Transcriptional Profiling Reveals Ribosome Biogenesis, Microtubule Dynamics and Expression of Specific lncRNAs to be Part of a Common Response to Cell-Penetrating Peptides
Abstract
1. Introduction
2. Material and Methods
2.1. Preparation of Cell Penetrating Peptides
2.2. Tissue Culture and Peptide Treatment
2.3. High Content Phenotypic Profiling
2.4. RNA-Seq Library Preparation, Sequencing, and Analysis
2.5. Quantitative RT-PCR
2.6. Cell Proliferation Assay
2.7. Statistical Analysis
3. Results
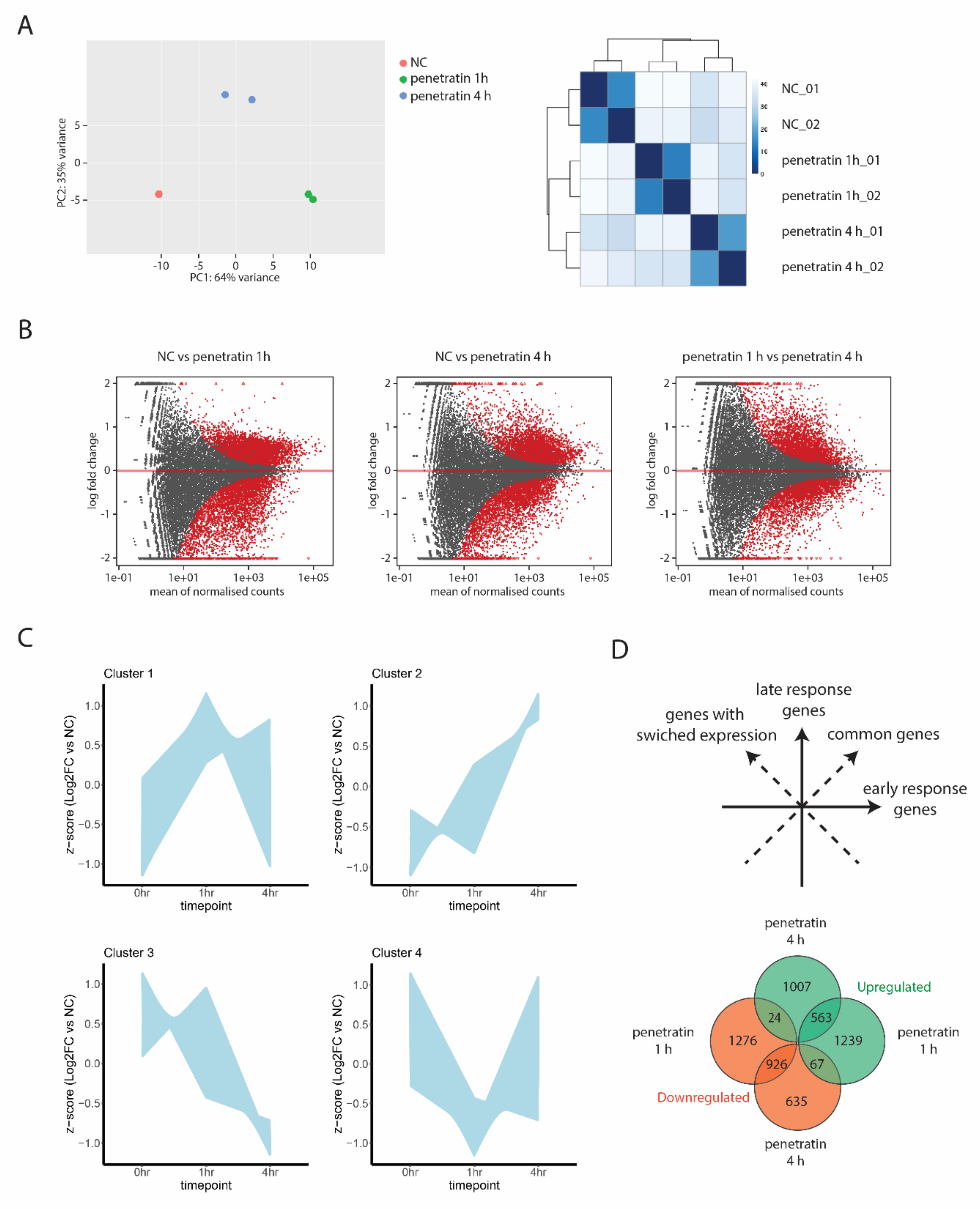
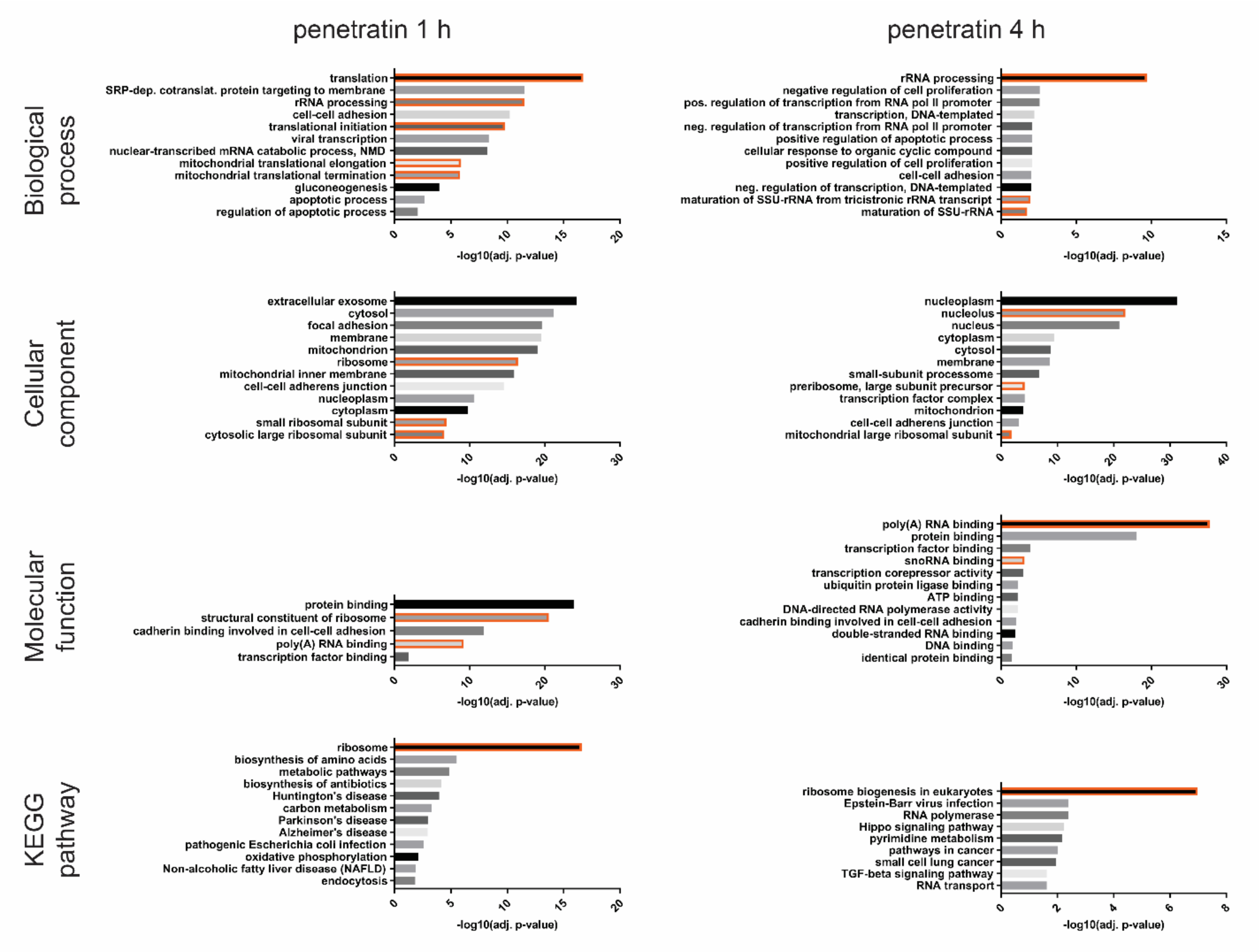
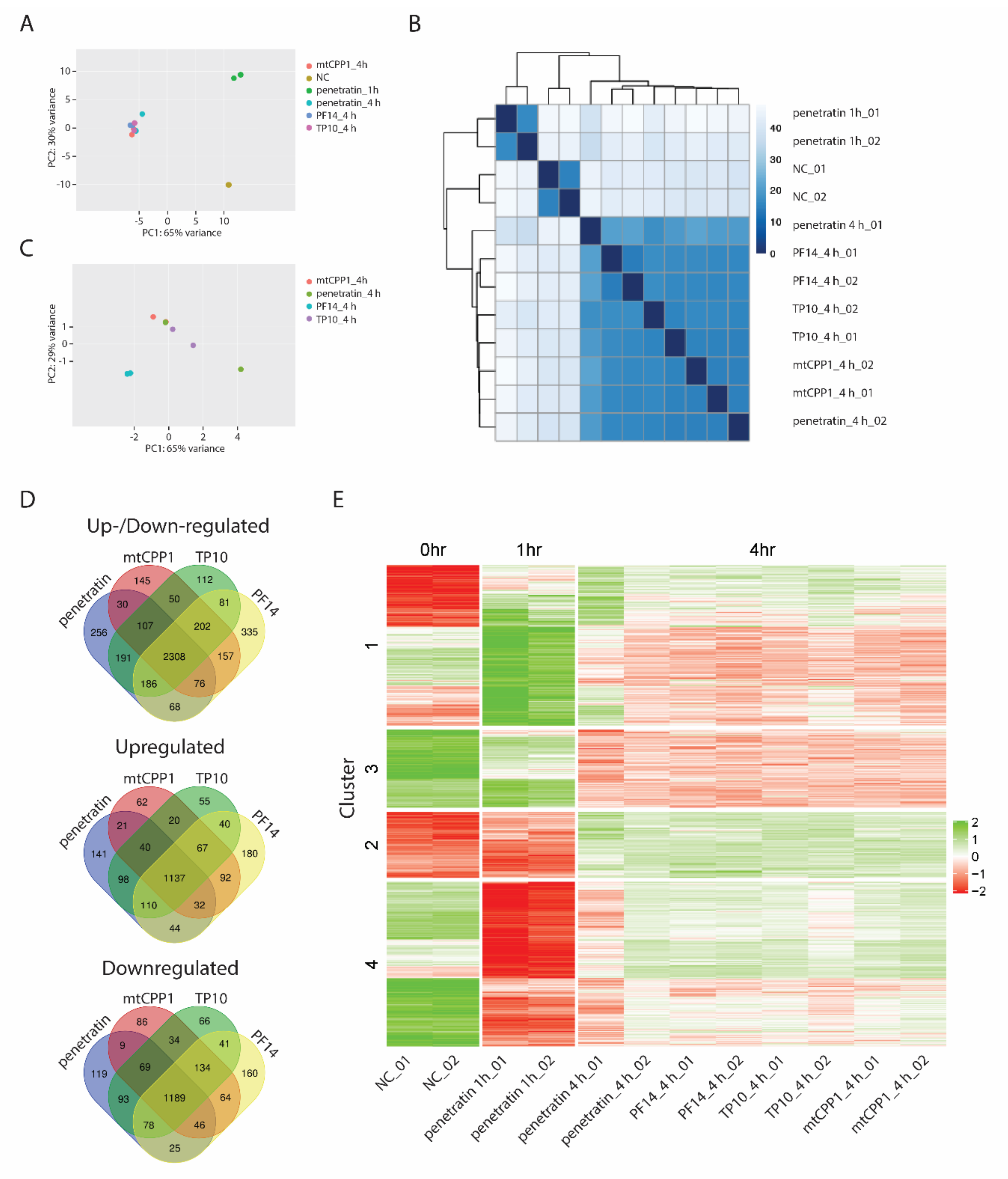
3.1. Transcriptional Profiling Reveals Time Dependent Cellular Response to CPPs
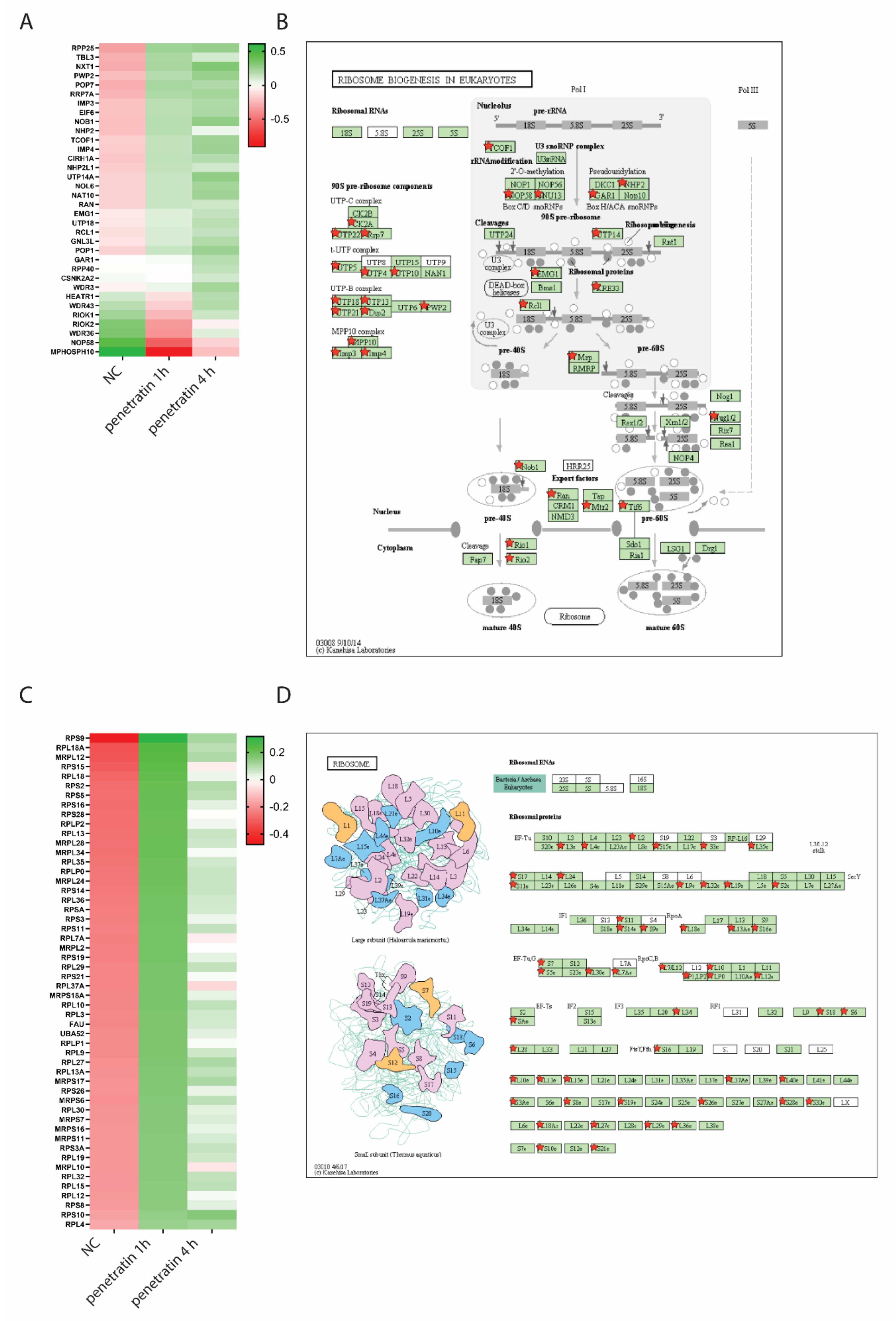
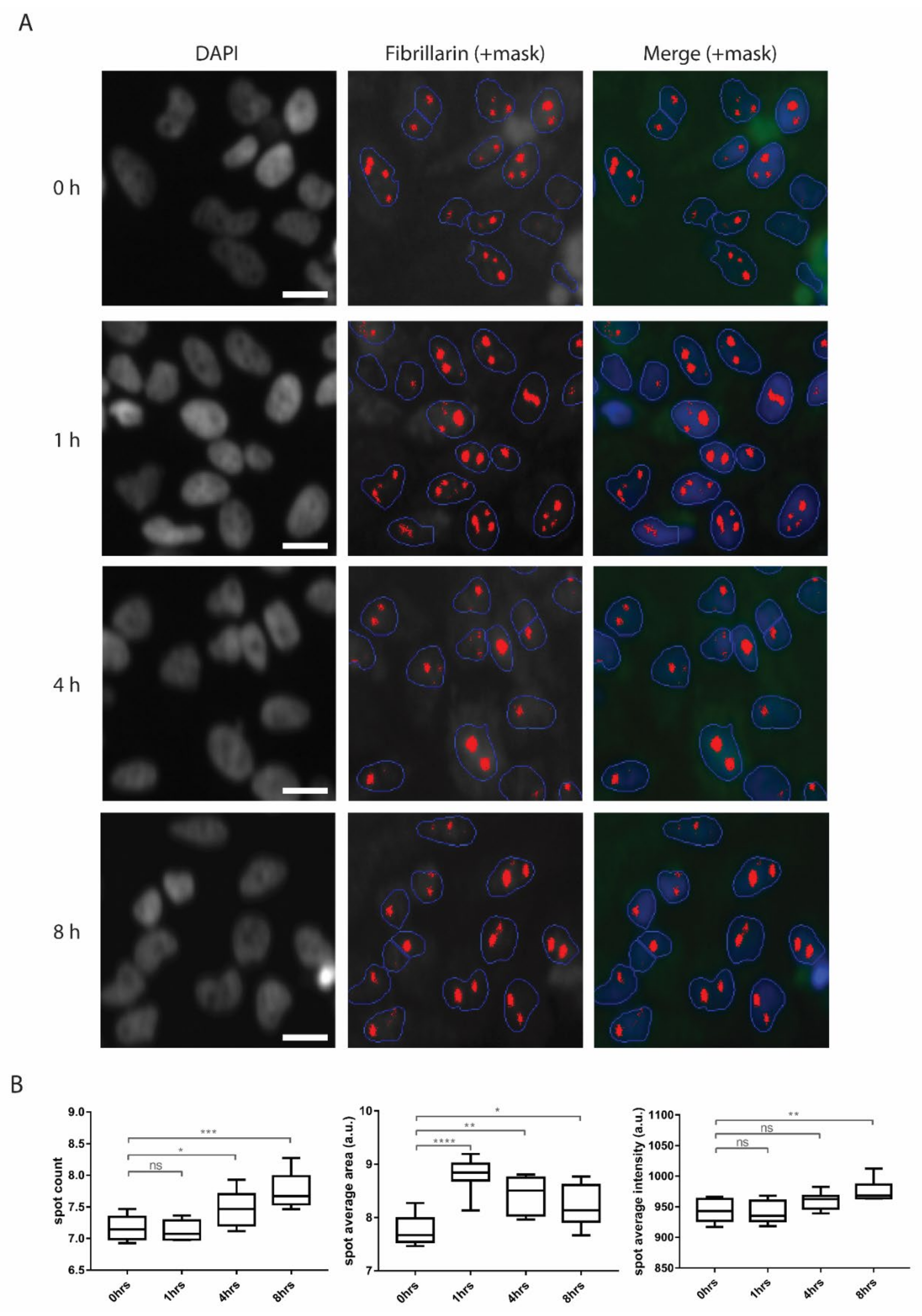
3.2. Increased Nucleolar Size Suggests Increased Levels of Ribosome Biogenesis Upon Penetratin Treatment
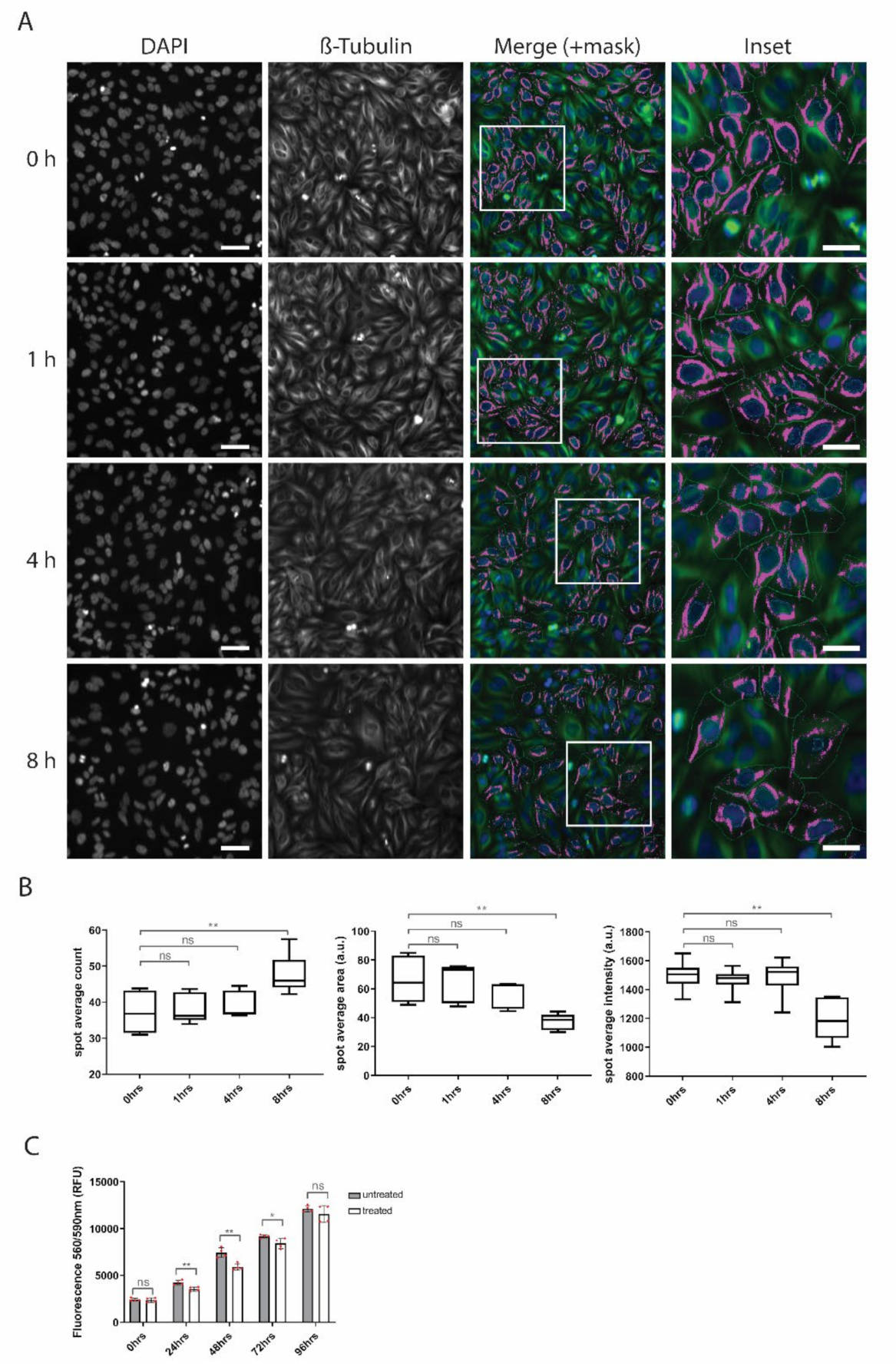
3.3. Misregulation of Microtubule- and Centrosome-Related Genes Upon Penetratin Treatment Is Accompanied by a Dispersion of Microtubules in Cells
3.4. Penetratin Treatment Decreases Proliferative Potential of Cells
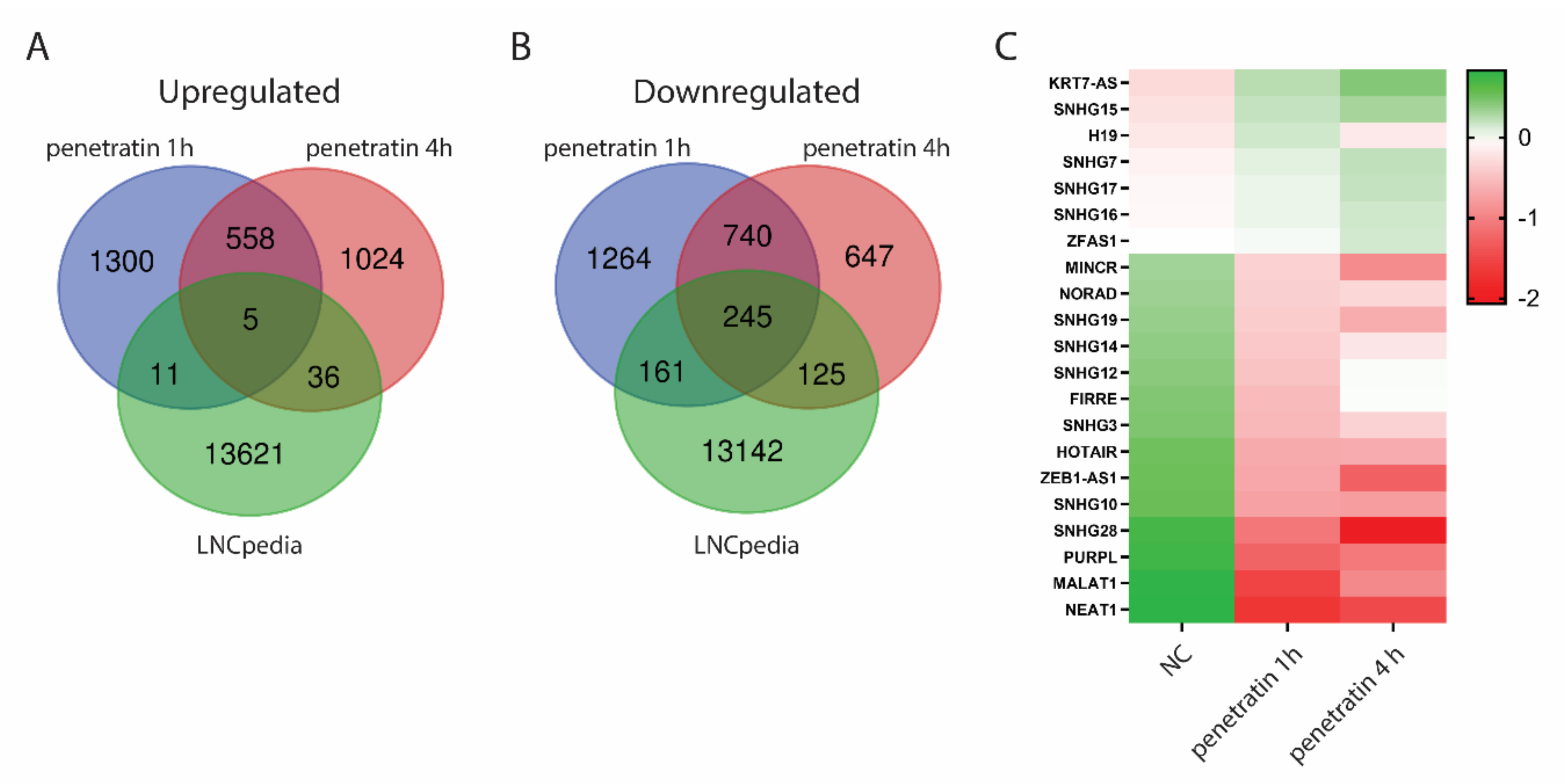
3.5. Treatment with Penetratin Affects Expression of Long Non-Coding RNAs
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bohmova, E.; Machova, D.; Pechar, M.; Pola, R.; Venclikova, K.; Janouskova, O.; Etrych, T. Cell-penetrating peptides: A useful tool for the delivery of various cargoes into cells. Physiol. Res. 2018, 67, S267–S279. [Google Scholar] [CrossRef] [PubMed]
