Notch3 in Development, Health and Disease
Abstract
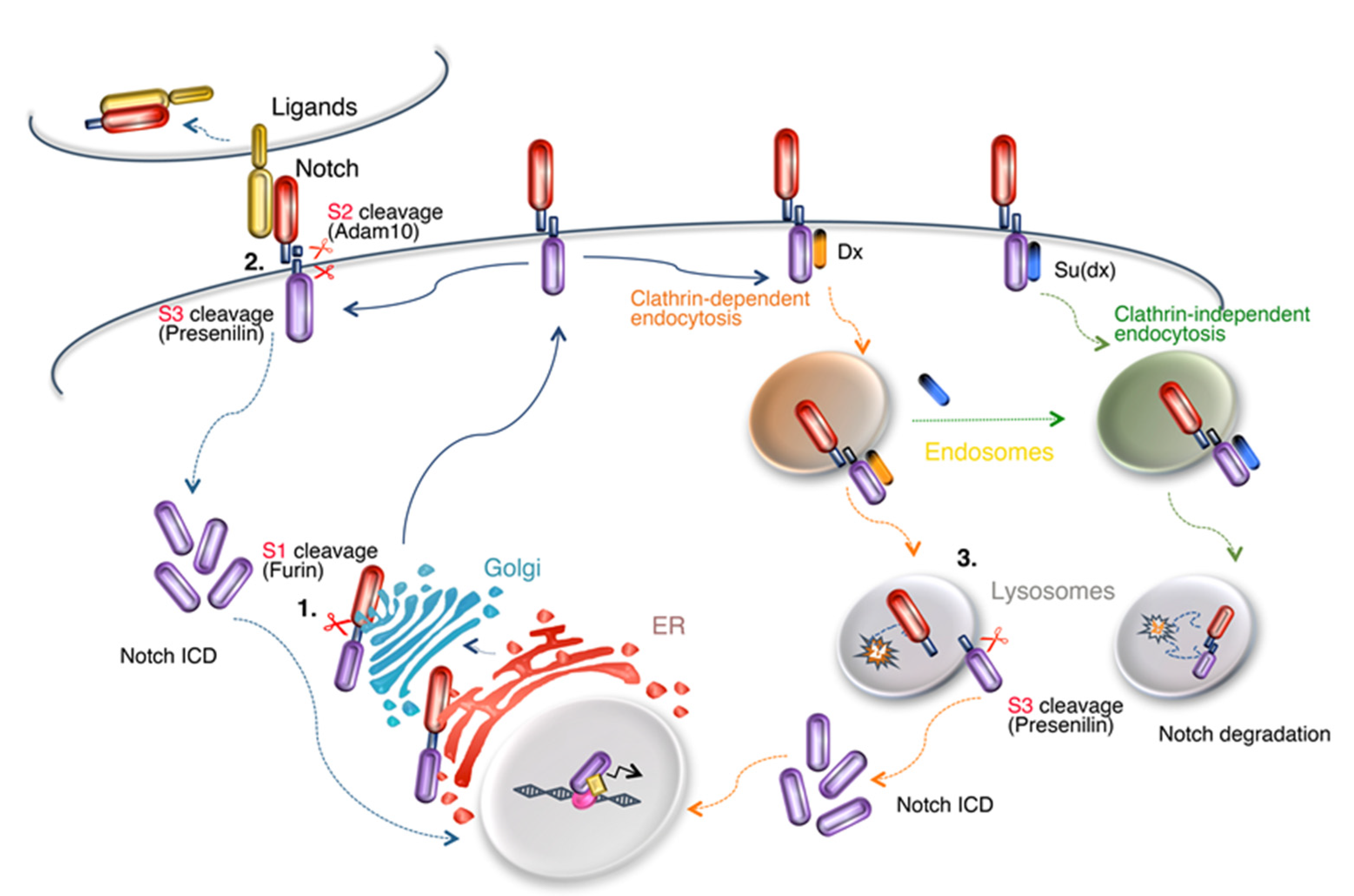
1. Introduction
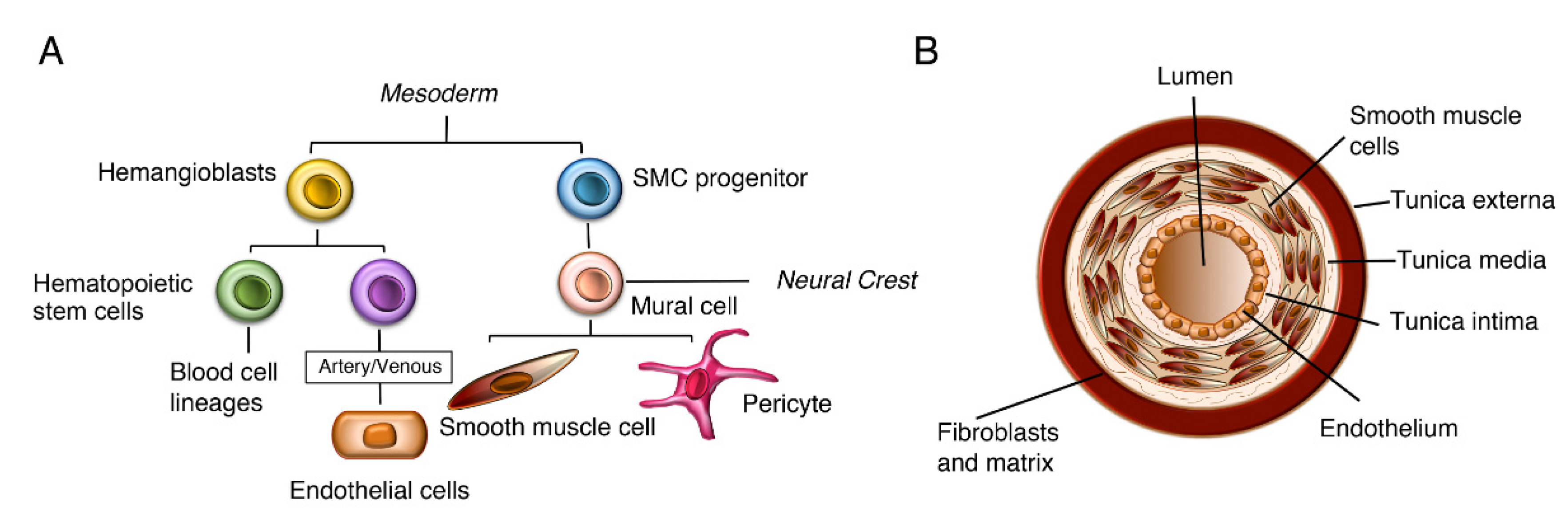
2. Developmental Roles of Notch3
3. Roles of Notch3 in the Adult Organism
4. Notch3 and Disease
4.1. Notch3 and Cancer
4.2. Notch3 and Cerebral Autosomal Dominant Arteriopathy with Subcortical Infarcts and Leukoencephalopathy (CADASIL)
4.3. Notch3 and Pulmonary Hypertension
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Kopan, R.; Ilagan, M.X. The canonical Notch signaling pathway: Unfolding the activation mechanism. Cell 2009, 137, 216–233. [Google Scholar] [CrossRef] [PubMed]
- Bray, S.J. Notch signalling in context. Nat. Rev. Mol. Cell Biol. 2016, 17, 722–735. [Google Scholar] [CrossRef] [PubMed]
- Baron, M. Combining genetic and biophysical approaches to probe the structure and function relationships of the Notch receptor. Mol. Membr. Biol. 2017, 34, 33–49. [Google Scholar] [CrossRef] [PubMed]
- Wharton, K.A.; Johansen, K.M.; Xu, T.; Artavanis-Tsakonas, S. Nucleotide sequence from the neurogenic locus Notch implies a gene product that shares homology with proteins containing EGF-like repeats. Cell 1985, 43, 567–581. [Google Scholar] [CrossRef]
- Bellavia, D.; Checquolo, S.; Campese, A.F.; Felli, M.P.; Gulino, A.; Screpanti, I. Notch3: From subtle structural differences to functional diversity. Oncogene 2008, 27, 5092–5098. [Google Scholar] [CrossRef]
- Hori, K.; Sen, A.; Artavanis-Tsakonas, S. Notch signaling at a glance. J. Cell Sci. 2013, 126, 2135–2140. [Google Scholar] [CrossRef]
- Gordon, W.R.; Vardar-Ulu, D.; Histen, G.; Sanchez-Irizarry, C.; Aster, J.C.; Blacklow, S.C. Structural basis for autoinhibition of Notch. Nat. Struct. Mol. Biol. 2007, 14, 295–300. [Google Scholar] [CrossRef]
- Gordon, W.R.; Vardar-Ulu, D.; L’Heureux, S.; Ashworth, T.; Malecki, M.J.; Sanchez-Irizarry, C.; McArthur, D.G.; Histen, G.; Mitchell, J.L.; Aster, J.C.; et al. Effects of S1 cleavage on the structure, surface export, and signaling activity of human Notch1 and Notch2. PLoS ONE 2009, 4, e6613. [Google Scholar] [CrossRef]
- Stephenson, N.L.; Avis, J.M. Direct observation of proteolytic cleavage at the S2 site upon forced unfolding of the Notch negative regulatory region. Proc. Natl. Acad. Sci. USA 2012, 109, E2757–E2765. [Google Scholar] [CrossRef]
- Gordon, W.R.; Zimmerman, B.; He, L.; Miles, L.J.; Huang, J.; Tiyanont, K.; McArthur, D.G.; Aster, J.C.; Perrimon, N.; Loparo, J.J.; et al. Mechanical allostery: Evidence for a force requirement in the proteolytic activation of Notch. Dev. Cell 2015, 33, 729–736. [Google Scholar] [CrossRef]
