng. C 2013, 33, 4568–4574. [Google Scholar] [CrossRef]
- Mihály, J.; Gombás, V.; Afishah, A.; Mink, J. FT-Raman investigation of human dental enamel surfaces. J. Raman Spectrosc. Int. J. Orig.Work Asp. Raman Spectrosc. Incl. Higher Order Process. Brillouin Rayleigh Scatt. 2009, 40, 898–902. [Google Scholar] [CrossRef]
- Kim, I.-H.; Son, J.S.; Min, B.K.; Kim, Y.K.; Kim, K.-H.; Kwon, T.-Y. A simple, sensitive and non-destructive technique for characterizing bovine dental enamel erosion: Attenuated total reflection Fourier transform infrared spectroscopy. Int. J. Oral Sci. 2016, 8, 54–60. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Yao, X.; Liu, Y.; Wang, Y. A Fourier transform infrared spectroscopy analysis of carious dentin from transparent zone to normal zone. Caries Res. 2014, 48, 320–329. [Google Scholar] [CrossRef] [Green Version]
- Rechmann, P.; White, J.M.; Cecchini, S.C.M.; Hennig, T. Fourier transform infrared spectroscopy (FTIR) of laser-irradiated cementum. In Proceedings of the Lasers in Dentistry IX, San Jose, CA, USA, 25–31 January 2003; pp. 115–121. [Google Scholar]
- Anwar Alebrahim, M.; Krafft, C.; Sekhaneh, W.; Sigusch, B.; Popp, J. ATR-FTIR and Raman spectroscopy of primary and permanent teeth. Biomed. Spectrosc. Imaging 2014, 3, 15–27. [Google Scholar] [CrossRef]
- Hędzelek, W.; Marcinkowska, A.; Domka, L.; Wachowiak, R. Infrared spectroscopic identification of chosen dental materials and natural teeth. Acta Phys. Pol. A 2008, 2, 471–484. [Google Scholar] [CrossRef]
- Sprawson, E.; Bury, F.W. On the chemical evidences of the organic content of human enamel. Proceedings of the Royal Society of London. Ser. B Contain. Pap. Biol. Character 1928, 102, 419–426. [Google Scholar]
- Ghadimi, E.; Eimar, H.; Marelli, B.; Nazhat, S.N.; Asgharian, M.; Vali, H.; Tamimi, F. Trace elements can influence the physical properties of tooth enamel. SpringerPlus 2013, 2, 499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Annegarn, H.; Jodaikin, A.; Cleaton-Jones, P.; Sellschop, J.; Madiba, C.; Bibby, D. PIXE analysis of caries related trace elements in tooth enamel. Nucl. Instrum. Methods 1981, 181, 323–326. [Google Scholar] [CrossRef]
- Hare, D.; Austin, C.; Doble, P.; Arora, M. Elemental bio-imaging of trace elements in teeth using laser ablation-inductively coupled plasma-mass spectrometry. J. Dent. 2011, 39, 397–403. [Google Scholar] [CrossRef] [Green Version]
- Brudevold, F.; Steadman, L.T.; Spinelli, M.A.; Amdur, B.H.; Grøn, P. A study of zinc in human teeth. Arch. Oral Biol. 1963, 8, 135–144. [Google Scholar] [CrossRef]
- Brown, C.J.; Chenery, S.R.; Smith, B.; Mason, C.; Tomkins, A.; Roberts, G.J.; Sserunjogi, L.; Tiberindwa, J.V. Environmental influences on the trace element content of teeth—implications for disease and nutritional status. Arch. Oral Biol. 2004, 49, 705–717. [Google Scholar] [CrossRef]
- Webb, E.; Amarasiriwardena, D.; Tauch, S.; Green, E.F.; Jones, J.; Goodman, A.H. Inductively coupled plasma-mass (ICP-MS) and atomic emission spectrometry (ICP-AES): Versatile analytical techniques to identify the archived elemental information in human teeth. Microchem. J. 2005, 81, 201–208. [Google Scholar] [CrossRef]
- Jernvall, J.; Thesleff, I. Tooth shape formation and tooth renewal: Evolving with the same signals. Development 2012, 139, 3487–3497. [Google Scholar] [CrossRef] [Green Version]
- Moradian-Oldak, J. Protein-mediated enamel mineralization. Front. Biosci. 2012, 17, 1996–2023. [Google Scholar] [CrossRef] [Green Version]