- Green, M.; Loewenstein, P.M. Autonomous functional domains of chemically synthesized human immunodeficiency virus tat trans-activator protein. Cell 1988, 55, 1179–1188. [Google Scholar] [CrossRef] [PubMed]
- Kurrikoff, K.; Aphkhazava, D.; Langel, U. The future of peptides in cancer treatment. Curr. Opin. Pharmacol. 2019, 47, 27–32. [Google Scholar] [CrossRef] [PubMed]
- Kurrikoff, K.; Langel, U. Recent CPP-based applications in medicine. Expert Opin. Drug Deliv. 2019, 16, 1183–1191. [Google Scholar] [CrossRef]
- Ruseska, I.; Zimmer, A. Internalization mechanisms of cell-penetrating peptides. Beilstein J. Nanotechnol. 2020, 11, 101–123. [Google Scholar] [CrossRef]
- de la Fuente, J.M.; Berry, C.C. Tat peptide as an efficient molecule to translocate gold nanoparticles into the cell nucleus. Bioconjug. Chem. 2005, 16, 1176–1180. [Google Scholar] [CrossRef]
- Ding, Y.; Sun, D.; Wang, G.L.; Yang, H.G.; Xu, H.F.; Chen, J.H.; Xie, Y.; Wang, Z.Q. An efficient PEGylated liposomal nanocarrier containing cell-penetrating peptide and pH-sensitive hydrazone bond for enhancing tumor-targeted drug delivery. Int. J. Nanomedicine 2015, 10, 6199–6214. [Google Scholar] [CrossRef]
- Golan, M.; Feinshtein, V.; David, A. Conjugates of HA2 with octaarginine-grafted HPMA copolymer offer effective siRNA delivery and gene silencing in cancer cells. Eur. J. Pharm. Biopharm. 2016, 109, 103–112. [Google Scholar] [CrossRef]
- Goswami, D.; Vitorino, H.A.; Machini, M.T.; Esposito, B.P. Self-assembled penetratin-deferasirox micelles as potential carriers for hydrophobic drug delivery. Biopolymers 2015, 104, 712–719. [Google Scholar] [CrossRef]