- Struhl, G.; Adachi, A. Requirements for presenilin-dependent cleavage of Notch and other transmembrane proteins. Mol. Cell 2000, 6, 625–636. [Google Scholar] [CrossRef]
- Wilkin, M.B.; Carbery, A.M.; Fostier, M.; Aslam, H.; Mazaleyrat, S.L.; Higgs, J.; Myat, A.; Evans, D.A.; Cornell, M.; Baron, M. Regulation of Notch endosomal sorting and signaling by Drosophila Nedd4 family proteins. Curr. Biol. 2004, 14, 2237–2244. [Google Scholar] [CrossRef] [PubMed]
- Wilkin, M.; Tongngok, P.; Gensch, N.; Clemence, S.; Motoki, M.; Yamada, K.; Hori, K.; Taniguchi-Kanai, M.; Franklin, E.; Matsuno, K.; et al. Drosophila HOPS and AP-3 complex genes are required for a Deltex-regulated activation of notch in the endosomal trafficking pathway. Dev. Cell 2008, 15, 762–772. [Google Scholar] [CrossRef] [PubMed]
- Hori, K.; Sen, A.; Kirchhausen, T.; Artavanis-Tsakonas, S. Synergy between the ESCRT-III complex and Deltex defines a ligand-independent Notch signal. J. Cell Biol. 2011, 195, 1005–1015. [Google Scholar] [CrossRef]
- Mukherjee, T.; Kim, W.S.; Mandal, L.; Banerjee, U. Interaction between Notch and Hif-alpha in development and survival of Drosophila blood cells. Science 2011, 332, 1210–1213. [Google Scholar] [CrossRef]
- Zheng, L.; Saunders, C.A.; Sorensen, E.B.; Waxmonsky, N.C.; Conner, S.D. Notch signaling from the endosome requires a conserved dileucine motif. Mol. Biol. Cell 2013, 24, 297–307. [Google Scholar] [CrossRef]
- Nemetschke, L.; Knust, E. Drosophila Crumbs prevents ectopic Notch activation in developing wings by inhibiting ligand-independent endocytosis. Development 2016, 143, 4543–4553. [Google Scholar] [CrossRef]
- Steinbuck, M.P.; Winandy, S. A review of notch processing with new insights into ligand-independent notch signaling in T-Cells. Front. Immunol. 2018, 9, 1230. [Google Scholar] [CrossRef]
- Schneider, M.; Troost, T.; Grawe, F.; Martinez-Arias, A.; Klein, T. Activation of Notch in lgd mutant cells requires the fusion of late endosomes with the lysosome. J. Cell Sci. 2013, 126, 645–656. [Google Scholar] [CrossRef]
- Shimizu, H.; Woodcock, S.A.; Wilkin, M.B.; Trubenová, B.; Monk, N.A.; Baron, M. Compensatory flux changes within an endocytic trafficking network maintain thermal robustness of Notch signaling. Cell 2014, 157, 1160–1174. [Google Scholar] [CrossRef]
- Shimizu, K.; Chiba, S.; Saito, T.; Kumano, K.; Hirai, H. Physical interaction of Delta1, Jagged1, and Jagged2 with Notch1 and Notch3 receptors. Biochem. Biophys. Res. Commun. 2000, 276, 385–389. [Google Scholar] [CrossRef] [PubMed]
