USP25 Regulates EGFR Fate by Modulating EGF-Induced Ubiquitylation Dynamics
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents and Constructs
2.2. Cell Lines
2.3. siRNA Transfection
- USP25
- S1 CAGGAGGAGACAACUUACUACCAAA;
- AMSH
- S1 CGCUCUGGAGUUGAGAUUAUCCGAA;
- USP8
- S1 UCGUGAUGAGGAAAGGGCCUAUGUA.
2.4. Biochemistry
2.5. ELISA-Based Dissociation-Enhanced Lanthanide Fluorescence Immunoassay (DELFIA)
2.6. Immunofluorescence
2.7. EGF Internalization Assay and Measurement of Surface EGFRs
2.8. TCGA Cohort’s Analysis
3. Results
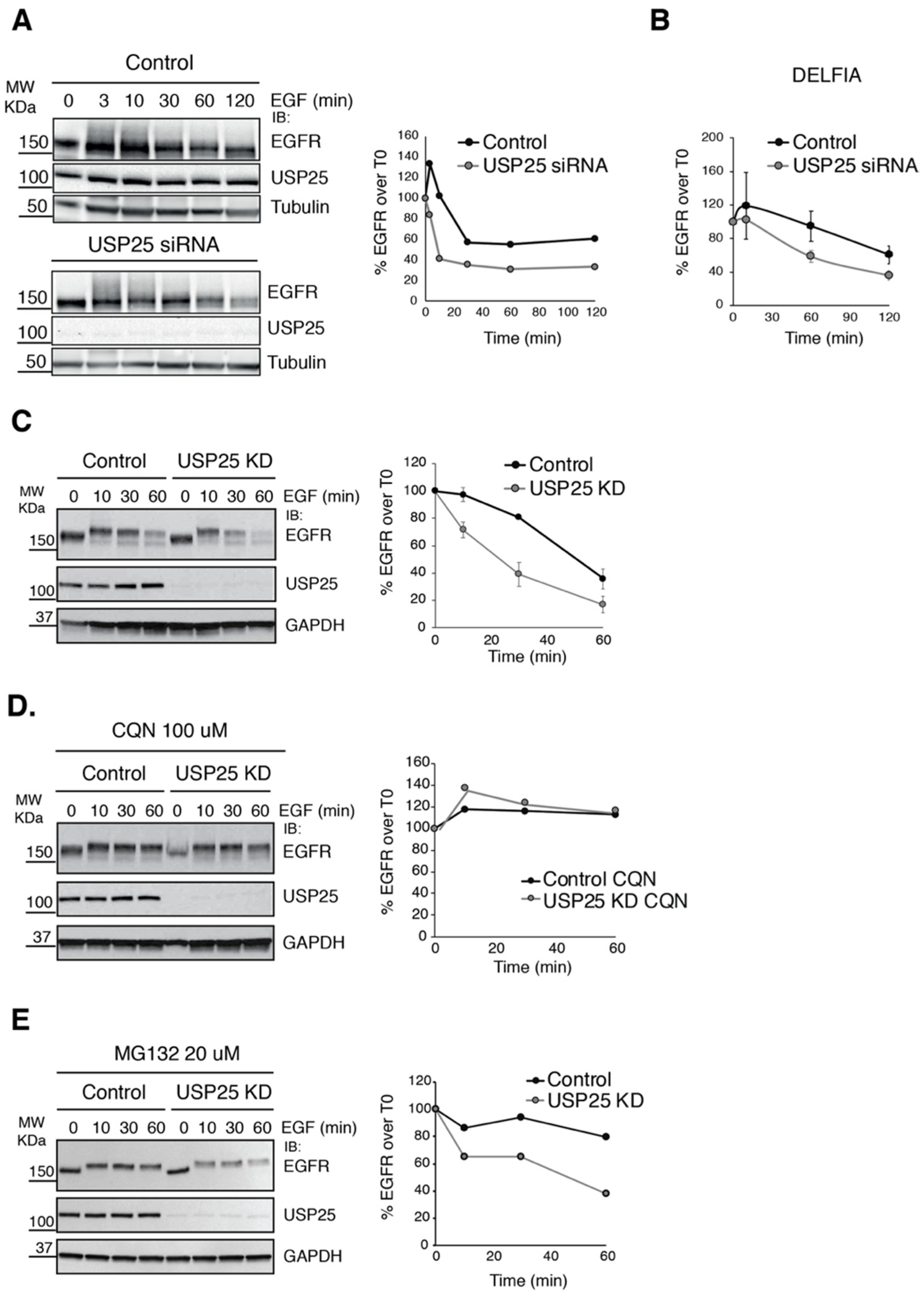
3.1. EGFR Degradation Occurs Faster upon USP25 Knock-Down
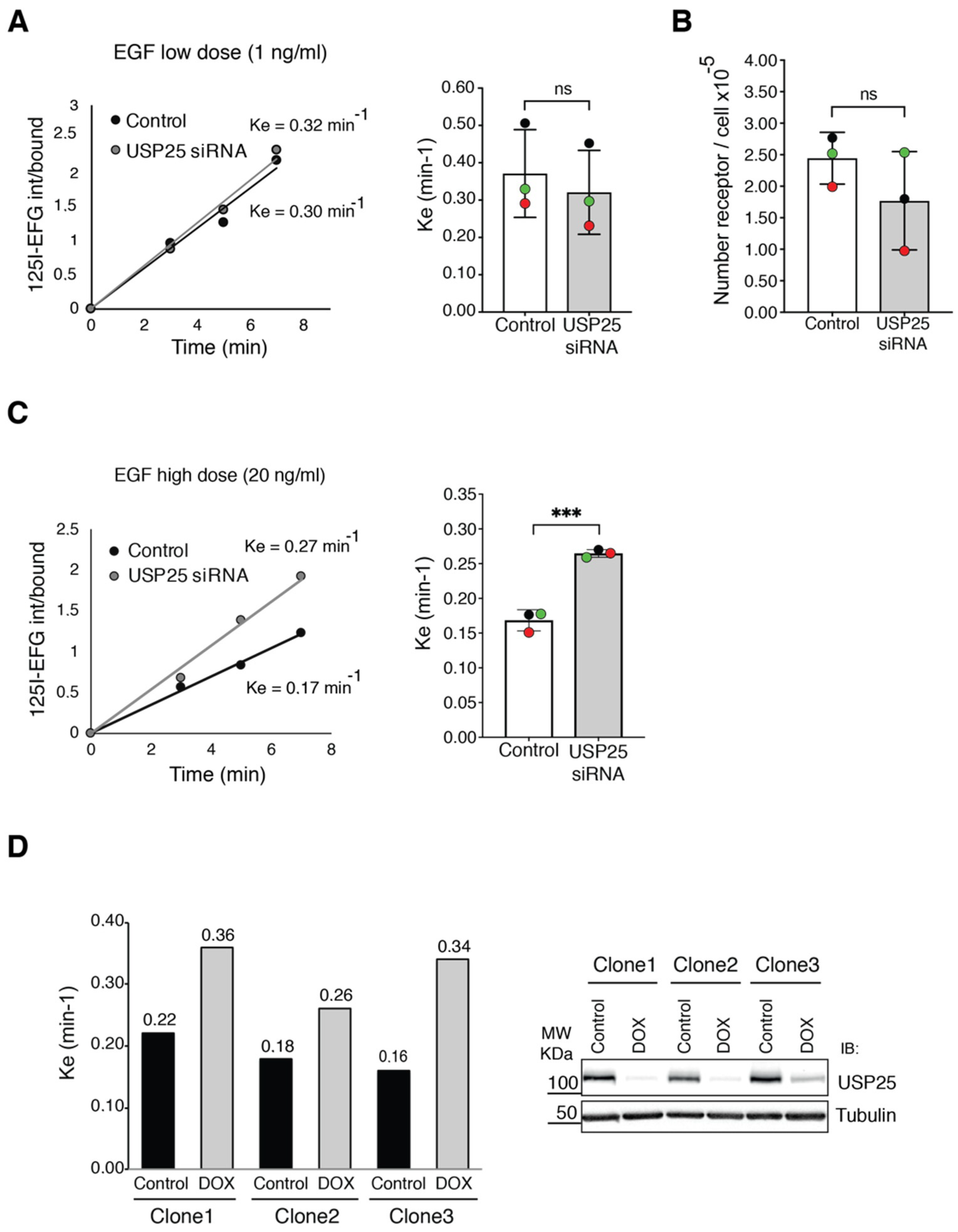
3.2. EGFR Internalization Is Enhanced upon USP25 Knock-Down
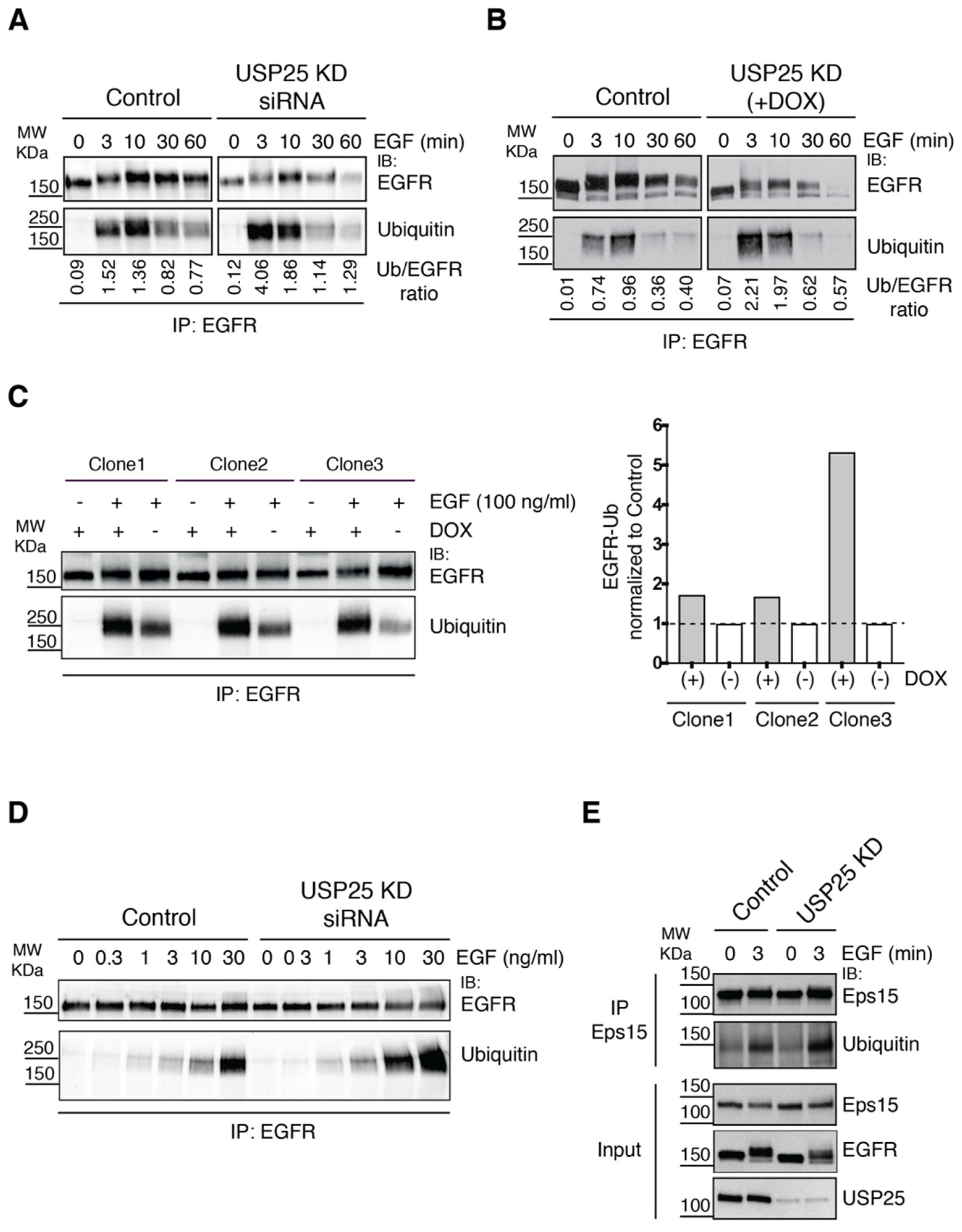
3.3. EGFR Ubiquitylation Is Accelerated upon USP25 Knock-Down
3.4. EGFR Phosphorylation Sites and c-Cbl Interaction Are Altered upon USP25 Knock-Down
3.5. USP25 and EGFR Expression Levels Correlate in Cancer Patients
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lemmon, M.A.; Schlessinger, J.; Ferguson, K.M. The EGFR Family: Not So Prototypical Receptor Tyrosine Kinases. Cold Spring Harb. Perspect. Biol. 2014, 6, a020768. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zeng, F.; Forrester, S.J.; Eguchi, S.; Zhang, M.-Z.; Harris, R.C. Expression and Function of the Epidermal Growth Factor Receptor in Physiology and Disease. Physiol. Rev. 2016, 96, 1025–1069. [Google Scholar] [CrossRef] [PubMed]