- Shabanpoor, F.; McClorey, G.; Saleh, A.F.; Jarver, P.; Wood, M.J.; Gait, M.J. Bi-specific splice-switching PMO oligonucleotides conjugated via a single peptide active in a mouse model of Duchenne muscular dystrophy. Nucleic Acids Res. 2015, 43, 29–39. [Google Scholar] [CrossRef]
- Peng, J.; Rao, Y.; Yang, X.; Jia, J.; Wu, Y.; Lu, J.; Tao, Y.; Tu, W. Targeting neuronal nitric oxide synthase by a cell penetrating peptide Tat-LK15/siRNA bioconjugate. Neurosci. Lett. 2017, 650, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, P.P.; Arami, H.; Banga, I.; Gupta, J.; Gandhi, S. Cell penetrating peptides in preclinical and clinical cancer diagnosis and therapy. Oncotarget 2018, 9, 37252–37267. [Google Scholar] [CrossRef] [PubMed]
- Kalmouni, M.; Al-Hosani, S.; Magzoub, M. Cancer targeting peptides. Cell. Mol. Life Sci. 2019, 76, 2171–2183. [Google Scholar] [CrossRef]
- Jiang, B. Aerobic glycolysis and high level of lactate in cancer metabolism and microenvironment. Genes Dis. 2017, 4, 25–27. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, V.P.; Alves, D.S.; Scott, H.L.; Davis, F.L.; Barrera, F.N. A Novel Soluble Peptide with pH-Responsive Membrane Insertion. Biochemistry 2015, 54, 6567–6575. [Google Scholar] [CrossRef] [PubMed]
- Smith, K.T.; Workman, J.L. Chromatin proteins: Key responders to stress. PLoS Biol. 2012, 10, e1001371. [Google Scholar] [CrossRef]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef]
- Cerrato, C.P.; Pirisinu, M.; Vlachos, E.N.; Langel, U. Novel cell-penetrating peptide targeting mitochondria. FASEB J. 2015, 29, 4589–4599. [Google Scholar] [CrossRef]
- Lindgren, M.; Gallet, X.; Soomets, U.; Hallbrink, M.; Brakenhielm, E.; Pooga, M.; Brasseur, R.; Langel, U. Translocation properties of novel cell penetrating transportan and penetratin analogues. Bioconjug. Chem. 2000, 11, 619–626. [Google Scholar] [CrossRef]