- Peters, N.; Opherk, C.; Zacherle, S.; Capell, A.; Gempel, P.; Dichgans, M. CADASIL-associated Notch3 mutations have differential effects both on ligand binding and ligand-induced Notch3 receptor signaling through RBP-Jk. Exp. Cell Res. 2004, 299, 454–464. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Kennard, S.; Lilly, B. NOTCH3 expression is induced in mural cells through an autoregulatory loop that requires endothelial-expressed JAGGED1. Circ. Res. 2009, 104, 466–475. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Bhattacharyya, A.; Roszell, E.E.; Sandig, M.; Mequanint, K. The role of endothelial cell-bound Jagged1 in Notch3-induced human coronary artery smooth muscle cell differentiation. Biomaterials 2012, 33, 2462–2472. [Google Scholar] [CrossRef]
- Xu, X.; Choi, S.H.; Hu, T.; Tiyanont, K.; Habets, R.; Groot, A.J.; Vooijs, M.; Aster, J.C.; Chopra, R.; Fryer, C.; et al. Insights into autoregulation of notch3 from structural and functional studies of its negative regulatory region. Structure 2015, 23, 1227–1235. [Google Scholar] [CrossRef]
- Rauen, T.; Raffetseder, U.; Frye, B.C.; Djudjaj, S.; Mühlenberg, P.J.; Eitner, F.; Lendahl, U.; Bernhagen, J.; Dooley, S.; Mertens, P.R. YB-1 acts as a ligand for Notch-3 receptors and modulates receptor activation. J. Biol. Chem. 2009, 284, 26928–26940. [Google Scholar] [CrossRef]
- Gera, S.; Dighe, R.R. The soluble ligand Y box-1 activates Notch3 receptor by binding to epidermal growth factor like repeats 20–23. Arch. Biochem. Biophys. 2018, 660, 129–136. [Google Scholar] [CrossRef]
- Krebs, L.T.; Xue, Y.; Norton, C.R.; Sundberg, J.P.; Beatus, P.; Lendahl, U.; Joutel, A.; Gridley, T. Characterization of Notch3-deficient mice: Normal embryonic development and absence of genetic interactions with a Notch1 mutation. Genesis 2003, 37, 139–143. [Google Scholar] [CrossRef]
- Carmeliet, P. Angiogenesis in health and disease. Nat. Med. 2003, 9, 653–660. [Google Scholar] [CrossRef]
- Domenga, V.; Fardoux, P.; Lacombe, P.; Monet, M.; Maciazek, J.; Krebs, L.T.; Klonjkowski, B.; Berrou, E.; Mericskay, M.; Li, Z.; et al. Notch3 is required for arterial identity and maturation of vascular smooth muscle cells. Genes Dev. 2004, 18, 2730–2735. [Google Scholar] [CrossRef]
- Jin, S.; Hansson, E.M.; Tikka, S.; Lanner, F.; Sahlgren, C.; Farnebo, F.; Baumann, M.; Kalimo, H.; Lendahl, U. Notch signaling regulates platelet-derived growth factor receptor-beta expression in vascular smooth muscle cells. Circ. Res. 2008, 102, 1483–1491. [Google Scholar] [CrossRef] [PubMed]