- Roskoski, R. The ErbB/HER family of protein-tyrosine kinases and cancer. Pharmacol. Res. 2014, 79, 34–74. [Google Scholar] [CrossRef] [PubMed]
- Yarden, Y.; Pines, G. The ERBB network: At last, cancer therapy meets systems biology. Nat. Rev. Cancer 2012, 12, 553–563. [Google Scholar] [CrossRef] [PubMed]
- Chong, C.R.; Jänne, P.A. The quest to overcome resistance to EGFR-targeted therapies in cancer. Nat. Med. 2013, 19, 1389–1400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caldieri, G.; Malabarba, M.G.; Di Fiore, P.P.; Sigismund, S. EGFR Trafficking in Physiology and Cancer; Springer Science and Business Media LLC: Berlin, Germany, 2018; Volume 57, pp. 235–272. [Google Scholar]
- Acconcia, F.; Sigismund, S.; Polo, S. Ubiquitin in trafficking: The network at work. Exp. Cell Res. 2009, 315, 1610–1618. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Kirkpatrick, D.; Jiang, X.; Gygi, S.; Sorkin, A. Differential Regulation of EGF Receptor Internalization and Degradation by Multiubiquitination within the Kinase Domain. Mol. Cell 2006, 21, 737–748. [Google Scholar] [CrossRef]
- Huang, F.; Goh, L.K.; Sorkin, A. EGF receptor ubiquitination is not necessary for its internalization. Proc. Natl. Acad. Sci. USA 2007, 104, 16904–16909. [Google Scholar] [CrossRef] [Green Version]
- Sigismund, S.; Woelk, T.; Puri, C.; Maspero, E.; Tacchetti, C.; Transidico, P.; Di Fiore, P.P.; Polo, S. Clathrin-independent endocytosis of ubiquitinated cargos. Proc. Natl. Acad. Sci. USA 2005, 102, 2760–2765. [Google Scholar] [CrossRef] [Green Version]
