Nanobodies Right in the Middle: Intrabodies as Toolbox to Visualize and Modulate Antigens in the Living Cell
Abstract
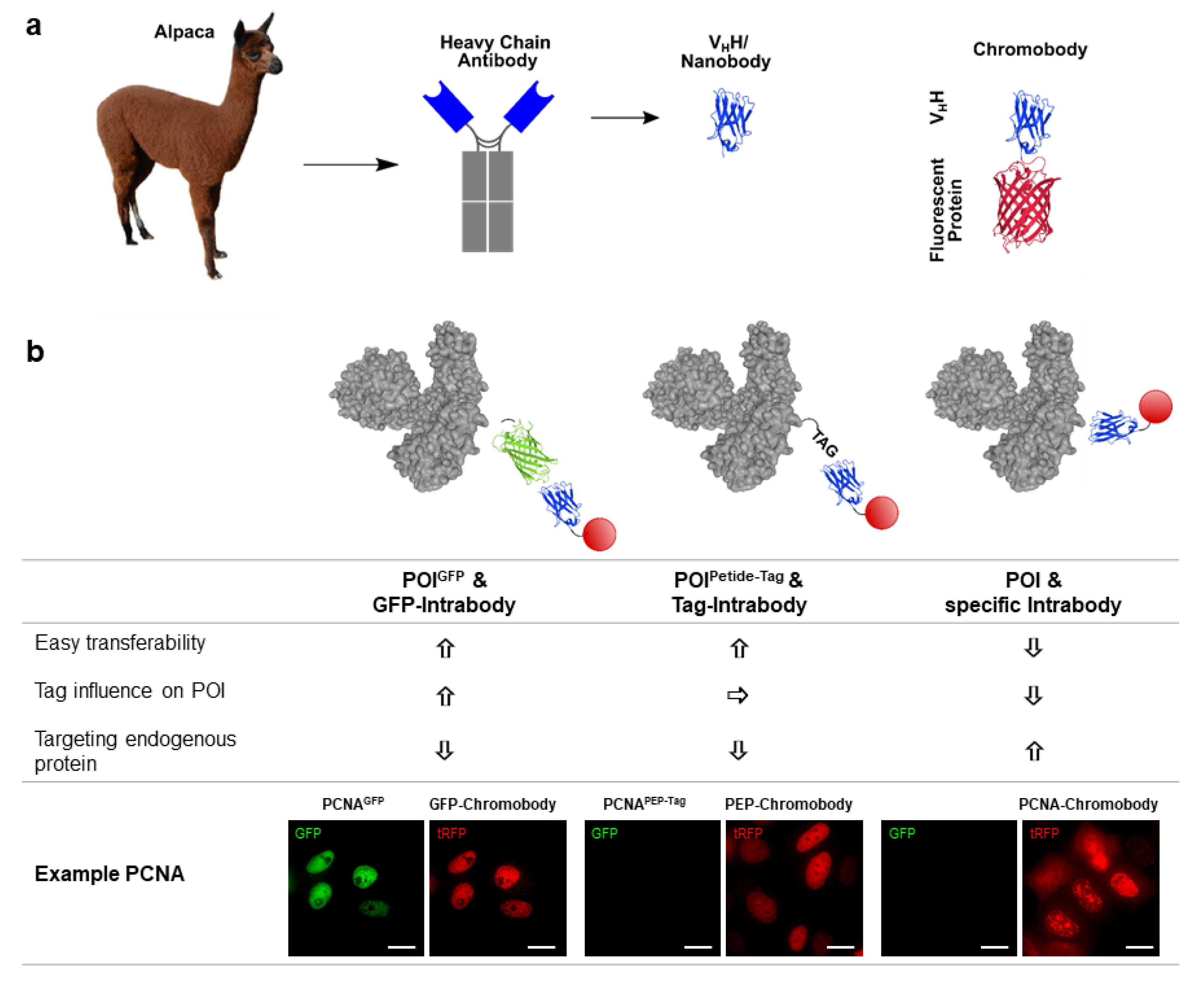
:1. Selection of Intracellular Functional Nanobodies
2. Delivery Systems of Intrabodies
3. Intrabodies to Visualize Antigens in Living Cells
3.1. Chromobody Technology
3.2. Tag-Specific Intrabodies
3.3. Intrabodies Targeting Endogenous Antigens
3.4. Intrabodies as Biosensors
4. Intrabodies to Modulate and Manipulate Intracellular Antigens
4.1. Intrabodies in Oncology
4.2. Intrabodies as Immune Modulators
4.3. Intrabodies to Address Neurological Disorders
4.4. Intrabodies Inhibiting Viral and Bacterial Pathogens in Live Cells
5. Switchable Intrabodies as Upcoming Tools
Funding
Acknowledgments
Conflicts of Interest
References
- Hamers-Casterman, C.; Atarhouch, T.; Muyldermans, S.; Robinson, G.; Hamers, C.; Songa, E.B.; Bendahman, N.; Hamers, R. Naturally occurring antibodies devoid of light chains. Nature 1993, 363, 446–448. [Google Scholar] [CrossRef] [PubMed]
- Muyldermans, S. Nanobodies: Natural single-domain antibodies. Annu. Rev. Biochem. 2013, 82, 775–797. [Google Scholar] [CrossRef] [Green Version]
- Peyvandi, F.; Scully, M.; Kremer Hovinga, J.A.; Cataland, S.; Knobl, P.; Wu, H.; Artoni, A.; Westwood, J.P.; Mansouri Taleghani, M.; Jilma, B.; et al. Caplacizumab for acquired thrombotic thrombocytopenic purpura. N. Engl. J. Med. 2016, 374, 511–522. [Google Scholar] [CrossRef] [PubMed]
- Morrison, C. Nanobody approval gives domain antibodies a boost. Nat. Rev. Drug Discov. 2019, 18, 485–487. [Google Scholar] [CrossRef] [PubMed]
- Dumoulin, M.; Conrath, K.; Van Meirhaeghe, A.; Meersman, F.; Heremans, K.; Frenken, L.G.; Muyldermans, S.; Wyns, L.; Matagne, A. Single-domain antibody fragments with high conformational stability. Protein Sci. 2002, 11, 500–515. [Google Scholar] [CrossRef] [PubMed]
- Kunz, P.; Zinner, K.; Mücke, N.; Bartoschik, T.; Muyldermans, S.; Hoheisel, J.D. The structural basis of nanobody unfolding reversibility and thermoresistance. Sci. Rep. 2018, 8, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, P.D.; Maier, J.; Traenkle, B.; Emele, F.; Rothbauer, U. Recent progress in generating intracellular functional antibody fragments to target and trace cellular components in living cells. Biochim. Biophys. Acta 2014, 1844, 1933–1942. [Google Scholar] [CrossRef]
- Helma, J.; Cardoso, M.C.; Muyldermans, S.; Leonhardt, H. Nanobodies and recombinant binders in cell biology. J. Cell Biol. 2015, 209, 633–644. [Google Scholar] [CrossRef] [Green Version]
- Traenkle, B.; Rothbauer, U. Under the microscope: Single-domain antibodies for live-cell imaging and super-resolution microscopy. Front. Immunol. 2017, 8, 1030. [Google Scholar] [CrossRef] [Green Version]
- Moutel, S.; Bery, N.; Bernard, V.; Keller, L.; Lemesre, E.; de Marco, A.; Ligat, L.; Rain, J.C.; Favre, G.; Olichon, A.; et al. Nali-h1: A universal synthetic library of humanized nanobodies providing highly functional antibodies and intrabodies. eLife 2016, 5, e16228. [Google Scholar] [CrossRef]
- Wörn, A.; Plückthun, A. Mutual stabilization of vl and vh in single-chain antibody fragments, investigated with mutants engineered for stability. Biochemistry 1998, 37, 13120–13127. [Google Scholar] [CrossRef] [PubMed]
- Proba, K.; WoÈrn, A.; Honegger, A.; PluÈckthun, A. Antibody scfv fragments without disulfide bonds, made by molecular evolution. J. Mol. Biol. 1998, 275, 245–253. [Google Scholar] [CrossRef] [PubMed]
- Olichon, A.; Surrey, T. Selection of genetically encoded fluorescent single domain antibodies engineered for efficient expression in escherichia coli. J. Biol. Chem. 2007, 282, 36314–36320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kvam, E.; Sierks, M.R.; Shoemaker, C.B.; Messer, A. Physico-chemical determinants of soluble intrabody expression in mammalian cell cytoplasm. Protein Eng. Des. Sel. 2010, 23, 489–498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joshi, S.N.; Butler, D.C.; Messer, A. Fusion to a highly charged proteasomal retargeting sequence increases soluble cytoplasmic expression and efficacy of diverse anti-synuclein intrabodies. In MAbs; Taylor & Francis: London, UK, 2012; pp. 686–693. [Google Scholar] [CrossRef] [Green Version]
- Kabayama, H.; Takeuchi, M.; Tokushige, N.; Muramatsu, S.-I.; Kabayama, M.; Fukuda, M.; Yamada, Y.; Mikoshiba, K. An ultra-stable cytoplasmic antibody engineered for in vivo applications. Nat. Commun. 2020, 11, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Bourgeois, J.P.; Celli, S.; Glacial, F.; Le Sourd, A.M.; Mecheri, S.; Weksler, B.; Romero, I.; Couraud, P.O.; Rougeon, F.; et al. Cell-penetrating anti-gfap vhh and corresponding fluorescent fusion protein vhh-gfp spontaneously cross the blood-brain barrier and specifically recognize astrocytes: Application to brain imaging. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2012, 26, 3969–3979. [Google Scholar] [CrossRef]