- Ando, K.; Wang, W.; Peng, D.; Chiba, A.; Lagendijk, A.K.; Barske, L.; Crump, J.G.; Stainier, D.Y.R.; Lendahl, U.; Koltowska, K.; et al. Peri-arterial specification of vascular mural cells from naïve mesenchyme requires Notch signaling. Development 2019, 146, dev165589. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Zhao, N.; Kennard, S.; Lilly, B. Notch2 and Notch3 function together to regulate vascular smooth muscle development. PLoS ONE 2012, 7, e37365. [Google Scholar] [CrossRef] [PubMed]
- Baeten, J.T.; Lilly, B. Differential regulation of NOTCH2 and NOTCH3 contribute to their unique functions in vascular smooth muscle cells. J. Biol. Chem. 2015, 290, 16226–16237. [Google Scholar] [CrossRef] [PubMed]
- Boucher, J.M.; Harrington, A.; Rostama, B.; Lindner, V.; Liaw, L. A receptor-specific function for Notch2 in mediating vascular smooth muscle cell growth arrest through cyclin-dependent kinase inhibitor 1B. Circ. Res. 2013, 113, 975–985. [Google Scholar] [CrossRef]
- Kofler, N.M.; Cuervo, H.; Uh, M.K.; Murtomäki, A.; Kitajewski, J. Combined deficiency of Notch1 and Notch3 causes pericyte dysfunction, models CADASIL, and results in arteriovenous malformations. Sci. Rep. 2015, 5, 16449. [Google Scholar] [CrossRef]
- Wang, Y.; Pan, L.; Moens, C.B.; Appel, B. Notch3 establishes brain vascular integrity by regulating pericyte number. Development 2014, 141, 307–317. [Google Scholar] [CrossRef]
- Volz, K.S.; Jacobs, A.H.; Chen, H.I.; Poduri, A.; McKay, A.S.; Riordan, D.P.; Kofler, N.; Kitajewski, J.; Weissman, I.; Red-Horse, K. Pericytes are progenitors for coronary artery smooth muscle. Elife 2015, 4, e10036. [Google Scholar] [CrossRef]
- Qiu, X.; Lim, C.H.; Ho, S.H.; Lee, K.H.; Jiang, Y.J. Temporal Notch activation through Notch1a and Notch3 is required for maintaining zebrafish rhombomere boundaries. Dev. Genes Evol. 2009, 219, 339–351. [Google Scholar] [CrossRef]
- Belin de Chantemèle, E.J.; Retailleau, K.; Pinaud, F.; Vessières, E.; Bocquet, A.; Guihot, A.L.; Lemaire, B.; Domenga, V.; Baufreton, C.; Loufrani, L.; et al. Notch3 is a major regulator of vascular tone in cerebral and tail resistance arteries. Arterioscler. Thromb. Vasc. Biol. 2008, 28, 2216–2224. [Google Scholar] [CrossRef] [PubMed]
- Frismantiene, A.; Philippova, M.; Erne, P.; Resink, T.J. Smooth muscle cell-driven vascular diseases and molecular mechanisms of VSMC plasticity. Cell Signal. 2018, 52, 48–64. [Google Scholar] [CrossRef]
- Loerakker, S.; Stassen, O.M.J.A.; Ter Huurne, F.M.; Boareto, M.; Bouten, C.V.C.; Sahlgren, C.M. Mechanosensitivity of Jagged-Notch signaling can induce a switch-type behavior in vascular homeostasis. Proc. Natl. Acad. Sci. USA 2018, 115, E3682–E3691. [Google Scholar] [CrossRef] [PubMed]