- Sigismund, S.; Algisi, V.; Nappo, G.; Conte, A.; Pascolutti, R.; Cuomo, A.; Bonaldi, T.; Argenzio, E.; Verhoef, L.G.G.C.; Maspero, E.; et al. Threshold-controlled ubiquitination of the EGFR directs receptor fate. EMBO J. 2013, 32, 2140–2157. [Google Scholar] [CrossRef] [Green Version]
- Capuani, F.; Conte, A.; Argenzio, E.; Marchetti, L.; Priami, C.; Polo, S.; Di Fiore, P.P.; Sigismund, S.; Ciliberto, A. Quantitative analysis reveals how EGFR activation and downregulation are coupled in normal but not in cancer cells. Nat. Commun. 2015, 6, 7999. [Google Scholar] [CrossRef] [PubMed]
- Levkowitz, G.; Waterman, H.; Ettenberg, S.A.; Katz, M.; Tsygankov, A.Y.; Alroy, I.; Lavi, S.; Iwai, K.; Reiss, Y.; Ciechanover, A.; et al. Ubiquitin ligase activity and tyrosine phosphorylation underlie suppression of growth factor signaling by c-Cbl/Sli-1. Mol. Cell 1999, 4, 1029–1040. [Google Scholar] [CrossRef]
- Waterman, H.; Katz, M.; Rubin, C.; Shtiegman, K.; Lavi, S.; Elson, A.; Elson, A.; Jovin, T.; Yarden, Y. A mutant EGF-receptor defective in ubiquitylation and endocytosis unveils a role for Grb2 in negative signaling. EMBO J. 2002, 21, 303–313. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Huang, F.; Marusyk, A.; Sorkin, A. Grb2 regulates internalization of EGF receptors through clathrin-coated pits. Mol. Biol. Cell 2003, 14, 858–870. [Google Scholar] [CrossRef] [Green Version]
