Validation of Omega Subunit of RNA Polymerase as a Functional Entity
Abstract
:1. Introduction
2. The Transcription Machinery of the Cells
3. RNA Polymerase Machinery as a Whole
4. The Role of ω Subunit
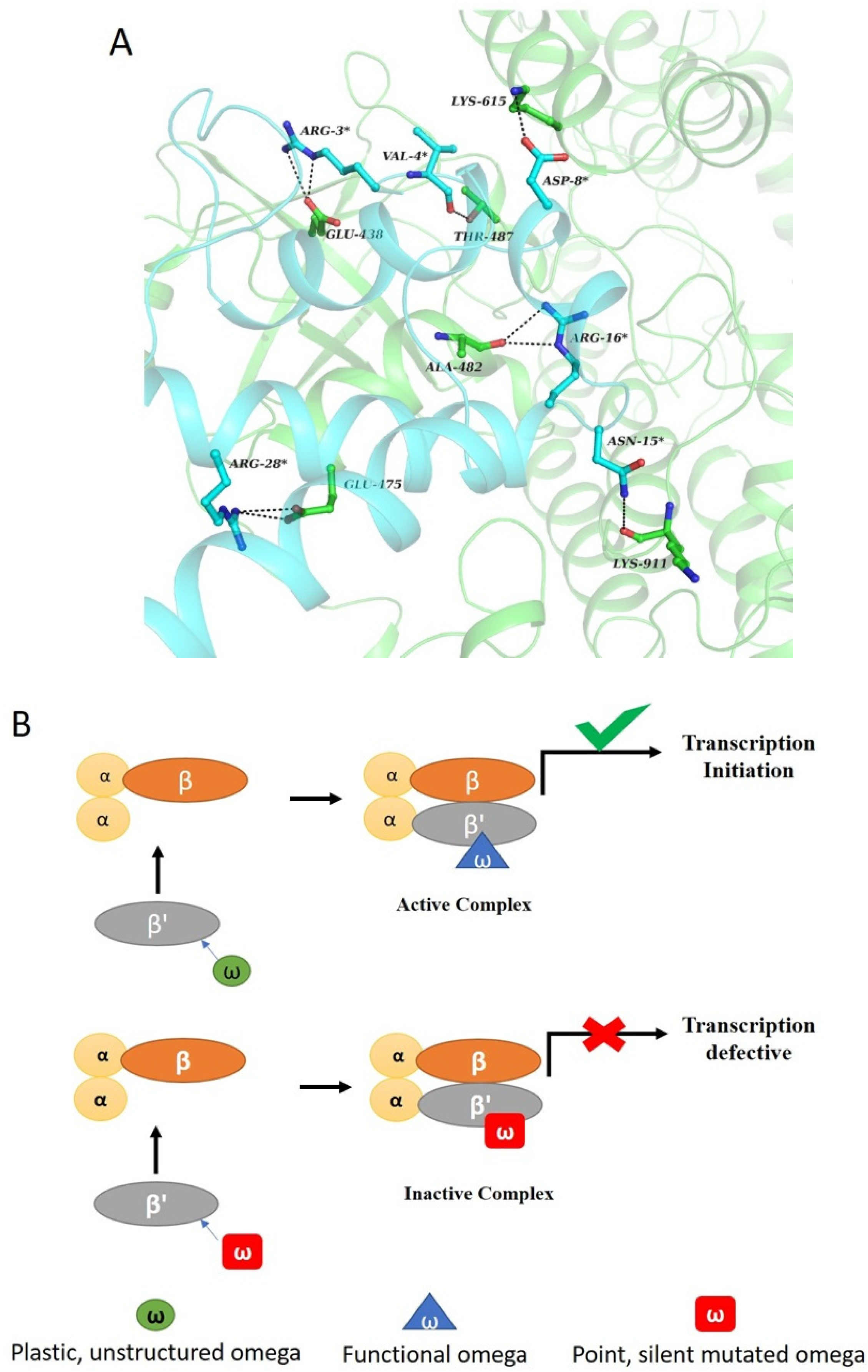
5. Functions of ω Subunit in RNAP
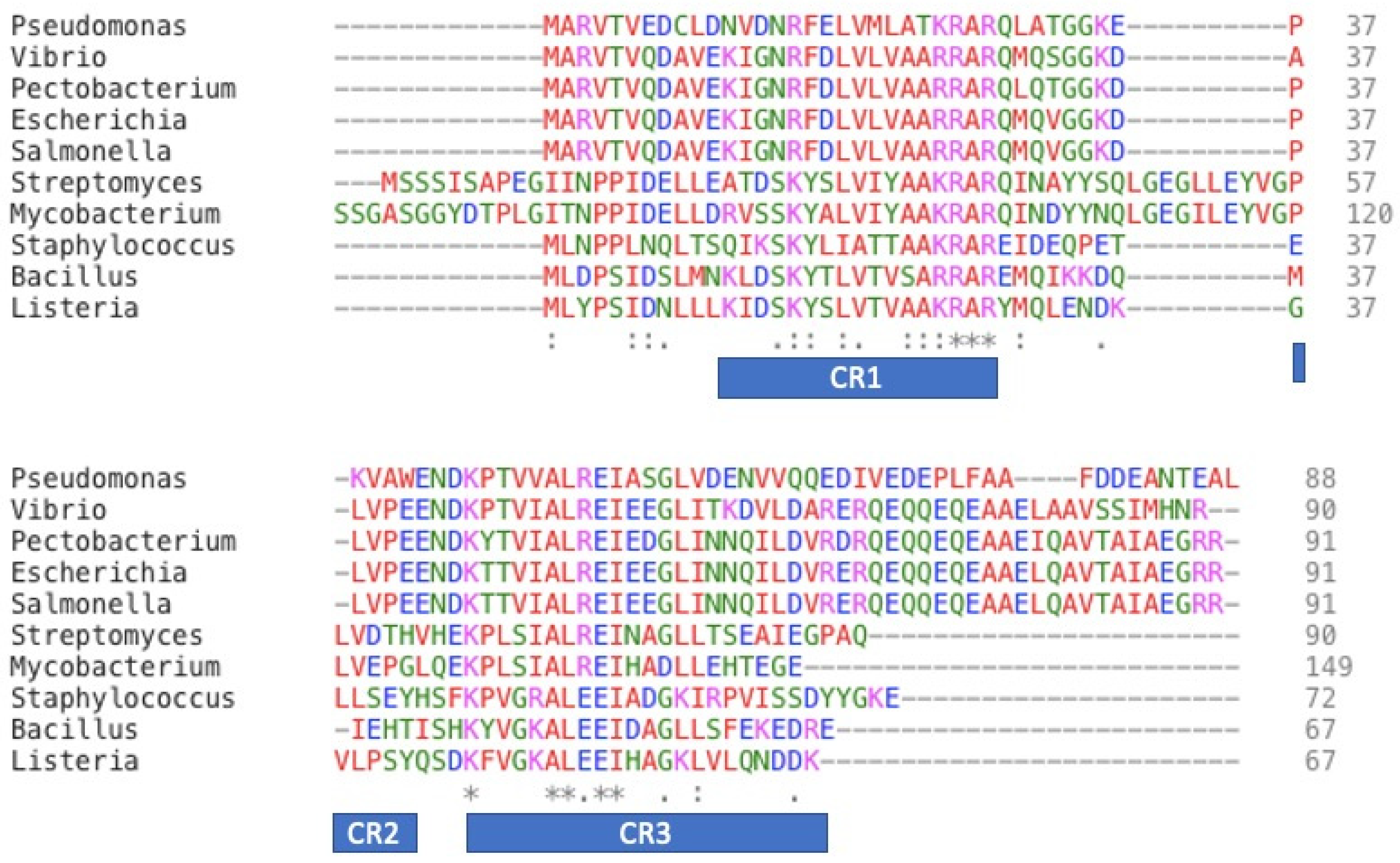
6. Structure–Function Relationship in ω-Subunit
7. Role of ω-Subunit in Stringent Response
Funding
Acknowledgments
Conflicts of Interest
References
- Russo, F.D.; Silhavy, T.J. Alpha: The Cinderella subunit of RNA polymerase. J. Biol. Chem. 1992, 267, 14515–14518. [Google Scholar] [PubMed]
- Burgess, R.R. Separation and characterization of the subunits of ribonucleic acid polymerase. J. Biol. Chem. 1969, 244, 6168–6176. [Google Scholar] [PubMed]
- Gross, C.A.; Lonetto, M.; Losick, R. Bacterial sigma factors. In Transcriptional Regulation; McKnight, S.L., Yamamoto, K.R., Eds.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1992; pp. 129–178. [Google Scholar]
- Weiss, S.B.; Gladstone, L. A mammalian system for the incorporation of cytidine triphosphate into ribonucleic acid. J. Am. Chem. Soc. 1959, 81, 4118–4119. [Google Scholar] [CrossRef]
- Ochoa, S.; Burma, D.P.; Kroger, H.; Weill, J.D. Deoxyribonucleic acid-dependent incorporation of nucleotides from nucleoside triphosphates into ribonucleic acid. Proc. Natl. Acad. Sci. USA 1961, 47, 670–679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Furth, J.J.; Hurwitz, J.; Goldmann, M. The directing role of DNA in RNA synthesis. Biochem. Biophys. Res. Commun. 1961, 4, 362–367. [Google Scholar] [CrossRef]
- Hurwitz, J.; Bresler, A.; Diringer, R. The enzymic incorporation of ribonucleotides into polyribonucleotides and the effect of DNA. Biochem. Biophys. Res. Commun. 1960, 3, 15–19. [Google Scholar] [CrossRef]
- Stevens, A. Incorporation of the adenine ribonucleotide into RNA by cell fractions from E. coli B. Biochem. Biophys. Res. Commun. 1960, 3, 92–96. [Google Scholar] [CrossRef]
- Stevens, A. Net Formation of Polyribonucleotides with Base Compositions Analogous to Deoxyribonucleic Acid. J. Biol. Chem. 1961, 236, PC43–PC45. [Google Scholar]
- Huang, R.C.; Maheshwari, N.; Bonner, J. Enzymatic synthesis of RNA. Biochem. Biophys. Res. Commun. 1960, 3, 689–694. [Google Scholar] [CrossRef]