- Fields, S.; Song, O. A novel genetic system to detect protein-protein interactions. Nature 1989, 340, 245–246. [Google Scholar] [CrossRef]
- Young, K. Yeast two-hybrid: So many interactions, (in) so little time…. Biol. Reprod. 1998, 58, 302–311. [Google Scholar] [CrossRef] [Green Version]
- Brückner, A.; Polge, C.; Lentze, N.; Auerbach, D.; Schlattner, U. Yeast two-hybrid, a powerful tool for systems biology. Int. J. Mol. Sci. 2009, 10, 2763–2788. [Google Scholar] [CrossRef] [Green Version]
- Pellis, M.; Muyldermans, S.; Vincke, C. Bacterial two hybrid: A versatile one-step intracellular selection method. Methods Mol. Biol. 2012, 911, 135–150. [Google Scholar]
- Tanaka, T.; Rabbitts, T.H. Intracellular antibody capture (iac) methods for single domain antibodies. In Single Domain Antibodies; Springer: Cham, Switzerland, 2012; pp. 151–173. [Google Scholar]
- Visintin, M.; Tse, E.; Axelson, H.; Rabbitts, T.H.; Cattaneo, A. Selection of antibodies for intracellular function using a two-hybrid in vivo system. Proc. Natl. Acad. Sci. USA 1999, 96, 11723–11728. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Visintin, M.; Settanni, G.; Maritan, A.; Graziosi, S.; Marks, J.D.; Cattaneo, A. The intracellular antibody capture technology (iact): Towards a consensus sequence for intracellular antibodies. J. Mol. Biol. 2002, 317, 73–83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanaka, T.; Lobato, M.N.; Rabbitts, T.H. Single domain intracellular antibodies: A minimal fragment for direct in vivo selection of antigen-specific intrabodies. J. Mol. Biol. 2003, 331, 1109–1120. [Google Scholar] [CrossRef]
- Pellis, M.; Pardon, E.; Zolghadr, K.; Rothbauer, U.; Vincke, C.; Kinne, J.; Dierynck, I.; Hertogs, K.; Leonhardt, H.; Messens, J.; et al. A bacterial-two-hybrid selection system for one-step isolation of intracellularly functional nanobodies. Arch. Biochem. Biophys. 2012, 526, 114–123. [Google Scholar] [CrossRef] [PubMed]
- Zolghadr, K.; Mortusewicz, O.; Rothbauer, U.; Kleinhans, R.; Goehler, H.; Wanker, E.E.; Cardoso, M.C.; Leonhardt, H. A fluorescent two-hybrid assay for direct visualization of protein interactions in living cells. Mol. Cell. Proteom. MCP 2008, 7, 2279–2287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmidt, F.I.; Hanke, L.; Morin, B.; Brewer, R.; Brusic, V.; Whelan, S.P.; Ploegh, H.L. Phenotypic lentivirus screens to identify functional single domain antibodies. Nat. Microbiol. 2016, 1, 16080. [Google Scholar] [CrossRef] [Green Version]
- Mazuc, E.; Guglielmi, L.; Bec, N.; Parez, V.; Hahn, C.S.; Mollevi, C.; Parrinello, H.; Desvignes, J.-P.; Larroque, C.; Jupp, R. In-cell intrabody selection from a diverse human library identifies c12orf4 protein as a new player in rodent mast cell degranulation. PLoS ONE 2014, 9, e104998. [Google Scholar] [CrossRef]
- Lee, S.; Kaku, Y.; Inoue, S.; Nagamune, T.; Kawahara, M. Growth signalobody selects functional intrabodies in the mammalian cytoplasm. Biotechnol. J. 2016, 11, 565–573. [Google Scholar] [CrossRef]
- Keller, B.-M.; Maier, J.; Weldle, M.; Segan, S.; Traenkle, B.; Rothbauer, U. A strategy to optimize the generation of stable chromobody cell lines for visualization and quantification of endogenous proteins in living cells. Antibodies 2019, 8, 10. [Google Scholar] [CrossRef] [Green Version]
- Wegner, W.; Ilgen, P.; Gregor, C.; Van Dort, J.; Mott, A.C.; Steffens, H.; Willig, K.I. In vivo mouse and live cell sted microscopy of neuronal actin plasticity using far-red emitting fluorescent proteins. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Chatterjee, D.; Bhatt, M.; Butler, D.; De Genst, E.; Dobson, C.M.; Messer, A.; Kordower, J.H. Proteasome-targeted nanobodies alleviate pathology and functional decline in an α-synuclein-based parkinson’s disease model. npj Park. Dis. 2018, 4, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Hammond, C.; Helenius, A. Quality control in the secretory pathway: Retention of a misfolded viral membrane glycoprotein involves cycling between the er, intermediate compartment, and golgi apparatus. J. Cell Biol. 1994, 126, 41–52. [Google Scholar] [CrossRef]
- Slastnikova, T.A.; Ulasov, A.V.; Rosenkranz, A.A.; Sobolev, A.S. Targeted intracellular delivery of antibodies: The state of the art. Front. Pharmacol. 2018, 9, 1208. [Google Scholar] [CrossRef] [Green Version]
- Singh, K.; Ejaz, W.; Dutta, K.; Thayumanavan, S. Antibody delivery for intracellular targets: Emergent therapeutic potential. Bioconjugate Chem. 2019, 30, 1028–1041. [Google Scholar] [CrossRef]
- Li, Y.; Li, P.; Li, R.; Xu, Q. Intracellular antibody delivery mediated by lipids, polymers, and inorganic nanomaterials for therapeutic applications. Adv. Ther. 2020, 2000178. [Google Scholar] [CrossRef]