- Ezzat, K.; Andaloussi, S.E.; Zaghloul, E.M.; Lehto, T.; Lindberg, S.; Moreno, P.M.; Viola, J.R.; Magdy, T.; Abdo, R.; Guterstam, P.; et al. PepFect 14, a novel cell-penetrating peptide for oligonucleotide delivery in solution and as solid formulation. Nucleic Acids Res. 2011, 39, 5284–5298. [Google Scholar] [CrossRef]
- Lehto, T.; Vasconcelos, L.; Margus, H.; Figueroa, R.; Pooga, M.; Hallbrink, M.; Langel, U. Saturated Fatty Acid Analogues of Cell-Penetrating Peptide PepFect14: Role of Fatty Acid Modification in Complexation and Delivery of Splice-Correcting Oligonucleotides. Bioconjug. Chem. 2017, 28, 782–792. [Google Scholar] [CrossRef] [PubMed]
- Montanaro, L.; Trere, D.; Derenzini, M. Nucleolus, ribosomes, and cancer. Am. J. Pathol. 2008, 173, 301–310. [Google Scholar] [CrossRef] [PubMed]
- Muroyama, A.; Lechler, T. Microtubule organization, dynamics and functions in differentiated cells. Development 2017, 144, 3012–3021. [Google Scholar] [CrossRef] [PubMed]
- Djebali, S.; Davis, C.A.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F.; et al. Landscape of transcription in human cells. Nature 2012, 489, 101–108. [Google Scholar] [CrossRef]
- Batista, P.J.; Chang, H.Y. Long noncoding RNAs: Cellular address codes in development and disease. Cell 2013, 152, 1298–1307. [Google Scholar] [CrossRef]
- Raveh, E.; Matouk, I.J.; Gilon, M.; Hochberg, A. The H19 Long non-coding RNA in cancer initiation, progression and metastasis—A proposed unifying theory. Mol. Cancer 2015, 14, 184. [Google Scholar] [CrossRef]