- Driessen, R.C.H.; Stassen, O.M.J.A.; Sjöqvist, M.; Suarez Rodriguez, F.; Grolleman, J.; Bouten, C.V.C.; Sahlgren, C.M. Shear stress induces expression, intracellular reorganization and enhanced Notch activation potential of Jagged1. Integr. Biol. 2018, 10, 719–726. [Google Scholar] [CrossRef] [PubMed]
- Kawai, H.; Kawaguchi, D.; Kuebrich, B.D.; Kitamoto, T.; Yamaguchi, M.; Gotoh, Y.; Furutachi, S. Area-Specific Regulation of Quiescent Neural Stem Cells by Notch3 in the Adult Mouse Subependymal Zone. J. Neurosci. 2017, 37, 11867–11880. [Google Scholar] [CrossRef] [PubMed]
- Alunni, A.; Krecsmarik, M.; Bosco, A.; Galant, S.; Pan, L.; Moens, C.B.; Bally-Cuif, L. Notch3 signaling gates cell cycle entry and limits neural stem cell amplification in the adult pallium. Development 2013, 140, 3335–3347. [Google Scholar] [CrossRef] [PubMed]
- Kitamoto, T.; Hanaoka, K. Notch3 null mutation in mice causes muscle hyperplasia by repetitive muscle regeneration. Stem Cells 2010, 28, 2205–2216. [Google Scholar] [CrossRef]
- Conboy, I.M.; Rando, T.A. The regulation of Notch signaling controls satellite cell activation and cell fate determination in postnatal myogenesis. Dev. Cell 2002, 3, 397–409. [Google Scholar] [CrossRef]
- Lafkas, D.; Rodilla, V.; Huyghe, M.; Mourao, L.; Kiaris, H.; Fre, S. Notch3 marks clonogenic mammary luminal progenitor cells in vivo. J. Cell Biol. 2013, 203, 47–56. [Google Scholar] [CrossRef]
- Ohashi, S.; Natsuizaka, M.; Yashiro-Ohtani, Y.; Kalman, R.A.; Nakagawa, M.; Wu, L.; Klein-Szanto, A.J.; Herlyn, M.V.; Diehl, J.A.; Katz, J.P.; et al. NOTCH1 and NOTCH3 coordinate esophageal squamous differentiation through a CSL-dependent transcriptional network. Gastroenterology 2010, 139, 2113–2123. [Google Scholar] [CrossRef]
- Ellisen, L.W.; Bird, J.; West, D.C.; Soreng, A.L.; Reynolds, T.C.; Smith, S.D.; Sklar, J. TAN-1, the human homolog of the Drosophila Notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell 1991, 66, 649–661. [Google Scholar] [CrossRef]
- Weng, A.P.; Ferrando, A.A.; Lee, W.; Morris, J.P., 4th; Silverman, L.B.; Sanchez-Irizarry, C.; Blacklow, S.C.; Look, A.T.; Aster, J.C. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science 2004, 306, 269–271. [Google Scholar] [CrossRef] [PubMed]
- Bernasconi-Elias, P.; Hu, T.; Jenkins, D.; Firestone, B.; Gans, S.; Kurth, E.; Capodieci, P.; Deplazes-Lauber, J.; Petropoulos, K.; Thiel, P.; et al. Characterization of activating mutations of NOTCH3 in T-cell acute lymphoblastic leukemia and anti-leukemic activity of NOTCH3 inhibitory antibodies. Oncogene 2016, 35, 6077–6086. [Google Scholar] [CrossRef] [PubMed]
