Interactions between Avibactam and Ceftazidime-Hydrolyzing Class D β-Lactamases
Abstract
:1. Introduction
2. Materials and Methods
2.1. Compounds
2.2. Enzymes
2.3. Kinetic Measurements
2.4. Mass Spectrometry
3. Results
3.1. OXA-24
3.1.1. Interaction with Ceftazidime and Nitrocefin
3.1.2. Inactivation by Avibactam in the Presence of 70 µM Nitrocefin
3.1.3. Reactivation Rate
3.1.4. Global Equilibrium Constant
3.2. OXA-163
3.2.1. Interaction of OXA-163 with Ceftazidime
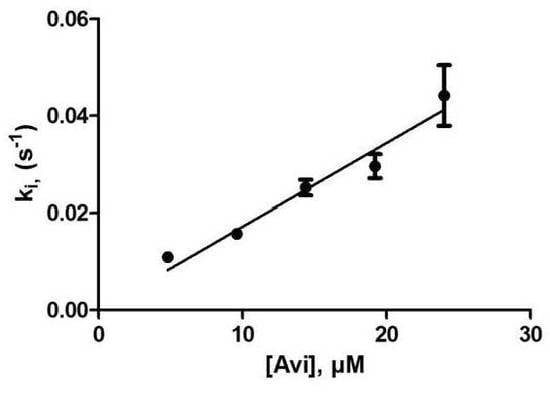
3.2.2. Titration by Avibactam
3.2.3. Inactivation of OXA-163 by Avibactam in the Presence of 80 µM Ceftazidime
3.2.4. Reactivation Rate
3.2.5. Global Equilibrium Constant
3.3. OXA-427
3.3.1. Hydrolysis of Ceftazidime and Cephalothin
Ceftazidime
Cephalothin
3.3.2. Inactivation by Avibactam with Ceftazidime as Reporter Substrate
In the Absence of NaHCO3
In the Presence of 40 mM NaHCO3
3.3.3. With Cephalothin as a Reporter Substrate
3.3.4. Reactivation
4. Mass Spectrometry
4.1. OXA-427
4.2. OXA-163
5. Discussion
6. Conclusion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A


| Amount of Avibactam (nmol) | Residual Activity (%) |
|---|---|
| 0 | 100 |
| 0.025 | 74 |
| 0.05 | 49 |
| 0.075 | 24 |
| 0.1 | <2 |
References
- Bush, K. Past and present perspectives on β-lactamases. Antimicrob. Agents Chemother. 2018, 62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bush, K.; Bradford, P.A. Interplay between β-lactamases and β-lactamase inhibitors. Nat. Rev. Microbiol. 2019, 17, 295–306. [Google Scholar] [CrossRef] [PubMed]
- Frère, J.M. (Ed.) Β-Lactamases; Nova Science Publishers: New York, NY, USA, 2012. [Google Scholar]
- Bunyak, J. β-Lactams as Unhibitors of β-Lactamases in Β-Lactamases; Frère, J.M., Ed.; Nova Science Publishers: New York, NY, USA, 2012; pp. 217–258. [Google Scholar]
- Prosperi-Meys, C.; Llabres, G.; de Seny, D.; Soto, R.P.; Valladares, M.H.; Frère, J.M.; Galleni, M. Interaction between class B β-lactamases and suicide substrates of active-site serine β-lactamases. FEBS Lett. 1999, 443, 109–111. [Google Scholar] [CrossRef] [Green Version]
- Dortet, L.; Oueslati, S.; Jeannot, K.; Tandé, D.; Naas, T.; Nordmann, P. Genetic and biochemical characterization of OXA-405, an OXA-48-type extended-spectrum β-lactamase without significant carbapenemase activity. Antimicrob. Agents Chemother. 2015, 59, 3823–3828. [Google Scholar] [CrossRef] [Green Version]
- Oueslati, S.; Nordmann, P.; Poirel, L. Heterogeneous hydrolytic features for OXA- 48-like β-lactamases. J. Antimicrob. Chemother. 2015, 70, 1059–1063. [Google Scholar] [CrossRef] [Green Version]
- Poirel, L.; Castanheira, M.; Carrër, A.; Rodriguez, C.P.; Jones, R.N.; Smayevsky, J.; Nordmann, P.L. OXA-163, an OXA-48-related class D β-lactamase with extended activity toward expanded-spectrum cephalosporins. Antimicrob. Agents Chemother. 2011, 55, 2546–2551. [Google Scholar] [CrossRef] [Green Version]
- Dortet, L.; Naas, T. Noncarbapenemase OXA-48 Variants (OXA-163 and OXA-405) Falsely Detected as Carbapenemases by the β Carba Test. J. Clin. Microbiol. 2017, 55, 654–655. [Google Scholar] [CrossRef] [Green Version]
- Arlet, G.; Decré, D.; Lavollay, M.; Podglajen, I. Reply to “Noncarbapenemase OXA-48 Variants (OXA-163 and OXA-405) Falsely Detected as Carbapenemases by the β Carba Test”. J. Clin. Microbiol. 2017, 55, 656–657. [Google Scholar] [CrossRef] [Green Version]