- Chen, S.; Gu, T.; Lu, Z.; Qiu, L.; Xiao, G.; Zhu, X.; Li, F.; Yu, H.; Li, G.; Liu, H. Roles of MYC-targeting long non-coding RNA MINCR in cell cycle regulation and apoptosis in non-small cell lung Cancer. Respir Res. 2019, 20, 202. [Google Scholar] [CrossRef]
- Liu, C.; Lin, J. Long noncoding RNA ZEB1-AS1 acts as an oncogene in osteosarcoma by epigenetically activating ZEB1. Am. J. Transl. Res. 2016, 8, 4095–4105. [Google Scholar]
- Li, X.L.; Subramanian, M.; Jones, M.F.; Chaudhary, R.; Singh, D.K.; Zong, X.; Gryder, B.; Sindri, S.; Mo, M.; Schetter, A.; et al. Long Noncoding RNA PURPL Suppresses Basal p53 Levels and Promotes Tumorigenicity in Colorectal Cancer. Cell Rep. 2017, 20, 2408–2423. [Google Scholar] [CrossRef]
- Deng, L.; Jiang, L.; Tseng, K.F.; Liu, Y.; Zhang, X.; Dong, R.; Lu, Z.; Wang, X. Aberrant NEAT1_1 expression may be a predictive marker of poor prognosis in diffuse large B cell lymphoma. Cancer Biomark. 2018, 23, 157–164. [Google Scholar] [CrossRef]
- Yoshimoto, R.; Mayeda, A.; Yoshida, M.; Nakagawa, S. MALAT1 long non-coding RNA in cancer. Biochim. Biophys. Acta 2016, 1859, 192–199. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.A.; Shah, N.; Wang, K.C.; Kim, J.; Horlings, H.M.; Wong, D.J.; Tsai, M.C.; Hung, T.; Argani, P.; Rinn, J.L.; et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010, 464, 1071–1076. [Google Scholar] [CrossRef] [PubMed]
- Hacisuleyman, E.; Goff, L.A.; Trapnell, C.; Williams, A.; Henao-Mejia, J.; Sun, L.; McClanahan, P.; Hendrickson, D.G.; Sauvageau, M.; Kelley, D.R.; et al. Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat. Struct Mol. Biol. 2014, 21, 198–206. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Kopp, F.; Chang, T.C.; Sataluri, A.; Chen, B.; Sivakumar, S.; Yu, H.; Xie, Y.; Mendell, J.T. Noncoding RNA NORAD Regulates Genomic Stability by Sequestering PUMILIO Proteins. Cell 2016, 164, 69–80. [Google Scholar] [CrossRef]