- Bellavia, D.; Campese, A.F.; Checquolo, S.; Balestri, A.; Biondi, A.; Cazzaniga, G.; Lendahl, U.; Fehling, H.J.; Hayday, A.C.; Frati, L.; et al. Combined expression of pTalpha and Notch3 in T cell leukemia identifies the requirement of preTCR for leukemogenesis. Proc. Natl. Acad. Sci. USA 2002, 99, 3788–3793. [Google Scholar] [CrossRef] [PubMed]
- Franciosa, G.; Diluvio, G.; Gaudio, F.D.; Giuli, M.V.; Palermo, R.; Grazioli, P.; Campese, A.F.; Talora, C.; Bellavia, D.; D’Amati, G.V.; et al. Prolyl-isomerase Pin1 controls Notch3 protein expression and regulates T-ALL progression. Oncogene 2016, 35, 4741–4751. [Google Scholar] [CrossRef]
- Choi, S.H.; Severson, E.; Pear, W.S.; Liu, X.S.; Aster, J.C.; Blacklow, S.C. The common oncogenomic program of NOTCH1 and NOTCH3 signaling in T-cell acute lymphoblastic leukemia. PLoS ONE 2017, 12, e0185762. [Google Scholar] [CrossRef]
- Wang, H.; Zang, C.; Taing, L.; Arnett, K.L.; Wong, Y.J.; Pear, W.S.; Blacklow, S.C.; Liu, X.S.; Aster, J.C. NOTCH1-RBPJ complexes drive target gene expression through dynamic interactions with superenhancers. Proc. Natl. Acad. Sci. USA 2014, 111, 705–710. [Google Scholar] [CrossRef]
- Tottone, L.; Zhdanovskaya, N.; Carmona Pestaña, Á; Zampieri, M.; Simeoni, F.; Lazzari, S.; Ruocco, V.; Pelullo, M.; Caiafa, P.; Felli, M.P.; et al. Histone Modifications Drive Aberrant Notch3 Expression/Activity and Growth in T-ALL. Front. Oncol. 2019, 9, 198. [Google Scholar] [CrossRef]
- Zampieri, M.; Ciccarone, F.; Palermo, R.; Cialfi, S.; Passananti, C.; Chiaretti, S.; Nocchia, D.; Talora, C.; Screpanti, I.; Caiafa, P. The epigenetic factor BORIS/CTCFL regulates the NOTCH3 gene expression in cancer cells. Biochim. Biophys. Acta 2014, 1839, 813–825. [Google Scholar] [CrossRef]
- Ghisi, M.; Corradin, A.; Basso, K.; Frasson, C.; Serafin, V.; Mukherjee, S.; Mussolin, L.; Ruggero, K.; Bonanno, L.; Guffanti, A.; et al. Modulation of microRNA expression in human T-cell development: Targeting of NOTCH3 by miR-150. Blood 2011, 117, 7053–7062. [Google Scholar] [CrossRef]
- Podshivalova, K.; Wang, E.A.; Hart, T.; Salomon, D.R. Expression of the miR-150 tumor suppressor is restored by and synergizes with rapamycin in a human leukemia T-cell line. Leuk. Res. 2018, 74, 1–9. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, B.; Wang, Y.; Zhao, Q.; Wu, W.; Zhang, P.; Miao, L.; Sun, S. NOTCH3 Overexpression and Posttranscriptional Regulation by miR-150 Were Associated With EGFR-TKI Resistance in Lung Adenocarcinoma. Oncol. Res. 2019, 27, 751–761. [Google Scholar] [CrossRef]