- Conic, S.; Desplancq, D.; Ferrand, A.; Fischer, V.; Heyer, V.; Reina San Martin, B.; Pontabry, J.; Oulad-Abdelghani, M.; Babu, N.K.; Wright, G.D.; et al. Imaging of native transcription factors and histone phosphorylation at high resolution in live cells. J. Cell Biol. 2018, 217, 1537–1552. [Google Scholar] [CrossRef] [Green Version]
- Dixon, C.R.; Platani, M.; Makarov, A.A.; Schirmer, E.C. Microinjection of antibodies targeting the lamin a/c histone-binding site blocks mitotic entry and reveals separate chromatin interactions with hp1, cenpb and pml. Cells 2017, 6, 9. [Google Scholar] [CrossRef] [Green Version]
- Klein, A.; Hank, S.; Raulf, A.; Joest, E.; Tissen, F.; Heilemann, M.; Wieneke, R.; Tampé, R. Live-cell labeling of endogenous proteins with nanometer precision by transduced nanobodies. Chem. Sci. 2018, 9, 7835–7842. [Google Scholar] [CrossRef] [Green Version]
- Frankel, A.D.; Pabo, C.O. Cellular uptake of the tat protein from human immunodeficiency virus. Cell 1988, 55, 1189–1193. [Google Scholar] [CrossRef]
- Green, M.; Loewenstein, P.M. Autonomous functional domains of chemically synthesized human immunodeficiency virus tat trans-activator protein. Cell 1988, 55, 1179–1188. [Google Scholar] [CrossRef]
- Joliot, A.; Pernelle, C.; Deagostini-Bazin, H.; Prochiantz, A. Antennapedia homeobox peptide regulates neural morphogenesis. Proc. Natl. Acad. Sci. USA 1991, 88, 1864–1868. [Google Scholar] [CrossRef] [Green Version]
- Futaki, S.; Suzuki, T.; Ohashi, W.; Yagami, T.; Tanaka, S.; Ueda, K.; Sugiura, Y. Arginine-rich peptides an abundant source of membrane-permeable peptides having potential as carriers for intracellular protein delivery. J. Biol. Chem. 2001, 276, 5836–5840. [Google Scholar] [CrossRef] [Green Version]
- Futaki, S. Arginine-rich peptides: Potential for intracellular delivery of macromolecules and the mystery of the translocation mechanisms. Int. J. Pharm. 2002, 245, 1–7. [Google Scholar] [CrossRef]
- Prochiantz, A. Messenger proteins: Homeoproteins, tat and others. Curr. Opin. Cell Biol. 2000, 12, 400–406. [Google Scholar] [CrossRef]
- Kaczmarczyk, S.J.; Sitaraman, K.; Young, H.A.; Hughes, S.H.; Chatterjee, D.K. Protein delivery using engineered virus-like particles. Proc. Natl. Acad. Sci. USA 2011, 108, 16998–17003. [Google Scholar] [CrossRef] [Green Version]
- Méndez, J.; Morales Cruz, M.; Delgado, Y.; Figueroa, C.M.; Orellano, E.A.; Morales, M.; Monteagudo, A.; Griebenow, K. Delivery of chemically glycosylated cytochrome c immobilized in mesoporous silica nanoparticles induces apoptosis in hela cancer cells. Mol. Pharm. 2014, 11, 102–111. [Google Scholar] [CrossRef] [Green Version]
- Ray, M.; Tang, R.; Jiang, Z.; Rotello, V.M. Quantitative tracking of protein trafficking to the nucleus using cytosolic protein delivery by nanoparticle-stabilized nanocapsules. Bioconjugate Chem. 2015, 26, 1004–1007. [Google Scholar] [CrossRef] [Green Version]
- Bruce, V.J.; Lopez-Islas, M.; McNaughton, B.R. Resurfaced cell-penetrating nanobodies: A potentially general scaffold for intracellularly targeted protein discovery. Protein Sci. 2016, 25, 1129–1137. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.; Ishii, T.; Kim, H.J.; Nishiyama, N.; Hayakawa, Y.; Itaka, K.; Kataoka, K. Efficient delivery of bioactive antibodies into the cytoplasm of living cells by charge-conversional polyion complex micelles. Angew. Chem. Int. Ed. 2010, 49, 2552–2555. [Google Scholar] [CrossRef]
- Sarker, S.R.; Hokama, R.; Takeoka, S. Intracellular delivery of universal proteins using a lysine headgroup containing cationic liposomes: Deciphering the uptake mechanism. Mol. Pharm. 2014, 11, 164–174. [Google Scholar] [CrossRef]
- Röder, R.; Helma, J.; Preiß, T.; Rädler, J.O.; Leonhardt, H.; Wagner, E. Intracellular delivery of nanobodies for imaging of target proteins in live cells. Pharm. Res. 2017, 34, 161–174. [Google Scholar] [CrossRef] [PubMed]
- Rothbauer, U.; Zolghadr, K.; Tillib, S.; Nowak, D.; Schermelleh, L.; Gahl, A.; Backmann, N.; Conrath, K.; Muyldermans, S.; Cardoso, M.C. Targeting and tracing antigens in live cells with fluorescent nanobodies. Nat. Methods 2006, 3, 887–889. [Google Scholar] [CrossRef] [PubMed]
- Prole, D.L.; Taylor, C.W. A genetically encoded toolkit of functionalized nanobodies against fluorescent proteins for visualizing and manipulating intracellular signalling. BMC Biol. 2019, 17, 1–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caussinus, E.; Kanca, O.; Affolter, M. Fluorescent fusion protein knockout mediated by anti-gfp nanobody. Nat. Struct. Mol. Biol. 2011, 19, 117–121. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.; Bates, J.A.; Wei, H.; Bartoschek, M.D.; Conradt, B.; Leonhardt, H. Tunable light and drug induced depletion of target proteins. Nat. Commun. 2020, 11, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Daniel, K.; Icha, J.; Horenburg, C.; Müller, D.; Norden, C.; Mansfeld, J. Conditional control of fluorescent protein degradation by an auxin-dependent nanobody. Nat. Commun. 2018, 9, 1–13. [Google Scholar] [CrossRef]