- Pasteran, F.; Denorme, L.; Ote, I.; Gomez, S.; De Belder, D.; Glupczynski, Y.; Bogaerts, P.; Ghiglione, B.; Power, P.; Mertens, P.; et al. Rapid Identification of OXA-48 and OXA-163 Subfamilies in Carbapenem-Resistant Gram-Negative Bacilli with a Novel Immunochromatographic Lateral Flow Assay. J. Clin. Microbiol. 2016, 54, 2832–2836. [Google Scholar] [CrossRef] [Green Version]
- Abdelaziz, M.O.; Bonura, C.; Aleo, A.; El-Domany, R.A.; Fasciana, T.; Mammina, C. OXA-163-producing Klebsiella pneumoniae in Cairo, Egypt, in 2009 and 2010. J. Clin. Microbiol. 2012, 50, 2489–2491. [Google Scholar] [CrossRef] [Green Version]
- Bogaerts, P.; Naas, T.; Saegeman, V.; Bonnin, R.A.; Schuermans, A.; Evrard, S.; Bouchahrouf, W.; Jove, T.; Tande, D.; de Bolle, X.; et al. OXA-427, a new plasmid-borne carbapenem-hydrolysing class D β-lactamase in Enterobacteriaceae. J. Antimicrob. Chemother. 2017, 72, 2469–2477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Desmet, S.; Nepal, S.; van Dijl, J.M.; Van Ranst, M.; Chlebowicz, M.A.; Rossen, J.W.; Van Houdt, J.K.J.; Maes, P.; Lagrou, K.; Bathoorn, E. Antibiotic Resistance Plasmids Cointegrated into a Megaplasmid Harboring the blaOXA-427 Carbapenemase Gene. Antimicrob. Agents Chemother. 2018, 62, e01448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Laveleye, M.; Huang, T.D.; Bogaerts, P.; Berhin, C.; Bauraing, C.; Sacré, P.; Noel, A.; Glupczynski, Y. multicenter study group. Increasing incidence of carbapenemase-producing Escherichia coli and Klebsiella pneumoniae in Belgian hospitals. Eur. J. Clin. Microbiol. Infect. Dis. 2017, 36, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Ehmann, D.E.; Jahic, H.; Ross, P.L.; Gu, R.F.; Hu, J.; Durand-Réville, T.F.; Lahiri, S.; Thresher, J.; Livchak, S.; Gao, N.; et al. Kinetics of avibactam inhibition against Class A, C, and D β-lactamases. J. Biol. Chem. 2013, 288, 27960–27971. [Google Scholar] [CrossRef] [Green Version]
- Lahiri, S.D.; Mangani, S.; Jahić, H.; Benvenuti, M.; Durand-Reville, T.F.; De Luca, F.; Ehmann, D.E.; Rossolini, G.M.; Alm, R.A.; Docquier, J.D. Molecular basis of selective inhibition and slow reversibility of avibactam against class D carbapenemases: A structure-guided study of OXA-24 and OXA-48. ACS Chem. Biol. 2015, 10, 591–600. [Google Scholar] [CrossRef]
- Bou, G.; Oliver, A.; Martínez-Beltrán, J. OXA-24, a novel class D β-lactamase with carbapenemase activity in an Acinetobacter baumannii clinical strain. Antimicrob. Agents Chemother. 2000, 44, 1556–1561. [Google Scholar] [CrossRef] [Green Version]
- De Meester, F.B.; Joris, B.; Reckinger, G.; Bellefroid-Bourguignon, C.; Frère, J.M.; Waley, S.G. Automated analysis of enzyme inactivation phenomena. Application to -lactamases and D-peptidases. Biochem. Pharmacol. 1987, 36, 2393–2403. [Google Scholar] [CrossRef]
- Nukaga, M.; Papp-Wallace, K.M.; Hoshino, T.; Lefurgy, S.T.; Bethel, C.R.; Barnes, M.D.; Zeiser, E.T.; Johnson, J.K.; Bonomo, R.A. Probing the Mechanism of Inactivation of the FOX-4 Cephamycinase by Avibactam. Antimicrob. Agents Chemother. 2018, 62, e02371. [Google Scholar] [CrossRef] [Green Version]
- Ruggiero, M.; Papp-Wallace, K.M.; Taracila, M.A.; Mojica, M.F.; Bethel, C.R.; Rudin, S.D.; Zeiser, E.T.; Gutkind, G.; Bonomo, R.A.; Power, P. Exploring the Landscape of Diazabicyclooctane (DBO) Inhibition: Avibactam Inactivation of PER-2 β-Lactamase. Antimicrob. Agents Chemother. 2017, 61. [Google Scholar] [CrossRef] [Green Version]
- Ehmann, D.E.; Jahić, H.; Ross, P.L.; Gu, R.F.; Hu, J.; Kern, G.; Walkup, G.K.; Fisher, S.L. Avibactam is a covalent, reversible, non-β-lactam β-lactamase inhibitor. Proc. Natl. Acad. Sci. USA 2012, 109, 11663–11668. [Google Scholar] [CrossRef] [Green Version]
- Yoshizumi, A.; Ishii, Y.; Aoki, K.; Testa, R.; Nichols, W.W.; Tateda, K. In vitro susceptibility of characterized β-lactamase-producing Gram-negative bacteria isolated in Japan to ceftazidime-, ceftaroline-, and aztreonam-avibactam combinations. J. Infect. Chemother. 2015, 21, 148–151. [Google Scholar] [CrossRef] [PubMed]
- Vázquez-Ucha, J.C.; Maneiro, M.; Martínez-Guitián, M.; Buynak, J.; Bethel, C.R.; Bonomo, R.A.; Bou, G.; Poza, M.; González-Bello, C.; Beceiro, A. Activity of the β-Lactamase Inhibitor LN-1-255 against Carbapenem-Hydrolyzing Class D β-Lactamases from Acinetobacter baumannii. Antimicrob. Agents Chemother. 2017, 61, e01172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hall, L.M.; Livermore, D.M.; Gur, D.; Akova, M.; Akalin, H.E. OXA-11, an extended- spectrum variant of OXA-10 (PSE-2) β-lactamase from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 1993, 37, 1637–1644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaitany, K.C.; Klinger, N.V.; June, C.M.; Ramey, M.E.; Bonomo, R.A.; Powers, R.A.; Leonard, D.A. Structures of the class D Carbapenemases OXA-23 and OXA-146: Mechanistic basis of activity against carbapenems, extended-spectrum cephalosporins, and aztreonam. Antimicrob. Agents Chemother. 2013, 57, 4848–4855. [Google Scholar] [CrossRef] [PubMed] [Green Version]





| Enzyme | Interaction with | Interaction with Ceftazidime | References | ||
|---|---|---|---|---|---|
| k2/K (M−1s−1) | t1/2 (s) † | k−2 (s−1) * | kcat/Km (M−1s−1) | ||
| OXA-10 | 11 ± 1 | 7000 | <0.16 × 10−5 | ND # | [16,25] |
| OXA-23 | 300 ± 20 | 230 | (0.8 ± 0.4) × 10−5 | ND | [17,26] |
| OXA-24 | 50 ± 5 | 1400 | (0.3-0.6) × 10−5 | <8 M−1s−1 | This work, [17] |
| OXA-48 | 1400 ± 100 | 50 | (1.2 ± 0.4) × 10−5 | ND | [16] |
| OXA-163 | 1720 ± 75 | 40 | (0.5 ± 0.4) × 10−5 | 57,000 ± 5000 | This work |
| OXA-427 | 870 ± 200 | 80 | (1.3 ± 0.7) × 10−5 | 85,000 ± 13,000 | This work |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Frère, J.-M.; Bogaerts, P.; Huang, T.-D.; Stefanic, P.; Moray, J.; Bouillenne, F.; Brans, A. Interactions between Avibactam and Ceftazidime-Hydrolyzing Class D β-Lactamases. Biomolecules 2020, 10, 483. https://doi.org/10.3390/biom10030483
Frère J-M, Bogaerts P, Huang T-D, Stefanic P, Moray J, Bouillenne F, Brans A. Interactions between Avibactam and Ceftazidime-Hydrolyzing Class D β-Lactamases. Biomolecules. 2020; 10(3):483. https://doi.org/10.3390/biom10030483
Chicago/Turabian StyleFrère, Jean-Marie, Pierre Bogaerts, Te-Din Huang, Patrick Stefanic, Joël Moray, Fabrice Bouillenne, and Alain Brans. 2020. "Interactions between Avibactam and Ceftazidime-Hydrolyzing Class D β-Lactamases" Biomolecules 10, no. 3: 483. https://doi.org/10.3390/biom10030483
APA StyleFrère, J.-M., Bogaerts, P., Huang, T.-D., Stefanic, P., Moray, J., Bouillenne, F., & Brans, A. (2020). Interactions between Avibactam and Ceftazidime-Hydrolyzing Class D β-Lactamases. Biomolecules, 10(3), 483. https://doi.org/10.3390/biom10030483