- Clague, M.J.; Liu, H.; Urbé, S. Governance of Endocytic Trafficking and Signaling by Reversible Ubiquitylation. Dev. Cell 2012, 23, 457–467. [Google Scholar] [CrossRef] [Green Version]
- McCann, A.P.; Scott, C.J.; Van Schaeybroeck, S.; Burrows, J.F. Deubiquitylating enzymes in receptor endocytosis and trafficking. Biochem. J. 2016, 473, 4507–4525. [Google Scholar] [CrossRef]
- McCullough, J.; Clague, M.J.; Urbé, S. AMSH is an endosome-associated ubiquitin isopeptidase. J. Cell Biol. 2004, 166, 487–492. [Google Scholar] [CrossRef] [Green Version]
- Bowers, K.; Piper, S.C.; Edeling, M.A.; Gray, S.R.; Owen, D.J.; Lehner, P.J.; Luzio, J.P. Degradation of Endocytosed Epidermal Growth Factor and Virally Ubiquitinated Major Histocompatibility Complex Class I Is Independent of Mammalian ESCRTII. J. Biol. Chem. 2005, 281, 5094–5105. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Zanata, S.M.; Kim, J.; Peterson, M.; Di Vizio, L.; Chirieac, L.R.; Pyne, S.; Agostini, M.; Freeman, M.R.; Loda, M. The ubiquitin-specific protease USP2a prevents endocytosis-mediated EGFR degradation. Oncogene 2012, 32, 1660–1669. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Han, B.; Lu, H.; Zhao, Y.; Chen, X.; Meng, Q.; Cao, M.; Cai, L.; Hu, J. USP22 promotes resistance to EGFR-TKIs by preventing ubiquitination-mediated EGFR degradation in EGFR-mutant lung adenocarcinoma. Cancer Lett. 2018, 433, 186–198. [Google Scholar] [CrossRef]