- Clift, D.; McEwan, W.A.; Labzin, L.I.; Konieczny, V.; Mogessie, B.; James, L.C.; Schuh, M. A method for the acute and rapid degradation of endogenous proteins. Cell 2017, 171, 1692–1706.e18. [Google Scholar] [CrossRef] [Green Version]
- Tang, J.C.; Rudolph, S.; Dhande, O.S.; Abraira, V.E.; Choi, S.; Lapan, S.W.; Drew, I.R.; Drokhlyansky, E.; Huberman, A.D.; Regehr, W.G.; et al. Cell type-specific manipulation with gfp-dependent cre recombinase. Nat. Neurosci. 2015, 18, 1334–1341. [Google Scholar] [CrossRef] [Green Version]
- Tang, J.C.; Szikra, T.; Kozorovitskiy, Y.; Teixiera, M.; Sabatini, B.L.; Roska, B.; Cepko, C.L. A nanobody-based system using fluorescent proteins as scaffolds for cell-specific gene manipulation. Cell 2013, 154, 928–939. [Google Scholar] [CrossRef] [Green Version]
- Bothma, J.P.; Norstad, M.R.; Alamos, S.; Garcia, H.G. Llamatags: A versatile tool to image transcription factor dynamics in live embryos. Cell 2018, 173, 1810–1822.e16. [Google Scholar] [CrossRef]
- Harmansa, S.; Alborelli, I.; Bieli, D.; Caussinus, E.; Affolter, M. A nanobody-based toolset to investigate the role of protein localization and dispersal in drosophila. eLife 2017, 6, e22549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hosein, R.E.; Williams, S.A.; Haye, K.; Gavin, R.H. Expression of gfp-actin leads to failure of nuclear elongation and cytokinesis in tetrahymena thermophila. J. Eukaryot. Microbiol. 2003, 50, 403–408. [Google Scholar] [CrossRef] [PubMed]
- Snapp, E.L. Fluorescent proteins: A cell biologist’s user guide. Trends Cell Biol. 2009, 19, 649–655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stadler, C.; Rexhepaj, E.; Singan, V.R.; Murphy, R.F.; Pepperkok, R.; Uhlen, M.; Simpson, J.C.; Lundberg, E. Immunofluorescence and fluorescent-protein tagging show high correlation for protein localization in mammalian cells. Nat. Methods 2013, 10, 315–323. [Google Scholar] [CrossRef]
- De Genst, E.; Silence, K.; Decanniere, K.; Conrath, K.; Loris, R.; Kinne, J.; Muyldermans, S.; Wyns, L. Molecular basis for the preferential cleft recognition by dromedary heavy-chain antibodies. Proc. Natl. Acad. Sci. USA 2006, 103, 4586–4591. [Google Scholar] [CrossRef] [Green Version]
- Pardon, E.; Laeremans, T.; Triest, S.; Rasmussen, S.G.; Wohlkonig, A.; Ruf, A.; Muyldermans, S.; Hol, W.G.; Kobilka, B.K.; Steyaert, J. A general protocol for the generation of nanobodies for structural biology. Nat. Protoc. 2014, 9, 674–693. [Google Scholar] [CrossRef]
- Nunes-Silva, S.; Gangnard, S.; Vidal, M.; Vuchelen, A.; Dechavanne, S.; Chan, S.; Pardon, E.; Steyaert, J.; Ramboarina, S.; Chêne, A. Llama immunization with full-length var2csa generates cross-reactive and inhibitory single-domain antibodies against the dbl1x domain. Sci. Rep. 2014, 4, 7373. [Google Scholar] [CrossRef]
- De Genst, E.J.; Guilliams, T.; Wellens, J.; O’Day, E.M.; Waudby, C.A.; Meehan, S.; Dumoulin, M.; Hsu, S.-T.D.; Cremades, N.; Verschueren, K.H. Structure and properties of a complex of α-synuclein and a single-domain camelid antibody. J. Mol. Biol. 2010, 402, 326–343. [Google Scholar] [CrossRef]
- Braun, M.B.; Traenkle, B.; Koch, P.A.; Emele, F.; Weiss, F.; Poetz, O.; Stehle, T.; Rothbauer, U. Peptides in headlock–a novel high-affinity and versatile peptide-binding nanobody for proteomics and microscopy. Sci. Rep. 2016, 6, 19211. [Google Scholar] [CrossRef] [Green Version]
- Virant, D.; Traenkle, B.; Maier, J.; Kaiser, P.D.; Bodenhofer, M.; Schmees, C.; Vojnovic, I.; Pisak-Lukats, B.; Endesfelder, U.; Rothbauer, U. A peptide tag-specific nanobody enables high-quality labeling for dstorm imaging. Nat. Commun. 2018, 9, 930. [Google Scholar] [CrossRef]
- Götzke, H.; Kilisch, M.; Martínez-Carranza, M.; Sograte-Idrissi, S.; Rajavel, A.; Schlichthaerle, T.; Engels, N.; Jungmann, R.; Stenmark, P.; Opazo, F. The alfa-tag is a highly versatile tool for nanobody-based bioscience applications. Nat. Commun. 2019, 10, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hulsik, D.L.; Liu, Y.-Y.; Strokappe, N.M.; Battella, S.; El Khattabi, M.; McCoy, L.E.; Sabin, C.; Hinz, A.; Hock, M.; Macheboeuf, P. A gp41 mper-specific llama vhh requires a hydrophobic cdr3 for neutralization but not for antigen recognition. PLoS Pathog. 2013, 9, e1003202. [Google Scholar]