- Kim, T.H.; Jeong, J.Y.; Park, J.Y.; Kim, S.W.; Heo, J.H.; Kang, H.; Kim, G.; An, H.J. miR-150 enhances apoptotic and anti-tumor effects of paclitaxel in paclitaxel-resistant ovarian cancer cells by targeting Notch3. Oncotarget 2017, 8, 72788–72800. [Google Scholar] [CrossRef] [PubMed]
- Pinazza, M.; Ghisi, M.; Minuzzo, S.; Agnusdei, V.; Fossati, G.; Ciminale, V.; Pezzè, L.; Ciribilli, Y.; Pilotto, G.; Venturoli, C.; et al. Histone deacetylase 6 controls Notch3 trafficking and degradation in T-cell acute lymphoblastic leukemia cells. Oncogene 2018, 37, 3839–3851. [Google Scholar] [CrossRef] [PubMed]
- Palermo, R.; Checquolo, S.; Giovenco, A.; Grazioli, P.; Kumar, V.; Campese, A.F.; Giorgi, A.; Napolitano, M.; Canettieri, G.; Ferrara, G.; et al. Acetylation controls Notch3 stability and function in T-cell leukemia. Oncogene 2012, 31, 3807–3817. [Google Scholar] [CrossRef] [PubMed]
- Indraccolo, S.; Minuzzo, S.; Masiero, M.; Pusceddu, I.; Persano, L.; Moserle, L.; Reboldi, A.; Favaro, E.; Mecarozzi, M.; Di Mario, G.; et al. Cross-talk between tumor and endothelial cells involving the Notch3-Dll4 interaction marks escape from tumor dormancy. Cancer Res. 2009, 69, 1314–1323. [Google Scholar] [CrossRef]
- Minuzzo, S.; Agnusdei, V.; Pusceddu, I.; Pinazza, M.; Moserle, L.; Masiero, M.; Rossi, E.; Crescenzi, M.; Hoey, T.; Ponzoni, M.; et al. DLL4 regulates NOTCH signaling and growth of T acute lymphoblastic leukemia cells in NOD/SCID mice. Carcinogenesis 2015, 36, 115–121. [Google Scholar] [CrossRef]
- Pelullo, M.; Quaranta, R.; Talora, C.; Checquolo, S.; Cialfi, S.; Felli, M.P.; te Kronnie, G.; Borga, C.; Besharat, Z.M.; Palermo, R.; et al. Notch3/Jagged1 circuitry reinforces Notch signaling and sustains T-ALL. Neoplasia 2014, 16, 1007–1017. [Google Scholar] [CrossRef]
- Choy, L.; Hagenbeek, T.J.; Solon, M.; French, D.; Finkle, D.; Shelton, A.; Venook, R.; Brauer, M.J.; Siebel, C.W. Constitutive NOTCH3 Signaling Promotes the Growth of Basal Breast Cancers. Cancer Res. 2017, 77, 1439–1452. [Google Scholar] [CrossRef]
- Leontovich, A.A.; Jalalirad, M.; Salisbury, J.L.; Mills, L.; Haddox, C.; Schroeder, M.; Tuma, A.; Guicciardi, M.E.; Zammataro, L.; Gambino, M.W.; et al. NOTCH3 expression is linked to breast cancer seeding and distant metastasis. Breast Cancer Res. 2018, 20, 105. [Google Scholar] [CrossRef]
- Wang, K.; Zhang, Q.; Li, D.; Ching, K.; Zhang, C.; Zheng, X.; Ozeck, M.; Shi, S.; