Cytokinin-Regulated Expression of Arabidopsis thaliana PAP Genes and Its Implication for the Expression of Chloroplast-Encoded Genes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material, Growth Conditions, and Cytokinin Treatment
2.2. Hypocotyl and Root Elongation Assays
2.3. Protein Retention Assay
2.4. Analysis of Transcript Levels by Quantitative Real Time (RT)-PCR
2.5. Protein Isolation and Western Blot Analysis
2.6. Hormone Extraction, Purification, and Determination
2.7. Statistical Data Processing
3. Results
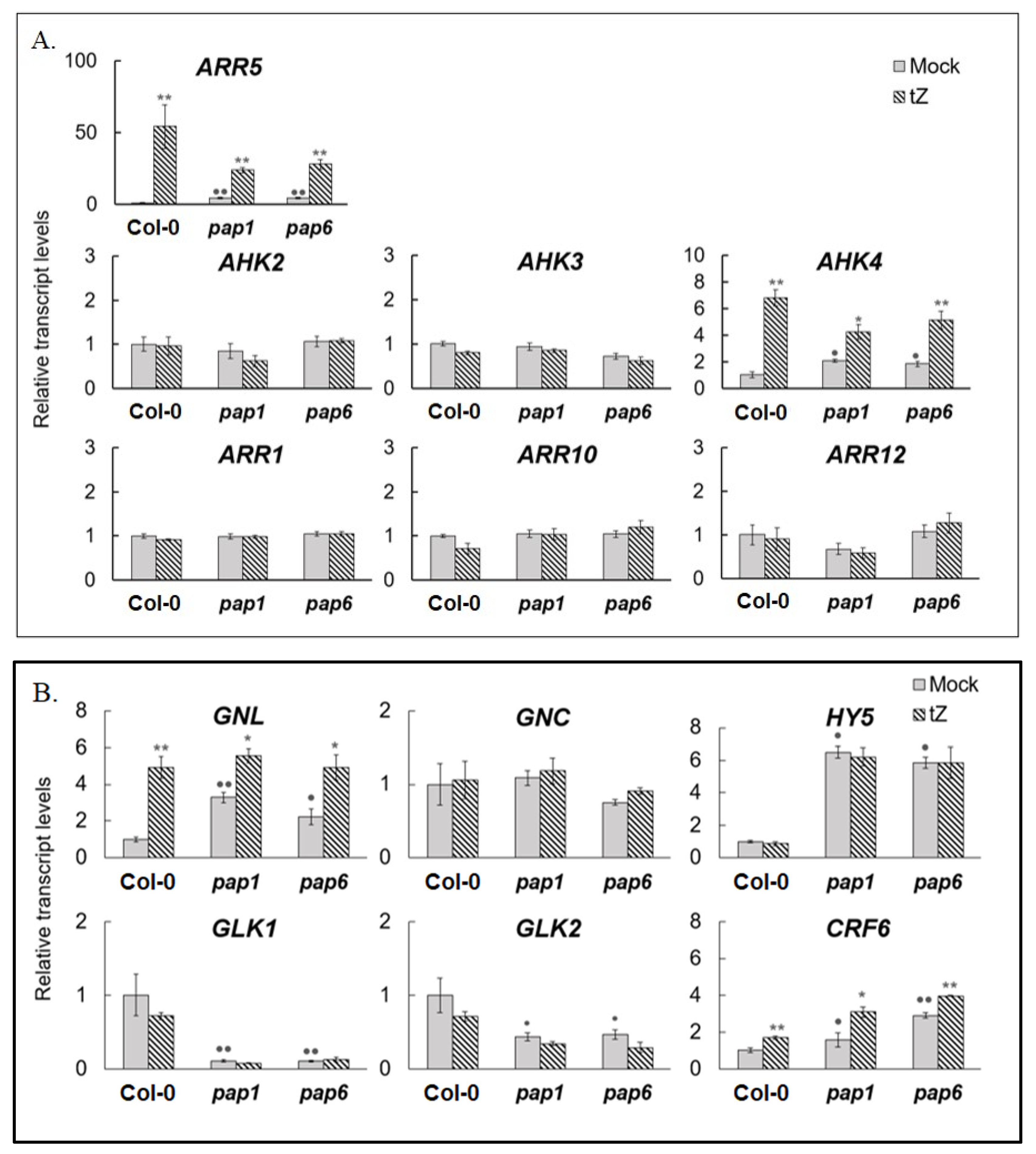
3.1. The Expression of PAP Genes Is Regulated by Cytokinin and Requires the Elements of CK Signaling Pathway
3.2. Loss of Function pap1 and pap6 Mutants Exhibited an Altered Response to Cytokinin Treatment
3.3. PAP Mutants Have Increased Cytokinin Content
3.4. CK-Regulated Expression of Chloroplast Encoded Genes and Nuclear Genes for Plastid Transcription Machinery Is Altered in the pap1 and pap6 Mutants
3.5. Pap 1 Mutant Is Affected in the CK-Dependent Accumulation of Plastid Proteins
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Brenner, W.G.; Ramireddy, E.; Heyl, A.; Schmülling, T. Gene regulation by cytokinin in Arabidopsis. Front. Plant Sci. 2012, 3, 8. [Google Scholar] [CrossRef] [Green Version]
- Bhargava, A.; Clabaugh, I.; To, J.P.; Maxwell, B.B.; Chiang, Y.H.; Schaller, G.E.; Kieber, J.J. Identification of cytokinin-responsive genes using microarray meta-analysis and RNA-Seq in Arabidopsis. Plant Physiol. 2013, 162, 272–294. [Google Scholar] [CrossRef] [Green Version]
- Brenner, W.G.; Schmülling, T. Summarizing and exploring data of a decade of cytokinin-related transcriptomics. Front. Plant Sci. 2015, 6, 29. [Google Scholar] [CrossRef] [Green Version]
- Rashotte, A.M.; Mason, M.G.; Hutchison, C.E.; Ferreira, F.J.; Schaller, G.E.; Kieber, J.J. A subset of Arabidopsis AP2 transcription factors mediates cytokinin responses in concert with a two-component pathway. Proc. Natl. Acad. Sci. USA 2006, 103, 11081–11085. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cortleven, A.; Schmülling, T. Regulation of chloroplast development and function by cytokinin. J. Exp. Bot. 2015, 66, 4999–5013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zubo, Y.O.; Blakley, I.C.; Franco-Zorrilla, J.M.; Yamburenko, M.V.; Solano, R.; Kieber, J.J.; Schaller, G.E. Coordination of chloroplast development through the action of the GNC and GLK transcription factor families. Plant Physiol. 2018, 178, 130–147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiang, Y.H.; Zubo, Y.O.; Tapken, W.; Kim, H.J.; Lavanway, A.M.; Howard, L.; Schaller, G.E. Functional characterization of the GATA transcription factors GNC and CGA1 reveals their key role in chloroplast development, growth, and division in Arabidopsis. Plant Physiol. 