- Boersma, S.; Khuperkar, D.; Verhagen, B.M.; Sonneveld, S.; Grimm, J.B.; Lavis, L.D.; Tanenbaum, M.E. Multi-color single-molecule imaging uncovers extensive heterogeneity in mrna decoding. Cell 2019, 178, 458–472.e19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Traenkle, B.; Segan, S.; Fagbadebo, F.O.; Kaiser, P.D.; Rothbauer, U. A novel epitope tagging system to visualize and monitor antigens in live cells with chromobodies. Sci. Rep. 2020, 10, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Mendez, M.G.; Kojima, S.; Goldman, R.D. Vimentin induces changes in cell shape, motility, and adhesion during the epithelial to mesenchymal transition. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2010, 24, 1838–1851. [Google Scholar] [CrossRef] [Green Version]
- Belin, B.J.; Goins, L.M.; Mullins, R.D. Comparative analysis of tools for live cell imaging of actin network architecture. Bioarchitecture 2014, 4, 189–202. [Google Scholar] [CrossRef]
- Lemieux, M.G.; Janzen, D.; Hwang, R.; Roldan, J.; Jarchum, I.; Knecht, D.A. Visualization of the actin cytoskeleton: Different f-actin-binding probes tell different stories. Cytoskeleton 2014, 71, 157–169. [Google Scholar] [CrossRef]
- Schmidthals, K.; Helma, J.; Zolghadr, K.; Rothbauer, U.; Leonhardt, H. Novel antibody derivatives for proteome and high-content analysis. Anal. Bioanal. Chem. 2010, 397, 3203–3208. [Google Scholar] [CrossRef] [Green Version]
- Panza, P.; Maier, J.; Schmees, C.; Rothbauer, U.; Sollner, C. Live imaging of endogenous protein dynamics in zebrafish using chromobodies. Development 2015, 142, 1879–1884. [Google Scholar] [CrossRef] [Green Version]
- Maier, J.; Traenkle, B.; Rothbauer, U. Real-time analysis of epithelial-mesenchymal transition using fluorescent single-domain antibodies. Sci. Rep. 2015, 5, 13402. [Google Scholar] [CrossRef] [Green Version]
- Zolghadr, K.; Gregor, J.; Leonhardt, H.; Rothbauer, U. Case study on live cell apoptosis-assay using lamin-chromobody cell-lines for high-content analysis. Methods Mol. Biol. 2012, 911, 569–575. [Google Scholar]
- Plessner, M.; Melak, M.; Chinchilla, P.; Baarlink, C.; Grosse, R. Nuclear f-actin formation and reorganization upon cell spreading. J. Biol. Chem. 2015, 290, 11209–11216. [Google Scholar] [CrossRef] [Green Version]
- Maier, J.; Traenkle, B.; Rothbauer, U. Visualizing epithelial-mesenchymal transition using the chromobody technology. Cancer Res. 2016, 76, 5592–5596. [Google Scholar] [CrossRef] [Green Version]
- Burgess, A.; Lorca, T.; Castro, A. Quantitative live imaging of endogenous DNA replication in mammalian cells. PLoS ONE 2012, 7, e45726. [Google Scholar] [CrossRef] [Green Version]
- Schorpp, K.; Rothenaigner, I.; Maier, J.; Traenkle, B.; Rothbauer, U.; Hadian, K. A multiplexed high-content screening approach using the chromobody technology to identify cell cycle modulators in living cells. J. Biomol. Screen. 2016, 21, 965–977. [Google Scholar] [CrossRef] [Green Version]
- Buchfellner, A.; Yurlova, L.; Nuske, S.; Scholz, A.M.; Bogner, J.; Ruf, B.; Zolghadr, K.; Drexler, S.E.; Drexler, G.A.; Girst, S.; et al. A new nanobody-based biosensor to study endogenous parp1 in vitro and in live human cells. PLoS ONE 2016, 11, e0151041. [Google Scholar] [CrossRef] [Green Version]
- Rajan, M.; Mortusewicz, O.; Rothbauer, U.; Hastert, F.D.; Schmidthals, K.; Rapp, A.; Leonhardt, H.; Cardoso, M.C. Generation of an alpaca-derived nanobody recognizing gamma-h2ax. FEBS Open Bio 2015, 5, 779–788. [Google Scholar] [CrossRef] [Green Version]
- Jullien, D.; Vignard, J.; Fedor, Y.; Bery, N.; Olichon, A.; Crozatier, M.; Erard, M.; Cassard, H.; Ducommun, B.; Salles, B.; et al. Chromatibody, a novel non-invasive molecular tool to explore and manipulate chromatin in living cells. J. Cell Sci. 2016, 129, 2673–2683. [Google Scholar] [CrossRef] [Green Version]
- Traenkle, B.; Emele, F.; Anton, R.; Poetz, O.; Haeussler, R.S.; Maier, J.; Kaiser, P.D.; Scholz, A.M.; Nueske, S.; Buchfellner, A.; et al. Monitoring interactions and dynamics of endogenous beta-catenin with intracellular nanobodies in living cells. Mol. Cell. Proteom. MCP 2015, 14, 707–723. [Google Scholar] [CrossRef] [Green Version]
- Dietrich, J.; Sommersdorf, C.; Gohlke, S.; Poetz, O.; Traenkle, B.; Rothbauer, U.; Hessel-Pras, S.; Lampen, A.; Braeuning, A. Okadaic acid activates wnt/beta-catenin-signaling in human heparg cells. Arch. Toxicol. 2019, 93, 1927–1939. [Google Scholar] [CrossRef]
- Keller, B.-M.; Maier, J.; Secker, K.-A.; Egetemaier, S.-M.; Parfyonova, Y.; Rothbauer, U.; Traenkle, B. Chromobodies to quantify changes of endogenous protein concentration in living cells. Mol. Cell. Proteom. 2018, 17, 2518–2533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, J.C.; Drokhlyansky, E.; Etemad, B.; Rudolph, S.; Guo, B.; Wang, S.; Ellis, E.G.; Li, J.Z.; Cepko, C.L. Detection and manipulation of live antigen-expressing cells using conditionally stable nanobodies. eLife 2016, 5, e15312. [Google Scholar] [CrossRef] [PubMed]
- Roebroek, T.; Duwé, S.; Vandenberg, W.; Dedecker, P. Reduced fluorescent protein switching fatigue by binding-induced emissive state stabilization. Int. J. Mol. Sci. 2017, 18, 2015. [Google Scholar] [CrossRef] [Green Version]
- Irannejad, R.; Tomshine, J.C.; Tomshine, J.R.; Chevalier, M.; Mahoney, J.P.; Steyaert, J.; Rasmussen, S.G.; Sunahara, R.K.; El-Samad, H.; Huang, B.; et al. Conformational biosensors reveal gpcr signalling from endosomes. Nature 2013, 495, 534–538. [Google Scholar] [CrossRef] [Green Version]
- Jakobs, B.D.; Spannagel, L.; Purvanov, V.; Uetz-von Allmen, E.; Matti, C.; Legler, D.F. Engineering of nanobodies recognizing the human chemokine receptor ccr7. Int. J. Mol. Sci. 2019, 20, 2597. [Google Scholar] [CrossRef] [Green Version]
- Stoeber, M.; Jullié, D.; Lobingier, B.T.; Laeremans, T.; Steyaert, J.; Schiller, P.W.; Manglik, A.; von Zastrow, M. A genetically encoded biosensor reveals location bias of opioid drug action. Neuron 2018, 98, 963–976.e5. [Google Scholar] [CrossRef] [Green Version]
- Bery, N.; Keller, L.; Soulié, M.; Gence, R.; Iscache, A.-L.; Cherier, J.; Cabantous, S.; Sordet, O.; Lajoie-Mazenc, I.; Pedelacq, J.-D. A targeted protein degradation cell-based screening for nanobodies selective toward the cellular rhob gtp-bound conformation. Cell Chem. Biol. 2019, 26, 1544–1558.e1546. [Google Scholar] [CrossRef]
- Cao, J.; Zhong, N.; Wang, G.; Wang, M.; Zhang, B.; Fu, B.; Wang, Y.; Zhang, T.; Zhang, Y.; Yang, K. Nanobody-based sandwich reporter system for living cell sensing influenza a virus infection. Sci. Rep. 2019, 9, 1–8. [Google Scholar] [CrossRef]
- Böldicke, T. Single domain antibodies for the knockdown of cytosolic and nuclear proteins. Protein Sci. 2017, 26, 925–945. [Google Scholar] [CrossRef] [Green Version]
- Lodish, M.B. Kinase inhibitors: Adverse effects related to the endocrine system. J. Clin. Endocrinol. Metab. 2013, 98, 1333–1342. [Google Scholar] [CrossRef] [Green Version]
- Ferguson, F.M.; Gray, N.S. Kinase inhibitors: The road ahead. Nat. Rev. Drug Discov. 2018, 17, 353. [Google Scholar] [CrossRef] [PubMed]
- Steels, A.; Verhelle, A.; Zwaenepoel, O.; Gettemans, J. Intracellular displacement of p53 using transactivation domain (p53 tad) specific nanobodies. In MAbs; Taylor & Francis: London, UK, 2018; pp. 1045–1059. [Google Scholar] [CrossRef]
- Steels, A.; Vannevel, L.; Zwaenepoel, O.; Gettemans, J. Nb-induced stabilisation of p53 in hpv-infected cells. Sci. Rep. 2019, 9, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gulati, S.; Jin, H.; Masuho, I.; Orban, T.; Cai, Y.; Pardon, E.; Martemyanov, K.A.; Kiser, P.D.; Stewart, P.L.; Ford, C.P. Targeting g protein-coupled receptor signaling at the g protein level with a selective nanobody inhibitor. Nat. Commun. 2018, 9, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Summanen, M.; Granqvist, N.; Tuominen, R.K.; Yliperttula, M.; Verrips, C.T.; Boonstra, J.; Blanchetot, C.; Ekokoski, E. Kinetics of pkcε activating and inhibiting llama single chain antibodies and their effect on pkcε translocation in hela cells. PLoS ONE 2012, 7, e35630. [Google Scholar] [CrossRef]
- Van Audenhove, I.; Boucherie, C.; Pieters, L.; Zwaenepoel, O.; Vanloo, B.; Martens, E.; Verbrugge, C.; Hassanzadeh-Ghassabeh, G.; Vandekerckhove, J.; Cornelissen, M. Stratifying fascin and cortactin function in invadopodium formation using inhibitory nanobodies and targeted subcellular delocalization. FASEB J. 2014, 28, 1805–1818. [Google Scholar] [CrossRef]
- Bertier, L.; Boucherie, C.; Zwaenepoel, O.; Vanloo, B.; Van Troys, M.; Van Audenhove, I.; Gettemans, J. Inhibitory cortactin nanobodies delineate the role of nta-and sh3-domain–specific functions during invadopodium formation and cancer cell invasion. FASEB J. 2017, 31, 2460–2476. [Google Scholar] [CrossRef] [Green Version]
- Bertier, L.; Hebbrecht, T.; Mettepenningen, E.; De Wit, N.; Zwaenepoel, O.; Verhelle, A.; Gettemans, J. Nanobodies targeting cortactin proline rich, helical and actin binding regions downregulate invadopodium formation and matrix degradation in scc-61 cancer cells. Biomed. Pharmacother. 2018, 102, 230–241. [Google Scholar] [CrossRef]
- Hebbrecht, T.; Van Audenhove, I.; Zwaenepoel, O.; Verhelle, A.; Gettemans, J. Vca nanobodies target n-wasp to reduce invadopodium formation and functioning. PLoS ONE 2017, 12, e0185076. [Google Scholar] [CrossRef] [Green Version]
- Van Impe, K.; Bethuyne, J.; Cool, S.; Impens, F.; Ruano-Gallego, D.; De Wever, O.; Vanloo, B.; Van Troys, M.; Lambein, K.; Boucherie, C. A nanobody targeting the f-actin capping protein capg restrains breast cancer metastasis. Breast Cancer Res. 2013, 15, R116. [Google Scholar] [CrossRef] [Green Version]
- Singh, S.; Murillo, G.; Chen, D.; Parihar, A.S.; Mehta, R.G. Suppression of breast cancer cell proliferation by selective single-domain antibody for intracellular stat3. Breast Cancer Basic Clin. Res. 2018, 12, 1178223417750858. [Google Scholar] [CrossRef]