- Iijima, M.; Fan, D.; Bromley, K.M.; Sun, Z.; Moradian-Oldak, J. Tooth enamel proteins enamelin and amelogenin cooperate to regulate the growth morphology of octacalcium phosphate crystals. Cryst. Growth Des. 2010, 10, 4815–4822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, L.; Han, S.; Wang, K.; Zheng, S.; Zheng, W.; Peng, X.; Niu, Y.; Li, W.; Zhang, L. Remineralization of enamel caries by an amelogenin-derived peptide and fluoride in vitro. Regen. Biomater. 2020, 7, 283–292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mukherjee, K.; Visakan, G.; Phark, J.-H.; Moradian-Oldak, J. Enhancing Collagen Mineralization with Amelogenin Peptide: Toward the Restoration of Dentin. ACS Biomater. Sci. Eng. 2020, 6, 2251–2262. [Google Scholar] [CrossRef]
- Park, E.-S.; Cho, H.-S.; Kwon, T.-G.; Jang, S.-N.; Lee, S.-H.; An, C.-H.; Shin, H.-I.; Kim, J.-Y.; Cho, J.-Y. Proteomics analysis of human dentin reveals distinct protein expression profiles. J. Proteome Res. 2009, 8, 1338–1346. [Google Scholar] [CrossRef] [PubMed]
- Jágr, M.; Eckhardt, A.; Pataridis, S.; Mikšík, I. Comprehensive proteomic analysis of human dentin. Eur. J. Oral Sci. 2012, 120, 259–268. [Google Scholar] [CrossRef] [PubMed]
- Eckhardt, A.; Jágr, M.; Pataridis, S.; Mikšík, I. Proteomic analysis of human tooth pulp: Proteomics of human tooth. J. Endod. 2014, 40, 1961–1966. [Google Scholar] [CrossRef] [PubMed]
- Chun, S.Y.; Lee, H.J.; Choi, Y.A.; Kim, K.M.; Baek, S.H.; Park, H.S.; Kim, J.-Y.; Ahn, J.-M.; Cho, J.-Y.; Cho, D.-W. Analysis of the soluble human tooth proteome and its ability to induce dentin/tooth regeneration. Tissue Eng. Part A 2011, 17, 181–191. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [Green Version]
- Lane, D.; Peach, D. Some observations on the trace element concentrations in human dental enamel. Biol. Trace Elem. Res. 1997, 60, 1–11. [Google Scholar] [CrossRef]
- Kampa, M.; Castanas, E. Human health effects of air pollution. Environ. Pollut. 2008, 151, 362–367. [Google Scholar] [CrossRef]
- Malczewska‒Toth, B. Phosphorus, Selenium, Tellurium, and Sulfur. Patty Toxicol. 2012, 841–884. [Google Scholar] [CrossRef]
- Cook, J.; Layrisse, M.; Martinez-Torres, C.; Walker, R.; Monsen, E.; Finch, C. Food iron absorption measured by an extrinsic tag. J. Clin. Investig. 1972, 51, 805–815. [Google Scholar] [CrossRef] [PubMed]
- Elkabouss, K.; Kacimi, M.; Ziyad, M.; Ammar, S.; Bozon-Verduraz, F. Cobalt-exchanged hydroxyapatite catalysts: Magnetic studies, spectroscopic investigations, performance in 2-butanol and ethane oxidative dehydrogenations. J. Catal. 2004, 226, 16–24. [Google Scholar] [CrossRef]
- Świetlicka, I.; Tomaszewska, E.; Muszyński, S.; Valverde Piedra, J.L.; Świetlicki, M.; Prószyński, A.; Cieślak, K.; Wiącek, D.; Szymańczyk, S.; Kamiński, D. The effect of cadmium exposition on the structure and mechanical properties of rat incisors. PLoS ONE 2019, 14, e0215370. [Google Scholar] [CrossRef]
- Borella, P.; Fantuzzi, G.; Aggazzotti, G. Trace elements in saliva and dental caries in young adults. Sci. Total Environ. 1994, 153, 219–224. [Google Scholar] [CrossRef]
- Sighinolfi, G.P.; Gorgoni, C.; Bonori, O.; Cantoni, E.; Martelli, M.; Simonetti, L. Comprehensive determination of trace elements in human saliva by ETA-AAS. Microchim. Acta 1989, 97, 171–179. [Google Scholar] [CrossRef]
- Little, M.F.; Steadman, L.T. Chemical and physical properties of altered and sound enamel—IV: Trace element composition. Arch. Oral Biol. 1966, 11, 273–278. [Google Scholar] [CrossRef]
- Alfrey, A.C.; LeGendre, G.R.; Kaehny, W.D. The dialysis encephalopathy syndrome. Possible aluminum intoxication. N. Engl. J. Med. 1976, 294, 184–188. [Google Scholar] [CrossRef]
- Lynch, R.J. Zinc in the mouth, its interactions with dental enamel and possible effects on caries; a review of the literature. Int. Dent. J. 2011, 61, 46–54. [Google Scholar] [CrossRef]
- Odanaka, H.; Obama, T.; Sawada, N.; Sugano, M.; Itabe, H.; Yamamoto, M. Comparison of protein profiles of the pellicle, gingival crevicular fluid, and saliva: Possible origin of pellicle proteins. Biol. Res. 2020, 53, 3. [Google Scholar] [CrossRef]