- Pareja, F.; Ferraro, D.A.; Rubin, C.; Cohen-Dvashi, H.; Zhang, F.; Aulmann, S.; Ben-Chetrit, N.; Pines, G.; Navon, R.; Crosetto, N.; et al. Deubiquitination of EGFR by Cezanne-1 contributes to cancer progression. Oncogene 2011, 31, 4599–4608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Savio, M.G.; Wollscheid, N.; Cavallaro, E.; Algisi, V.; Di Fiore, P.P.; Sigismund, S.; Maspero, E.; Polo, S. USP9X Controls EGFR Fate by Deubiquitinating the Endocytic Adaptor Eps15. Curr. Biol. 2016, 26, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Row, P.E.; Prior, I.A.; McCullough, J.; Clague, M.J.; Urbé, S. The Ubiquitin Isopeptidase UBPY Regulates Endosomal Ubiquitin Dynamics and Is Essential for Receptor Down-regulation. J. Biol. Chem. 2006, 281, 12618–12624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alwan, H.A.J.; Van Leeuwen, J.E.M. UBPY-mediated Epidermal Growth Factor Receptor (EGFR) De-ubiquitination Promotes EGFR Degradation. J. Biol. Chem. 2007, 282, 1658–1669. [Google Scholar] [CrossRef] [Green Version]
- Pascolutti, R.; Algisi, V.; Conte, A.; Raimondi, A.; Pasham, M.; Upadhyayula, S.; Gaudin, R.; Maritzen, T.; Barbieri, E.; Caldieri, G.; et al. Molecularly Distinct Clathrin-Coated Pits Differentially Impact EGFR Fate and Signaling. Cell Rep. 2019, 27, 3049–3061.e6. [Google Scholar] [CrossRef] [Green Version]
- He, F.; Wollscheid, H.-P.; Nowicka, U.; Biancospino, M.; Valentini, E.; Ehlinger, A.; Acconcia, F.; Magistrati, E.; Polo, S.; Walters, K.J. Myosin VI Contains a Compact Structural Motif that Binds to Ubiquitin Chains. Cell Rep. 2016, 14, 2683–2694. [Google Scholar] [CrossRef] [Green Version]