2012, 160, 332–348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhelyazkova, P.; Sharma, C.M.; Förstner, K.U.; Liere, K.; Vogel, J.; Börner, T. The primary transcriptome of barley chloroplasts: Numerous noncoding RNAs and the dominating role of the plastid-encoded RNA polymerase. Plant Cell 2012, 24, 123–136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pfannschmidt, T.; Blanvillain, R.; Merendino, L.; Courtois, F.; Chevalier, F.; Liebers, M.; Lerbs-Mache, S. Plastid RNA polymerases: Orchestration of enzymes with different evolutionary origins controls chloroplast biogenesis during the plant life cycle. J. Exp. Bot. 2015, 66, 6957–6973. [Google Scholar] [CrossRef] [PubMed]
- Grübler, B.; Merendino, L.; Twardziok, S.O.; Mininno, M.; Allorent, G.; Chevalier, F.; Ravanel, S. Light and plastid signals regulate different sets of genes in the albino mutant pap7-1. Plant Physiol. 2017, 175, 1203–1219. [Google Scholar] [CrossRef] [Green Version]
- Yagi, Y.; Shiina, T. Recent advances in the study of chloroplast gene expression and its evolution. Front. Plant Sci. 2014, 5, 61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Danilova, M.N.; Kudryakova, N.V.; Doroshenko, A.S.; Zabrodin, D.A.; Rakhmankulova, Z.F.; Oelmüller, R.; Kusnetsov, V.V. Opposite roles of the Arabidopsis cytokinin receptors AHK2 and AHK3 in the expression of plastid genes and genes for the plastid transcriptional machinery during senescence. Plant Mol. Biol. 2017, 93, 533–546. [Google Scholar] [CrossRef] [PubMed]
- Bastakis, E.; Hedtke, B.; Klermund, C.; Grimm, B.; Schwechheimer, C. LLM-domain B-GATA transcription factors play multifaceted roles in controlling greening in Arabidopsis. Plant Cell 2018, 30, 582–599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borsellino, L. Influence of Light and Cytokinin on Organellar Phage-Type RNA Polymerase Transcript Levels and Transcription of Organellar Genes in Arabidopsis thaliana. Ph.D. Thesis, Humbolt Universität zu Berlin, Berlin, Germany, 2012. [Google Scholar]
- Liebers, M.; Chevalier, F.; Blanvillain, R.; Pfannschmidt, T. PAP genes are tissue-and cell-specific markers of chloroplast development. Planta 2018, 248, 629–646. [Google Scholar] [CrossRef] [PubMed]
- Dobrev, P.I.; Kamınek, M. Fast and efficient separation of cytokinins from auxin and abscisic acid and their purification using mixed-mode solid-phase extraction. J. Chromatogr. A 2002, 950, 21–29. [Google Scholar] [CrossRef]
- Dobrev, P.I.; Vankova, R. Quantification of abscisic acid, cytokinin, and auxin content in salt-stressed plant tissues. Methods Mol. Biol. 2012, 913, 251–261. [Google Scholar]
- Steffens, N.O.; Galuschka, C.; Schindler, M.; Bülow, L.; Hehl, R. AthaMap web tools for database-assisted identification of combinatorial cis-regulatory elements and the display of highly conserved transcription factor binding sites in Arabidopsis thaliana. Nucleic Acids Res. 2005, 33, 397–402. [Google Scholar] [CrossRef]
- Yagi, Y.; Ishizaki, Y.; Nakahira, Y.; Tozawa, Y.; Shiina, T. Eukaryotic-type plastid nucleoid protein pTAC3 is essential for transcription by the bacterial-type plastid RNA polymerase. Proc. Natl. Acad. Sci. USA 2012, 109, 7541–7546. [Google Scholar] [CrossRef] [Green Version]
- Gilkerson, J.; Perez-Ruiz, J.M.; Chory, J.; Callis, J. The plastid-localized pfkB-type carbohydrate kinases fructokinase-like 1 and 2 are essential for growth and development of Arabidopsis thaliana. BMC Plant Biol. 2012, 12, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Danilova, M.N.; Kudryakova, N.V.; Andreeva, A.A.; Doroshenko, A.S.; Pojidaeva, E.S.; Kusnetsov, V.V. Differential impact of heat stress on the expression of chloroplast-encoded genes. Plant Physiol. Biochem. 2018, 129, 90–100. [Google Scholar] [CrossRef]
- He, L.; Zhang, S.; Qiu, Z.; Zhao, J.; Nie, W.; Lin, H.; Zhu, L. FRUCTOKINASE-LIKE PROTEIN 1 interacts with TRXz to regulate chloroplast development in rice. J. Integr. Plant Biol. 2018, 60, 94–111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Werner, T.; Schmuülling, T. Cytokinin action in plant development. Curr. Opin. Plant Biol. 2009, 12, 527–538. [Google Scholar] [CrossRef] [PubMed]
- Aloni, R.; Langhans, M.; Aloni, E.; Dreieicher, E.; Ullrich, C.I. Root-synthesized cytokinin in Arabidopsis is distributed in the shoot by the transpiration stream. J. Exp. Bot. 2005, 56, 1535–1544. [Google Scholar] [CrossRef] [PubMed]
- Miyawaki, K.; Tarkowski, P.; Matsumoto-Kitano, M.; Kato, T.; Sato, S.; Tarkowska, D.; Kakimoto, T. Roles of Arabidopsis ATP/ADP isopentenyltransferases and tRNA isopentenyltransferases in cytokinin biosynthesis. Proc. Natl. Acad. Sci. USA 2006, 103, 16598–16603. [Google Scholar] [CrossRef] [Green Version]
- Werner, T.; Motyka, V.; Laucou, V.; Smets, R.; Van Onckelen, H.; Schmülling, T. Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 2003, 15, 2532–2550. [Google Scholar] [CrossRef] [Green Version]
- Ha, S.; Vankova, R.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Tran, L.S.P. Cytokinins: Metabolism and function in plant adaptation to environmental stresses. Trends Plant Sci. 2012, 17, 172–179. [Google Scholar] [CrossRef]
- Chang, S.H.; Lee, S.; Um, T.Y.; Kim, J.K.; Do Choi, Y.; Jang, G. pTAC10, a key subunit of plastid-encoded RNA polymerase, promotes chloroplast development. Plant Physiol. 2017, 174, 435–449. [Google Scholar] [CrossRef] [Green Version]
- Arsova, B.; Hoja, U.; Wimmelbacher, M.; Greiner, E.; Üstün, Ş.; Melzer, M.; Börnke, F. Plastidial thioredoxin z interacts with two fructokinase-like proteins in a thiol-dependent manner: Evidence for an essential role in chloroplast development in Arabidopsis and Nicotiana benthamiana. Plant Cell 2010, 22, 1498–1515. [Google Scholar] [CrossRef] [Green Version]
- Kusnetsov, V.V.; Oelmüller, R.; Sarwat, M.I.; Porfirova, S.A.; Cherepneva, G.N.; Herrmann, R.G.; Kulaeva, O.N. Cytokinins, abscisic acid and light affect accumulation of chloroplast proteins in Lupinus luteus cotyledons without notable effect on steady-state mRNA levels. Planta 1994, 194, 318–327. [Google Scholar] [CrossRef]
- Shevtsov, S.; Nevo-Dinur, K.; Faigon, L.; Sultan, L.D.; Zmudjak, M.; Markovits, M.; Ostersetzer-Biran, O. Control of organelle gene expression by the mitochondrial transcription termination factor mTERF22 in Arabidopsis thaliana plants. PLoS ONE 2018, 13, e0201631. [Google Scholar] [CrossRef] [Green Version]
- Yamburenko, M.V.; Zubo, Y.O.; Vanková, R.; Kusnetsov, V.V.; Kulaeva, O.N.; Börner, Th. Abscisic acid represses the transcription of chloroplast genes. J. Exp. Bot. 2013, 64, 4491–4502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, M.; Chen, H.; Huang, L.; O’Neil, R.C.; Shokhirev, M.N.; Ecker, J.R. A B-ARR-mediated cytokinin transcriptional network directs hormone cross-regulation and shoot development. Nat. Commun. 2018, 9, 1604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- MacQuarrie, K.L.; Fong, A.P.; Morse, R.H.; Tapscott, S.J. Genome-wide transcription factor binding: Beyond direct target regulation. Trends Genet. 2011, 27, 141–148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; He, H.; Wang, X.; Wang, X.; Yang, X.; Li, L.; Deng, X.W. Genome-wide mapping of the HY5-mediated genenetworks in Arabidopsis that involve both transcriptional and post-transcriptional regulation. Plant J. 2011, 65, 346–358. [Google Scholar] [CrossRef]
- Doroshenko, A.S.; Danilova, M.N.; Andreeva, A.A.; Kudryakova, N.V.; Kuznetsov, V.V.; Kusnetsov, V.V. The Transcription Factor HY5 Is Involved in the Cytokinin-Dependent Regulation of the Expression of Genes Encoding Proteins Associated with Bacterial Plastid RNA-Polymerase during De-Etiolation of Arabidopsis thaliana. Dokl. Biochem. Biophys. 2020, 492, 124–129. [Google Scholar] [CrossRef]
- Kulaeva, O.N.; Burkhanova, E.A.; Karavaiko, N.N.; Selivankina, S.Y.; Porfirova, S.A.; Maslova, G.G.; Börner, T. Chloroplasts affect the leaf response to cytokinin. J. Plant. Physiol. 2002, 159, 1309–1316. [Google Scholar] [CrossRef]
- Chory, J.; Reinecke, D.; Sim, S.; Washburn, T.; Brenner, M. A role for cytokinins in de-etiolation in Arabidopsis (det mutants have an altered response to cytokinins). Plant. Physiol. 1994, 104, 339–347. [Google Scholar] [CrossRef] [Green Version]
- Burman, N.; Khurana, J.P. Photoregulation of Chloroplast Development: Retrograde Signaling. In Plastid Development in Leaves during Growth and Senescence; Springer: Dordrecht, The Netherlands, 2013; pp. 569–588. [Google Scholar]






| Line | DZ | tZR | iPR | cZR | CK Ribosides | ZRMPt | iPRMP | cZRMP | CK Phosphates |
|---|---|---|---|---|---|---|---|---|---|
| Col-0 | 1.40 ± 0.60 | 20.37 ± 4.74 | 1.04 ± 0.60 | 1.23 ± 0.62 | 22.64 ± 5.76 | 9.69 ± 4.94 | 12.38 ± 3.44 | 5.63 ± 1.96 | 27.70 ± 9.07 |
| pap1 | 0.43 ± 0.43 | 31.28 ± 3.42 * | 4.36 ± 1.75 * | 2.87 ± 2.13 | 38.50 ± 4.77 * | 71.57 ± 9.95 ** | 97.40 ± 26.79 ** | 5.48 ± 2.81 | 174.44 ± 32.57 ** |
| pap6 | 2.2 ± 0.38 | 23.43 ± 4.46 | 9.59 ± 2.95 ** | 0.62 ± 0.42 | 33.64 ± 6.57 | 71.53 ± 17.82 ** | 89.84 ± 32.28 ** | 6.98 ± 3.48 | 168.35 ± 51.31 ** |
| Line | tZROG | cZROG | CK-O glucosides | tZ7G | tZ9G | iP7G | iP9G | cZ7G | CK N glucosides |
| Col-0 | 4.20 ± 1.70 | 10.16 ± 2.11 | 14.35 ± 3.56 | 143.11 ± 31.95 | 33.39 ± 13.33 | 372.59 ± 134.58 | 12.23 ± 5.54 | 82.66 ± 25.47 | 643.99 ± 191.84 |
| pap1 | 6.71 ± 0.84 | 10.12 ± 4.49 | 16.83 ± 4.48 | 114.11 ± 20.62 | 33.22 ± 4.39 | 418.30 ± 36.34 | 12.89 ± 5.30 | 75.44 ± 38.30 | 653.97 ± 56.98 |
| pap6 | 5.90 ± 1.22 | 7.94 ± 1.45 | 13.84 ± 2.60 | 80.08 ± 11.72 * | 19.31 ± 8.17 | 359.95.95 ± 53.39 | 10.93 ± 3.09 | 30.66 ± 13.89 * | 500.93 ± 69.43 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Andreeva, A.A.; Vankova, R.; Bychkov, I.A.; Kudryakova, N.V.; Danilova, M.N.; Lacek, J.; Pojidaeva, E.S.; Kusnetsov, V.V. Cytokinin-Regulated Expression of Arabidopsis thaliana PAP Genes and Its Implication for the Expression of Chloroplast-Encoded Genes. Biomolecules 2020, 10, 1658. https://doi.org/10.3390/biom10121658
Andreeva AA, Vankova R, Bychkov IA, Kudryakova NV, Danilova MN, Lacek J, Pojidaeva ES, Kusnetsov VV. Cytokinin-Regulated Expression of Arabidopsis thaliana PAP Genes and Its Implication for the Expression of Chloroplast-Encoded Genes. Biomolecules. 2020; 10(12):1658. https://doi.org/10.3390/biom10121658
Chicago/Turabian StyleAndreeva, Aleksandra A., Radomira Vankova, Ivan A. Bychkov, Natalia V. Kudryakova, Maria N. Danilova, Jozef Lacek, Elena S. Pojidaeva, and Victor V. Kusnetsov. 2020. "Cytokinin-Regulated Expression of Arabidopsis thaliana PAP Genes and Its Implication for the Expression of Chloroplast-Encoded Genes" Biomolecules 10, no. 12: 1658. https://doi.org/10.3390/biom10121658
APA StyleAndreeva, A. A., Vankova, R., Bychkov, I. A., Kudryakova, N. V., Danilova, M. N., Lacek, J., Pojidaeva, E. S., & Kusnetsov, V. V. (2020). Cytokinin-Regulated Expression of Arabidopsis thaliana PAP Genes and Its Implication for the Expression of Chloroplast-Encoded Genes. Biomolecules, 10(12), 1658. https://doi.org/10.3390/biom10121658