- Grummt, I. The nucleolus-guardian of cellular homeostasis and genome integrity. Chromosoma 2013, 122, 487–497. [Google Scholar] [CrossRef]
- Weeks, S.E.; Metge, B.J.; Samant, R.S. The nucleolus: A central response hub for the stressors that drive cancer progression. Cell. Mol. Life Sci. 2019, 76, 4511–4524. [Google Scholar] [CrossRef]
- Harrison, D.E.; Strong, R.; Sharp, Z.D.; Nelson, J.F.; Astle, C.M.; Flurkey, K.; Nadon, N.L.; Wilkinson, J.E.; Frenkel, K.; Carter, C.S.; et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature 2009, 460, 392–395. [Google Scholar] [CrossRef]
- Schosserer, M.; Minois, N.; Angerer, T.B.; Amring, M.; Dellago, H.; Harreither, E.; Calle-Perez, A.; Pircher, A.; Gerstl, M.P.; Pfeifenberger, S.; et al. Methylation of ribosomal RNA by NSUN5 is a conserved mechanism modulating organismal lifespan. Nat. Commun. 2015, 6, 6158. [Google Scholar] [CrossRef]
- Watanabe-Susaki, K.; Takada, H.; Enomoto, K.; Miwata, K.; Ishimine, H.; Intoh, A.; Ohtaka, M.; Nakanishi, M.; Sugino, H.; Asashima, M.; et al. Biosynthesis of ribosomal RNA in nucleoli regulates pluripotency and differentiation ability of pluripotent stem cells. Stem Cells 2014, 32, 3099–3111. [Google Scholar] [CrossRef]
- Qu, J.; Bishop, J.M. Nucleostemin maintains self-renewal of embryonic stem cells and promotes reprogramming of somatic cells to pluripotency. J. Cell Biol. 2012, 197, 731–745. [Google Scholar] [CrossRef]