- Chamberlin, M.; Berg, P. Deoxyribo ucleic acid-directed synthesis of ribonucleic acid by an enzyme from Escherichia coli. Proc. Natl. Acad. Sci. USA 1962, 48, 81–94. [Google Scholar] [CrossRef] [Green Version]
- Burgess, R.R.; Jendrisak, J.J. A procedure for the rapid, large-scall purification of Escherichia coli DNA-dependent RNA polymerase involving Polymin P precipitation and DNA-cellulose chromatography. Biochemistry 1975, 14, 4634–4638. [Google Scholar] [CrossRef]
- Burgess, R.R. A new method for the large scale purification of Escherichia coli deoxyribonucleic acid-dependent ribonucleic acid polymerase. J. Biol. Chem. 1969, 244, 6160–6167. [Google Scholar] [PubMed]
- Nüsslein, C.; Heyden, B. Chromatography of RNA polymerase from Escherichia coli on single stranded DNA-agarose columns. Biochem. Biophys. Res. Commun. 1972, 47, 282–289. [Google Scholar] [CrossRef]
- Mathew, R.; Chatterji, D. The evolving story of the omega subunit of bacterial RNA polymerase. Trends Microbiol. 2006, 14, 450–455. [Google Scholar] [CrossRef] [PubMed]
- Kurkela, J.; Fredman, J.; Salminen, T.A.; Tyystjärvi, T. Revealing secrets of the enigmatic omega subunit of bacterial RNA polymerase. Mol. Microbiol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Heil, A.; Zillig, W. Reconstitution of bacterial DNA-dependent RNA-polymerase from isolated subunits as a tool for the elucidation of the role of the subunits in transcription. FEBS Lett. 1970, 11, 165–168. [Google Scholar] [CrossRef] [Green Version]
- Burgess, R.R. RNA polymerase. Annu. Rev. Biochem. 1971, 40, 711–740. [Google Scholar] [CrossRef]
- Duffy, J.J.; Geiduschek, E.P. RNA polymerase from phage SP01-infected and uninfected Bacillus subtilis. J. Biol. Chem. 1975, 250, 4530–4541. [Google Scholar]
- Keller, A.N.; Yang, X.; Wiedermannová, J.; Delumeau, O.; Krásný, L.; Lewis, P.J. ε, a new subunit of RNA polymerase found in gram-positive bacteria. J. Bacteriol. 2014, 19, 3622–3632. [Google Scholar] [CrossRef] [Green Version]
- Gentry, D.R.; Burgess, R.R. Cross-linking of Escherichia coli RNA polymerase subunits: Identification of beta’ as the binding site of omega. Biochemistry 1993, 32, 11224–11227. [Google Scholar] [CrossRef]
- Dove, S.L.; Hochschild, A. Conversion of the omega subunit of Escherichia coli RNA polymerase into a transcriptional activator or an activation target. Genes Dev. 1998, 12, 745–754. [Google Scholar] [CrossRef] [Green Version]
- Mukherjee, K.; Chatterji, D. Studies on the omega subunit of Escherichia coli RNA polymerase—Its role in the recovery of denatured enzyme activity. Eur. J. Biochem. 1997, 247, 884–889. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, K.; Nagai, H.; Shimamoto, N.; Chatterji, D. GroEL is involved in activation of Escherichia coli RNA polymerase devoid of the omega subunit in vivo. Eur. J. Biochem. 1999, 266, 228–235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mukherjee, K.; Chatterji, D. Alteration in template recognition by Escherichia coli RNA polymerase lacking the ω subunit: A mechanistic analysis through gel retardation and foot-printing studies. J. Biosci. 1999, 24, 453–459. [Google Scholar] [CrossRef]
- Houry, W.A.; Frishman, D.; Eckerskorn, C.; Lottspeich, F.; Hartl, F.U. Identification of in vivo substrates of the chaperonin GroEL. Nature 1999, 402, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Ishihama, A.; Ikeuchi, T.; Matsumoto, A.; Yamamoto, S. A novel adenosine triphosphatase isolated from RNA polymerase preparations of Escherichia coli. II. Enzymatic properties and molecular structure. J. Biochem. 1976, 79, 927–937. [Google Scholar] [CrossRef] [PubMed]
- Ishihama, A.; Ikeuchi, T.; Yura, T. A novel adenosine triphosphatase isolated from RNA polymerase preparations of Escherichia coli. I. Copurification and separation. J. Biochem. 1976, 79, 917–925. [Google Scholar] [CrossRef]
- Ghosh, P.; Ishihama, A.; Chatterji, D. Escherichia coli RNA polymerase subunit omega and its N-terminal domain bind full-length beta’ to facilitate incorporation into the alpha2beta subassembly. Eur. J. Biochem. 2001, 268, 4621–4627. [Google Scholar] [CrossRef]
- Zhang, G.; Campbell, E.A.; Minakhin, L.; Richter, C.; Severinov, K.; Darst, S.A. Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 A resolution. Cell 1999, 98, 811–824. [Google Scholar] [CrossRef] [Green Version]
- Minakhin, L.; Bhagat, S.; Brunning, A.; Campbell, E.A.; Darst, S.A.; Ebright, R.H.; Severinov, K. Bacterial RNA polymerase subunit omega and eukaryotic RNA polymerase subunit RPB6 are sequence, structural, and functional homologs and promote RNA polymerase assembly. Proc. Natl. Acad. Sci. USA 2001, 98, 892–897. [Google Scholar] [CrossRef] [Green Version]
- Mathew, R.; Ramakanth, M.; Chatterji, D. Deletion of the gene rpoZ, encoding the omega subunit of RNA polymerase, in Mycobacterium smegmatis results in fragmentation of the beta’ subunit in the enzyme assembly. J. Bacteriol. 2005, 187, 6565–6570. [Google Scholar] [CrossRef] [Green Version]
- Sarkar, P.; Sardesai, A.A.; Murakami, K.S.; Chatterji, D. Inactivation of the bacterial RNA polymerase due to acquisition of secondary structure by the ω subunit. J. Biol. Chem. 2013, 288, 25076–25087. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patel, U.R.; Gautam, S.; Chatterji, D. Unraveling the Role of Silent Mutation in the ω-Subunit of Escherichia coli RNA Polymerase: Structure Transition Inhibits Transcription. ACS Omega 2019, 4, 17714–17725. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zuo, Y.; Wang, Y.; Steitz, T.A. The mechanism of E. coli RNA polymerase regulation by ppGpp is suggested by the structure of their complex. Mol. Cell 2013, 50, 430–436. [Google Scholar] [CrossRef] [Green Version]
- Tompa, P. Intrinsically disordered proteins: A 10-year recap. Trends Biochem. Sci. 2012, 37, 509–516. [Google Scholar] [CrossRef] [PubMed]
- Sabareesh, V.; Sarkar, P.; Sardesai, A.A.; Chatterji, D. Identifying N60D mutation in ω subunit of Escherichia coli RNA polymerase by bottom-up proteomic approach. Analyst 2010, 135, 2723–2729. [Google Scholar] [CrossRef] [PubMed]
- Bhowmik, D.; Bhardwaj, N.; Chatterji, D. Influence of Flexible “ω” on the Activity of E. coli RNA Polymerase: A Thermodynamic Analysis. Biophys. J. 2017, 112, 901–910. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borukhov, S.; Nudler, E. RNA polymerase: The vehicle of transcription. Trends Microbiol. 2008, 16, 126–134. [Google Scholar] [CrossRef]
- Mustaev, A.; Kozlov, M.; Markovtsov, V.; Zaychikov, E.; Denissova, L.; Goldfarb, A. Modular organization of the catalytic center of RNA polymerase. Proc. Natl. Acad. Sci. USA 1997, 94, 6641–6645. [Google Scholar] [CrossRef] [Green Version]
- Murakami, K.S.; Masuda, S.; Darst, S.A. Structural basis of transcription initiation: RNA polymerase holoenzyme at 4 A resolution. Science 2002, 296, 1280–1284. [Google Scholar] [CrossRef]
- Tagami, S.; Sekine, S.-I.; Kumarevel, T.; Hino, N.; Murayama, Y.; Kamegamori, S.; Yamamoto, M.; Sakamoto, K.; Yokoyama, S. Crystal structure of bacterial RNA polymerase bound with a transcription inhibitor protein. Nature 2010, 468, 978–982. [Google Scholar] [CrossRef] [PubMed]
- Chatterji, D.; Ojha, A.K. Revisiting the stringent response, ppGpp and starvation signaling. Curr. Opin. Microbiol. 2001, 4, 160–165. [Google Scholar] [CrossRef]
- Cashel, M.; Gallant, J. Two Compounds implicated in the Function of the RC Gene of Escherichia coli. Nature 1969, 221, 838–841. [Google Scholar] [CrossRef] [PubMed]
- Cashel, M.; Gentry, D.; Hernandez, V.J.; Vinella, D. The stringent response. In Escherichia Coli and Salmonella Typhimurium. Cellular and Molecular Biology; Neidhardt, F.C., Ed.; ASM Press: Washington, DC, USA, 1996; Volume 2, pp. 1458–1496. [Google Scholar]
- Takahashi, K.; Kasai, K.; Ochi, K. Identification of the bacterial alarmone guanosine 5′-diphosphate 3′-diphosphate (ppGpp) in plants. Proc. Natl. Acad. Sci. USA 2004, 101, 4320–4324. [Google Scholar] [CrossRef] [Green Version]
- Srivatsan, A.; Wang, J.D. Control of bacterial transcription, translation and replication by (p)ppGpp. Curr. Opin. Microbiol. 2008, 11, 100–105. [Google Scholar] [CrossRef]
- Chatterji, D.; Fujita, N.; Ishihama, A. The mediator for stringent control, ppGpp, binds to the beta-subunit of Escherichia coli RNA polymerase. Genes Cells 1998, 3, 279–287. [Google Scholar] [CrossRef]
- Durfee, T.; Hansen, A.-M.; Zhi, H.; Blattner, F.R.; Jin, D.J. Transcription profiling of the stringent response in Escherichia coli. J. Bacteriol. 2008, 190, 1084–1096. [Google Scholar] [CrossRef] [Green Version]
- Sarubbi, E.; Rudd, K.E.; Xiao, H.; Ikehara, K.; Kalman, M.; Cashel, M. Characterization of the spoT gene of Escherichia coli. J. Biol. Chem. 1989, 264, 15074–15082. [Google Scholar]
- Igarashi, K.; Fujita, N.; Ishihama, A. Promoter selectivity of Escherichia coli RNA polymerase: Omega factor is responsible for the ppGpp sensitivity. Nucleic Acids Res. 1989, 17, 8755–8765. [Google Scholar] [CrossRef] [Green Version]
- Gentry, D.; Xiao, H.; Burgess, R.; Cashel, M. The omega subunit of Escherichia coli K-12 RNA polymerase is not required for stringent RNA control in vivo. J. Bacteriol. 1991, 173, 3901–3903. [Google Scholar] [CrossRef] [Green Version]
- Ross, W.; Vrentas, C.E.; Sanchez-Vazquez, P.; Gaal, T.; Gourse, R.L. The magic spot: A ppGpp binding site on E. coli RNA polymerase responsible for regulation of transcription initiation. Mol. Cell 2013, 50, 420–429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ross, W.; Sanchez-Vazquez, P.; Chen, A.Y.; Lee, J.-H.; Burgos, H.L.; Gourse, R.L. ppGpp Binding to a Site at the RNAP-DksA Interface Accounts for Its Dramatic Effects on Transcription Initiation during the Stringent Response. Mol. Cell 2016, 62, 811–823. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Molodtsov, V.; Sineva, E.; Zhang, L.; Huang, X.; Cashel, M.; Ades, S.E.; Murakami, K.S. Allosteric Effector ppGpp Potentiates the Inhibition of Transcript Initiation by DksA. Mol. Cell 2018, 69, 828–839.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geertz, M.; Travers, A.; Mehandziska, S.; Sobetzko, P.; Chandra-Janga, S.; Shimamoto, N.; Muskhelishvili, G. Structural coupling between RNA polymerase composition and DNA supercoiling in coordinating transcription: A global role for the omega subunit? mBio 2011, 2, e00034-11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamamoto, K.; Yamanaka, Y.; Shimada, T.; Sarkar, P.; Yoshida, M.; Bhardwaj, N.; Watanabe, H.; Taira, Y.; Chatterji, D.; Ishihama, A. Altered Distribution of RNA Polymerase Lacking the Omega Subunit within the Prophages along the Escherichia coli K-12 Genome. mSystems 2018, 3, e00172-17. [Google Scholar] [CrossRef] [Green Version]
- Bhardwaj, N.; Syal, K.; Chatterji, D. The role of ω-subunit of Escherichia coli RNA polymerase in stress response. Genes Cells 2018, 23, 357–369. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Patel, U.R.; Gautam, S.; Chatterji, D. Validation of Omega Subunit of RNA Polymerase as a Functional Entity. Biomolecules 2020, 10, 1588. https://doi.org/10.3390/biom10111588
Patel UR, Gautam S, Chatterji D. Validation of Omega Subunit of RNA Polymerase as a Functional Entity. Biomolecules. 2020; 10(11):1588. https://doi.org/10.3390/biom10111588
Chicago/Turabian StylePatel, Unnatiben Rajeshbhai, Sudhanshu Gautam, and Dipankar Chatterji. 2020. "Validation of Omega Subunit of RNA Polymerase as a Functional Entity" Biomolecules 10, no. 11: 1588. https://doi.org/10.3390/biom10111588
APA StylePatel, U. R., Gautam, S., & Chatterji, D. (2020). Validation of Omega Subunit of RNA Polymerase as a Functional Entity. Biomolecules, 10(11), 1588. https://doi.org/10.3390/biom10111588