- Wiley, H.S.; Cunningham, D.D. The endocytotic rate constant. A cellular parameter for quantitating receptor-mediated endocytosis. J. Biol. Chem. 1982, 257, 4222–4229. [Google Scholar]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [Green Version]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [Green Version]
- Zhong, B.; Liu, X.; Wang, X.; Chang, S.H.; Liu, X.; Wang, A.; Reynolds, J.M.; Dong, C. Negative regulation of IL-17-mediated signaling and inflammation by the ubiquitin-specific protease USP25. Nat. Immunol. 2012, 13, 1110–1117. [Google Scholar] [CrossRef]
- Zhong, B.; Liu, X.; Wang, X.; Li, H.; Darnay, B.G.; Lin, X.; Sun, S.-C.; Dong, C. Ubiquitin-Specific Protease 25 Regulates TLR4-Dependent Innate Immune Responses Through Deubiquitination of the Adaptor Protein TRAF3. Sci. Signal. 2013, 6, ra35. [Google Scholar] [CrossRef] [Green Version]
- Winkler, M.E.; O’Connor, L.; Winget, M.; Fendly, B. Epidermal growth factor and transforming growth factor .alpha. bind differently to the epidermal growth factor receptor. Biochemistry 1989, 28, 6373–6378. [Google Scholar] [CrossRef] [PubMed]
- Sigismund, S.; Argenzio, E.; Tosoni, D.; Cavallaro, E.; Polo, S.; Di Fiore, P.P. Clathrin-Mediated Internalization Is Essential for Sustained EGFR Signaling but Dispensable for Degradation. Dev. Cell 2008, 15, 209–219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Argenzio, E.; Bange, T.; Oldrini, B.; Bianchi, F.; Peesari, R.; Mari, S.; Di Fiore, P.P.; Mann, M.; Polo, S. Proteomic snapshot of the EGF-induced ubiquitin network. Mol. Syst. Biol. 2011, 7, 462. [Google Scholar] [CrossRef]