- Partanen, A.-M.; Thesleff, I. Transferrin and tooth morphogenesis: Retention of transferrin by mouse embryonic teeth in organ culture. Differentiation 1987, 34, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Kaur, J.; Reinhardt, D.P. Extracellular Matrix (ECM) Molecules. In Stem Cell Biology and Tissue Engineering in Dental Sciences; Elsevier: Amsterdam, The Netherlands, 2015; pp. 25–45. [Google Scholar]
- Sahlberg, C.; Aukhil, I.; Thesleff, I. Tenascin-C in developing mouse teeth: Expression of splice variants and stimulation by TGFβ and FGF. Eur. J. Oral Sci. 2001, 109, 114–124. [Google Scholar] [CrossRef] [PubMed]
- Tucker, R.P.; Degen, M. The Expression and Possible Functions of Tenascin-W During Development and Disease. Front. Cell Dev. Biol. 2019, 7. [Google Scholar] [CrossRef] [PubMed]
- Ueno, A.; Yamashita, K.; Nagata, T.; Tsurumi, C.; Miwa, Y.; Kitamura, S.; Inoue, H. cDNA cloning of bovine thrombospondin 1 and its expression in odontoblasts and predentin. Biochim. Biophys. Acta 1998, 1382, 17–22. [Google Scholar] [CrossRef]
- Yuasa, K.; Fukumoto, S.; Kamasaki, Y.; Yamada, A.; Fukumoto, E.; Kanaoka, K.; Saito, K.; Harada, H.; Arikawa-Hirasawa, E.; Miyagoe-Suzuki, Y. Laminin α2 is essential for odontoblast differentiation regulating dentin sialoprotein expression. J. Biol. Chem. 2004, 279, 10286–10292. [Google Scholar] [CrossRef] [Green Version]
- Hormia, M.; Sahlberg, C.; Thesleff, I.; Airenne, T. The epithelium-tooth interface--a basal lamina rich in laminin-5 and lacking other known laminin isoforms. J. Dent. Res. 1998, 77, 1479–1485. [Google Scholar] [CrossRef]
- Saito, K.; Fukumoto, E.; Yamada, A.; Yuasa, K.; Yoshizaki, K.; Iwamoto, T.; Saito, M.; Nakamura, T.; Fukumoto, S. Interaction between fibronectin and β1 integrin is essential for tooth development. PLoS ONE 2015, 10, e0121667. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Du, Y.; Ling, J.; Li, W.; Liao, Y.; Wei, X. Dickkopf-related protein 3 negatively regulates the osteogenic differentiation of rat dental follicle cells. Mol. Med. Rep. 2017, 15, 1673–1681. [Google Scholar] [CrossRef] [Green Version]
- Ablooglu, A.J.; Kang, J.; Handin, R.I.; Traver, D.; Shattil, S.J. The zebrafish vitronectin receptor: Characterization of integrin αV and β3 expression patterns in early vertebrate development. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 2007, 236, 2268–2276. [Google Scholar]
- Fabian, T.; Fejerdy, P.; Csermely, P. Salivary genomics, transcriptomics and proteomics: The emerging concept of the oral ecosystem and their use in the early diagnosis of cancer and other diseases. Curr. Genomics 2008, 9, 11–21. [Google Scholar] [CrossRef]
- Lamkin, M.S.; Oppenheim, F.G. Structural features of salivary function. Crit. Rev.Oral Biol. Med. 1993, 4, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Jungblut, P.R.; Holzhütter, H.G.; Apweiler, R.; Schlüter, H. The speciation of the proteome. Chem. Central J. 2008, 2, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schlüter, H.; Apweiler, R.; Holzhütter, H.G.; Jungblut, P.R. Finding one’s way in proteomics: A protein species nomenclature. Chem. Central J. 2009, 3, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duverger, O.; Beniash, E.; Morasso, M.I. Keratins as components of the enamel organic matrix. Matrix Biol. 2016, 52, 260–265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duverger, O.; Ohara, T.; Shaffer, J.R.; Donahue, D.; Zerfas, P.; Dullnig, A.; Crecelius, C.; Beniash, E.; Marazita, M.L.; Morasso, M.I. Hair keratin mutations in tooth enamel increase dental decay risk. J. Clin. Investig. 2014, 124, 5219–5224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohsaki, Y.; Nagata, K. Type III collagen is a major component of interodontoblastic fibers of the developing mouse molar root. Anat. Rec. 1994, 240, 308–313. [Google Scholar] [CrossRef]