- Le Bouteiller, M.; Souilhol, C.; Beck-Cormier, S.; Stedman, A.; Burlen-Defranoux, O.; Vandormael-Pournin, S.; Bernex, F.; Cumano, A.; Cohen-Tannoudji, M. Notchless-dependent ribosome synthesis is required for the maintenance of adult hematopoietic stem cells. J. Exp. Med. 2013, 210, 2351–2369. [Google Scholar] [CrossRef] [PubMed]
- Hetman, M.; Pietrzak, M. Emerging roles of the neuronal nucleolus. Trends Neurosci. 2012, 35, 305–314. [Google Scholar] [CrossRef] [PubMed]
- Gomes, C.; Smith, S.C.; Youssef, M.N.; Zheng, J.J.; Hagg, T.; Hetman, M. RNA polymerase 1-driven transcription as a mediator of BDNF-induced neurite outgrowth. J. Biol. Chem. 2011, 286, 4357–4363. [Google Scholar] [CrossRef] [PubMed]
- Yen, T.J.; Machlin, P.S.; Cleveland, D.W. Autoregulated instability of beta-tubulin mRNAs by recognition of the nascent amino terminus of beta-tubulin. Nature 1988, 334, 580–585. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Gasic, I.; Chandrasekaran, V.; Peters, N.; Shao, S.; Mitchison, T.J.; Hegde, R.S. TTC5 mediates autoregulation of tubulin via mRNA degradation. Science 2020, 367, 100–104. [Google Scholar] [CrossRef]
- Gasic, I.; Mitchison, T.J. Autoregulation and repair in microtubule homeostasis. Curr. Opin. Cell Biol. 2019, 56, 80–87. [Google Scholar] [CrossRef]
- Gasic, I.; Boswell, S.A.; Mitchison, T.J. Tubulin mRNA stability is sensitive to change in microtubule dynamics caused by multiple physiological and toxic cues. PLoS Biol. 2019, 17, e3000225. [Google Scholar] [CrossRef]
- Zhao, M.; Wang, S.; Li, Q.; Ji, Q.; Guo, P.; Liu, X. MALAT1: A long non-coding RNA highly associated with human cancers. Oncol. Lett. 2018, 16, 19–26. [Google Scholar] [CrossRef]