- Polo, S.; Sigismund, S.; Faretta, M.; Guidi, M.; Capua, M.R.; Bossi, G.; Chen, H.; De Camilli, P.; Di Fiore, P.P. A single motif responsible for ubiquitin recognition and monoubiquitination in endocytic proteins. Nat. Cell Biol. 2002, 416, 451–455. [Google Scholar] [CrossRef]
- Zhong, H.; Wang, D.; Fang, L.; Zhang, H.; Luo, R.; Shang, M.; Ouyang, C.; Ouyang, H.; Chen, H.; Xiao, S. Ubiquitin-Specific Proteases 25 Negatively Regulates Virus-Induced Type I Interferon Signaling. PLoS ONE 2013, 8, e80976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, D.; Liu, J.; Fu, T.; Shan, B.; Qian, L.; Pan, L.; Yuan, J. USP25 regulates Wnt signaling by controlling the stability of tankyrases. Genes Dev. 2017, 31, 1024–1035. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Tan, Q.; Yan, M.; Liu, L.; Lin, H.; Zhao, F.; Bao, G.; Kong, H.; Ge, C.; Zhang, F.; et al. miRNA-200c inhibits invasion and metastasis of human non-small cell lung cancer by directly targeting ubiquitin specific peptidase 25. Mol. Cancer 2014, 13, 166. [Google Scholar] [CrossRef] [Green Version]
- Harrigan, J.A.; Jacq, X.; Martin, N.M.; Jackson, S.P. Deubiquitylating enzymes and drug discovery: Emerging opportunities. Nat. Rev. Drug Discov. 2018, 17, 57–78. [Google Scholar] [CrossRef]


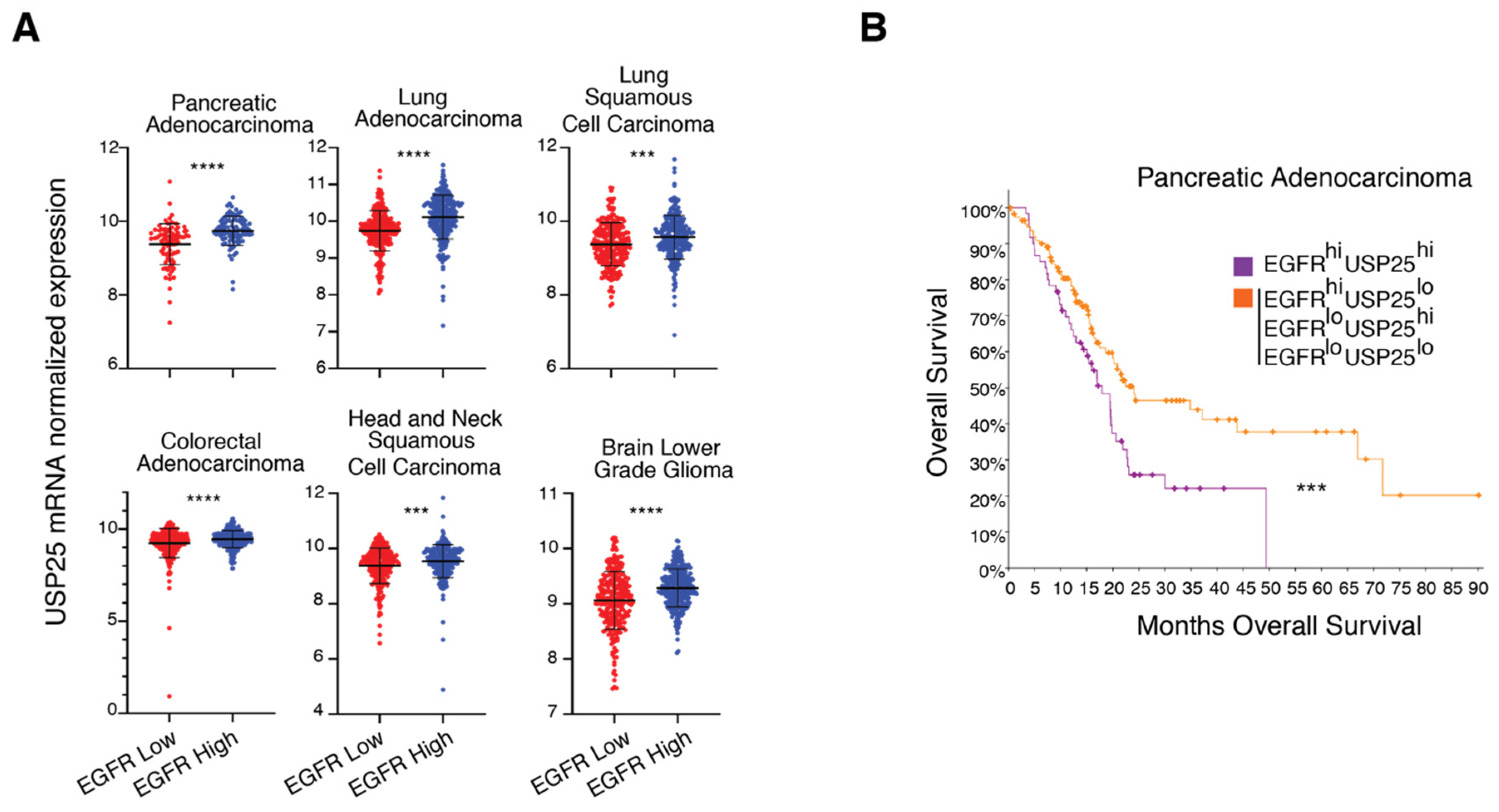
| USP25 Mean log2 in EGFR Low Group | USP25 Mean log2 in EGFR High Group | Standard Deviation in EGFR Low Group | Standard Deviation in EGFR High Group | p-Value | q-Value | |
|---|---|---|---|---|---|---|
| Pancreatic Adenocarcinoma | 9.38 | 9.74 | 0.55 | 0.40 | 1.248 × 10−6 | 1.875 × 10−5 |
| Lung squamous cell carcinoma | 9.37 | 9.57 | 0.58 | 0.59 | 2.678 × 10−4 | 1.622 × 10−3 |
| Lung Adenocarcinoma | 9.74 | 10.11 | 0.55 | 0.60 | 5.61 × 10−13 | 3.00 × 10−11 |
| Brain lower grade glioma | 9.06 | 9.29 | 0.52 | 0.35 | 1.91 × 10−8 | 1.44 × 10−7 |
| Colorectal Adenocarcinoma | 9.24 | 9.46 | 0.79 | 0.47 | 5.271 × 10−5 | 2.231 × 10−4 |
| Head and neck squamous cell carcinoma | 9.37 | 9.54 | 0.64 | 0.60 | 2.440 × 10−3 | 5.722 × 10−3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Niño, C.A.; Wollscheid, N.; Giangreco, G.; Maspero, E.; Polo, S. USP25 Regulates EGFR Fate by Modulating EGF-Induced Ubiquitylation Dynamics. Biomolecules 2020, 10, 1548. https://doi.org/10.3390/biom10111548
Niño CA, Wollscheid N, Giangreco G, Maspero E, Polo S. USP25 Regulates EGFR Fate by Modulating EGF-Induced Ubiquitylation Dynamics. Biomolecules. 2020; 10(11):1548. https://doi.org/10.3390/biom10111548
Chicago/Turabian StyleNiño, Carlos A., Nadine Wollscheid, Giovanni Giangreco, Elena Maspero, and Simona Polo. 2020. "USP25 Regulates EGFR Fate by Modulating EGF-Induced Ubiquitylation Dynamics" Biomolecules 10, no. 11: 1548. https://doi.org/10.3390/biom10111548