- Asaka, T.; Akiyama, M.; Domon, T.; Nishie, W.; Natsuga, K.; Fujita, Y.; Abe, R.; Kitagawa, Y.; Shimizu, H. Type XVII collagen is a key player in tooth enamel formation. Am. J. Pathol. 2009, 174, 91–100. [Google Scholar] [CrossRef] [Green Version]
- Huynh, N.C.-N.; Everts, V.; Ampornaramveth, R.S. Histone deacetylases and their roles in mineralized tissue regeneration. Bone Rep. 2017, 7, 33–40. [Google Scholar] [CrossRef]
- Assiry, A.A.; Albalawi, A.M.; Zafar, M.S.; Khan, S.D.; Ullah, A.; Almatrafi, A.; Ramzan, K.; Basit, S. KMT2C, a histone methyltransferase, is mutated in a family segregating non-syndromic primary failure of tooth eruption. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef]





| Teeth Type | C:I:M:P | C:M:I | C:I:P | M:I:P | C:I | C:M | P:I |
|---|---|---|---|---|---|---|---|
| Key proteins common in different teeth types | Collagen alpha-1(Q99715), Fibrillin-1 (P35555), Fibronectin (P02751), Collagen alpha-2(P08123), Hemopexin (P02790), Superoxide dismutase [Cu-Zn] (P00441), Serotransferrin (P02787), 60 kDa heat shock protein (P10809), Collagen alpha-1(P02452), Tenascin (P24821) | Alpha-amylase 2B precursor (P19961), Resistin precursor (Q9HD89), Alpha-amylase 1 precursor (P04745) | Versican core protein precursor (P13611), Vitronectin precursor (P04004), Prolow density lipoprotein receptor-related protein(Q07954), Thrombospondin-1 precursor (P07996), Collagen alpha-1(I) chain precursor (P02452), Filamin-A (P21333), Plasminogen precursor (P00747), Neutrophil defensin 3 precursor (P59666), Lactotransferrin precursor (P02788), Alpha-1-acid glycoprotein 1 precursor (P02763), Collagen alpha-1(XXVIII) chain precursor (Q2UY09), Neutrophil defensin 1 precursor (P59665), Tenascin-N precursor (Q9UQP3), Basement membrane-specific heparan sulfate (P98160), Prothrombin precursor (P00734) | Various subtypes of Keratins (P02538, P13647, P04259, O95678, P48668), Protein S100 (P06702), Laminin (P11047), Galectin-3-binding protein (Q08380) | Kininogen-1 precursor (P01042), Polyubiquitin-B precursor (P0CG47), Ubiquitin protein (P62987), Dickkopf-related protein 3 precursor (Q9UBP4) | Nucleolar RNA helicase 2 (Q9NR30) | Collagen alpha-2 (P08572), Aggrecan core protein(P16112), Filamin-B (O75369), Thrombospondin (Q6ZMP0), Zinc-alpha-2-glycoprotein (P25311) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sharma, V.; Rastogi, S.; Kumar Bhati, K.; Srinivasan, A.; Roychoudhury, A.; Nikolajeff, F.; Kumar, S. Mapping the Inorganic and Proteomic Differences among Different Types of Human Teeth: A Preliminary Compositional Insight. Biomolecules 2020, 10, 1540. https://doi.org/10.3390/biom10111540
Sharma V, Rastogi S, Kumar Bhati K, Srinivasan A, Roychoudhury A, Nikolajeff F, Kumar S. Mapping the Inorganic and Proteomic Differences among Different Types of Human Teeth: A Preliminary Compositional Insight. Biomolecules. 2020; 10(11):1540. https://doi.org/10.3390/biom10111540
Chicago/Turabian StyleSharma, Vaibhav, Simran Rastogi, Kaushal Kumar Bhati, Alagiri Srinivasan, Ajoy Roychoudhury, Fredrik Nikolajeff, and Saroj Kumar. 2020. "Mapping the Inorganic and Proteomic Differences among Different Types of Human Teeth: A Preliminary Compositional Insight" Biomolecules 10, no. 11: 1540. https://doi.org/10.3390/biom10111540
APA StyleSharma, V., Rastogi, S., Kumar Bhati, K., Srinivasan, A., Roychoudhury, A., Nikolajeff, F., & Kumar, S. (2020). Mapping the Inorganic and Proteomic Differences among Different Types of Human Teeth: A Preliminary Compositional Insight. Biomolecules, 10(11), 1540. https://doi.org/10.3390/biom10111540