- Liu, P.; Yang, H.; Zhang, J.; Peng, X.; Lu, Z.; Tong, W.; Chen, J. The lncRNA MALAT1 acts as a competing endogenous RNA to regulate KRAS expression by sponging miR-217 in pancreatic ductal adenocarcinoma. Sci. Rep. 2017, 7, 5186. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Venit, T.; Dowaidar, M.; Gestin, M.; Mahmood, S.R.; Langel, Ü.; Percipalle, P. Transcriptional Profiling Reveals Ribosome Biogenesis, Microtubule Dynamics and Expression of Specific lncRNAs to be Part of a Common Response to Cell-Penetrating Peptides. Biomolecules 2020, 10, 1567. https://doi.org/10.3390/biom10111567
Venit T, Dowaidar M, Gestin M, Mahmood SR, Langel Ü, Percipalle P. Transcriptional Profiling Reveals Ribosome Biogenesis, Microtubule Dynamics and Expression of Specific lncRNAs to be Part of a Common Response to Cell-Penetrating Peptides. Biomolecules. 2020; 10(11):1567. https://doi.org/10.3390/biom10111567
Chicago/Turabian StyleVenit, Tomas, Moataz Dowaidar, Maxime Gestin, Syed Raza Mahmood, Ülo Langel, and Piergiorgio Percipalle. 2020. "Transcriptional Profiling Reveals Ribosome Biogenesis, Microtubule Dynamics and Expression of Specific lncRNAs to be Part of a Common Response to Cell-Penetrating Peptides" Biomolecules 10, no. 11: 1567. https://doi.org/10.3390/biom10111567
APA StyleVenit, T., Dowaidar, M., Gestin, M., Mahmood, S. R., Langel, Ü., & Percipalle, P. (2020). Transcriptional Profiling Reveals Ribosome Biogenesis, Microtubule Dynamics and Expression of Specific lncRNAs to be Part of a Common Response to Cell-Penetrating Peptides. Biomolecules, 10(11), 1567. https://doi.org/10.3390/biom10111567