- Schmidt, F.I.; Lu, A.; Chen, J.W.; Ruan, J.; Tang, C.; Wu, H.; Ploegh, H.L. A single domain antibody fragment that recognizes the adaptor asc defines the role of asc domains in inflammasome assembly. J. Exp. Med. 2016, 213, 771–790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Clercq, S.; Zwaenepoel, O.; Martens, E.; Vandekerckhove, J.; Guillabert, A.; Gettemans, J. Nanobody-induced perturbation of lfa-1/l-plastin phosphorylation impairs mtoc docking, immune synapse formation and t cell activation. Cell. Mol. Life Sci. 2013, 70, 909–922. [Google Scholar] [CrossRef] [PubMed]
- De Clercq, S.; Boucherie, C.; Vandekerckhove, J.; Gettemans, J.; Guillabert, A. L-plastin nanobodies perturb matrix degradation, podosome formation, stability and lifetime in thp-1 macrophages. PLoS ONE 2013, 8, e78108. [Google Scholar] [CrossRef] [Green Version]
- Delanote, V.; Vanloo, B.; Catillon, M.; Friederich, E.; Vandekerckhove, J.; Gettemans, J. An alpaca single-domain antibody blocks filopodia formation by obstructing l-plastin-mediated f-actin bundling. FASEB J. 2010, 24, 105–118. [Google Scholar] [CrossRef]
- Van den Abbeele, A.; De Clercq, S.; De Ganck, A.; De Corte, V.; Van Loo, B.; Soror, S.H.; Srinivasan, V.; Steyaert, J.; Vandekerckhove, J.; Gettemans, J. A llama-derived gelsolin single-domain antibody blocks gelsolin–g-actin interaction. Cell. Mol. Life Sci. 2010, 67, 1519–1535. [Google Scholar] [CrossRef]
- Messer, A.; Butler, D.C. Optimizing intracellular antibodies (intrabodies/nanobodies) to treat neurodegenerative disorders. Neurobiol. Dis. 2020, 134, 104619. [Google Scholar] [CrossRef]
- Dong, J.-X.; Lee, Y.; Kirmiz, M.; Palacio, S.; Dumitras, C.; Moreno, C.M.; Sando, R.; Santana, L.F.; Südhof, T.C.; Gong, B. A toolbox of nanobodies developed and validated for use as intrabodies and nanoscale immunolabels in mammalian brain neurons. eLife 2019, 8, e48750. [Google Scholar] [CrossRef]
- Mahajan, S.P.; Meksiriporn, B.; Waraho-Zhmayev, D.; Weyant, K.B.; Kocer, I.; Butler, D.C.; Messer, A.; Escobedo, F.A.; DeLisa, M.P. Computational affinity maturation of camelid single-domain intrabodies against the nonamyloid component of alpha-synuclein. Sci. Rep. 2018, 8, 1–14. [Google Scholar] [CrossRef]
- Guilliams, T.; El-Turk, F.; Buell, A.K.; O’Day, E.M.; Aprile, F.A.; Esbjörner, E.K.; Vendruscolo, M.; Cremades, N.; Pardon, E.; Wyns, L. Nanobodies raised against monomeric α-synuclein distinguish between fibrils at different maturation stages. J. Mol. Biol. 2013, 425, 2397–2411. [Google Scholar] [CrossRef] [Green Version]
- El Turk, F.; De Genst, E.; Guilliams, T.; Fauvet, B.; Hejjaoui, M.; Di Trani, J.; Chiki, A.; Mittermaier, A.; Vendruscolo, M.; Lashuel, H.A. Exploring the role of post-translational modifications in regulating α-synuclein interactions by studying the effects of phosphorylation on nanobody binding. Protein Sci. 2018, 27, 1262–1274. [Google Scholar] [CrossRef] [Green Version]
- Butler, D.C.; Joshi, S.N.; Genst, E.D.; Baghel, A.S.; Dobson, C.M.; Messer, A. Bifunctional anti-non-amyloid component α-synuclein nanobodies are protective in situ. PLoS ONE 2016, 11, e0165964. [Google Scholar] [CrossRef] [Green Version]
- Gueorguieva, D.; Li, S.; Walsh, N.; Mukerji, A.; Tanha, J.; Pandey, S.; Gueorguieva, D.; Li, S.; Walsh, N.; Mukerji, A. Identification of single-domain, bax-specific intrabodies that confer resistance to mammalian cells against oxidative-stress-induced apoptosis. FASEB J. 2006, 20, 2636–2638. [Google Scholar] [CrossRef] [Green Version]
- Morgenstern, T.J.; Park, J.; Fan, Q.R.; Colecraft, H.M. A potent voltage-gated calcium channel inhibitor engineered from a nanobody targeted to auxiliary cavβ subunits. eLife 2019, 8, e49253. [Google Scholar] [CrossRef]
- Schenck, S.; Kunz, L.; Sahlender, D.; Pardon, E.; Geertsma, E.R.; Savtchouk, I.; Suzuki, T.; Neldner, Y.; Štefanić, S.A.; Steyaert, J. Generation and characterization of anti-vglut nanobodies acting as inhibitors of transport. Biochemistry 2017, 56, 3962–3971. [Google Scholar] [CrossRef]
- Dieleman, J.L.; Haakenstad, A.; Micah, A.; Moses, M.; Abbafati, C.; Acharya, P.; Adhikari, T.B.; Adou, A.K.; Kiadaliri, A.A.; Alam, K. Spending on health and hiv/aids: Domestic health spending and development assistance in 188 countries, 1995–2015. Lancet 2018, 391, 1799–1829. [Google Scholar] [CrossRef] [Green Version]
- Vercruysse, T.; Pardon, E.; Vanstreels, E.; Steyaert, J.; Daelemans, D. An intrabody based on a llama single-domain antibody targeting the n-terminal α-helical multimerization domain of hiv-1 rev prevents viral production. J. Biol. Chem. 2010, 285, 21768–21780. [Google Scholar] [CrossRef] [Green Version]
- Boons, E.; Li, G.; Vanstreels, E.; Vercruysse, T.; Pannecouque, C.; Vandamme, A.-M.; Daelemans, D. A stably expressed llama single-domain intrabody targeting rev displays broad-spectrum anti-hiv activity. Antivir. Res. 2014, 112, 91–102. [Google Scholar] [CrossRef]
- Matz, J.; Hérate, C.; Bouchet, J.; Dusetti, N.; Gayet, O.; Baty, D.; Benichou, S.; Chames, P. Selection of intracellular single-domain antibodies targeting the hiv-1 vpr protein by cytoplasmic yeast two-hybrid system. PLoS ONE 2014, 9, e113729. [Google Scholar] [CrossRef] [Green Version]
- Bouchet, J.; Basmaciogullari, S.E.; Chrobak, P.; Stolp, B.; Bouchard, N.; Fackler, O.T.; Chames, P.; Jolicoeur, P.; Benichou, S.; Baty, D. Inhibition of the nef regulatory protein of hiv-1 by a single-domain antibody. Blood 2011, 117, 3559–3568. [Google Scholar] [CrossRef] [Green Version]
- Bouchet, J.; Hérate, C.; Guenzel, C.A.; Vérollet, C.; Järviluoma, A.; Mazzolini, J.; Rafie, S.; Chames, P.; Baty, D.; Saksela, K. Single-domain antibody-sh3 fusions for efficient neutralization of hiv-1 nef functions. J. Virol. 2012, 86, 4856–4867. [Google Scholar] [CrossRef] [Green Version]
- Ashour, J.; Schmidt, F.I.; Hanke, L.; Cragnolini, J.; Cavallari, M.; Altenburg, A.; Brewer, R.; Ingram, J.; Shoemaker, C.; Ploegh, H.L. Intracellular expression of camelid single-domain antibodies specific for influenza virus nucleoprotein uncovers distinct features of its nuclear localization. J. Virol. 2015, 89, 2792–2800. [Google Scholar] [CrossRef] [Green Version]
- Hanke, L.; Knockenhauer, K.E.; Brewer, R.C.; van Diest, E.; Schmidt, F.I.; Schwartz, T.U.; Ploegh, H.L. The antiviral mechanism of an influenza a virus nucleoprotein-specific single-domain antibody fragment. MBio 2016, 7, e01569-16. [Google Scholar] [CrossRef] [Green Version]
- Glab-Ampai, K.; Malik, A.A.; Chulanetra, M.; Thanongsaksrikul, J.; Thueng-In, K.; Srimanote, P.; Tongtawe, P.; Chaicumpa, W. Inhibition of hcv replication by humanized-single domain transbodies to ns4b. Biochem. Biophys. Res. Commun. 2016, 476, 654–664. [Google Scholar] [CrossRef]
- Jittavisutthikul, S.; Thanongsaksrikul, J.; Thueng-In, K.; Chulanetra, M.; Srimanote, P.; Seesuay, W.; Malik, A.A.; Chaicumpa, W. Humanized-vhh transbodies that inhibit hcv protease and replication. Viruses 2015, 7, 2030–2056. [Google Scholar] [CrossRef] [Green Version]
- Thueng-In, K.; Thanongsaksrikul, J.; Srimanote, P.; Bangphoomi, K.; Poungpair, O.; Maneewatch, S.; Choowongkomon, K.; Chaicumpa, W. Cell penetrable humanized-vh/vhh that inhibit rna dependent rna polymerase (ns5b) of hcv. PLoS ONE 2012, 7, e49254. [Google Scholar] [CrossRef] [Green Version]
- Serruys, B.; Van Houtte, F.; Farhoudi-Moghadam, A.; Leroux-Roels, G.; Vanlandschoot, P. Production, characterization and in vitro testing of hbcag-specific vhh intrabodies. J. Gen. Virol. 2010, 91, 643–652. [Google Scholar] [CrossRef]
- Alzogaray, V.; Danquah, W.; Aguirre, A.; Urrutia, M.; Berguer, P.; Véscovi, E.G.; Haag, F.; Koch-Nolte, F.; Goldbaum, F.A. Single-domain llama antibodies as specific intracellular inhibitors of spvb, the actin adp-ribosylating toxin of salmonella typhimurium. FASEB J. 2011, 25, 526–534. [Google Scholar] [CrossRef]
- Tremblay, J.M.; Kuo, C.-L.; Abeijon, C.; Sepulveda, J.; Oyler, G.; Hu, X.; Jin, M.M.; Shoemaker, C.B. Camelid single domain antibodies (vhhs) as neuronal cell intrabody binding agents and inhibitors of clostridium botulinum neurotoxin (bont) proteases. Toxicon 2010, 56, 990–998. [Google Scholar] [CrossRef] [Green Version]
- Yu, D.; Lee, H.; Hong, J.; Jung, H.; Jo, Y.; Oh, B.-H.; Park, B.O.; Do Heo, W. Optogenetic activation of intracellular antibodies for direct modulation of endogenous proteins. Nat. Methods 2019, 16, 1095–1100. [Google Scholar] [CrossRef]
- Gil, A.A.; Carrasco-López, C.; Zhu, L.; Zhao, E.M.; Ravindran, P.T.; Wilson, M.Z.; Goglia, A.G.; Avalos, J.L.; Toettcher, J.E. Optogenetic control of protein binding using light-switchable nanobodies. Nat. Commun. 2020, 11, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Farrants, H.; Tarnawski, M.; Müller, T.G.; Otsuka, S.; Hiblot, J.; Koch, B.; Kueblbeck, M.; Kräusslich, H.-G.; Ellenberg, J.; Johnsson, K. Chemogenetic control of nanobodies. Nat. Methods 2020, 17, 279–282. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wagner, T.R.; Rothbauer, U. Nanobodies Right in the Middle: Intrabodies as Toolbox to Visualize and Modulate Antigens in the Living Cell. Biomolecules 2020, 10, 1701. https://doi.org/10.3390/biom10121701
Wagner TR, Rothbauer U. Nanobodies Right in the Middle: Intrabodies as Toolbox to Visualize and Modulate Antigens in the Living Cell. Biomolecules. 2020; 10(12):1701. https://doi.org/10.3390/biom10121701
Chicago/Turabian StyleWagner, Teresa R., and Ulrich Rothbauer. 2020. "Nanobodies Right in the Middle: Intrabodies as Toolbox to Visualize and Modulate Antigens in the Living Cell" Biomolecules 10, no. 12: 1701. https://doi.org/10.3390/biom10121701
APA StyleWagner, T. R., & Rothbauer, U. (2020). Nanobodies Right in the Middle: Intrabodies as Toolbox to Visualize and Modulate Antigens in the Living Cell. Biomolecules, 10(12), 1701. https://doi.org/10.3390/biom10121701





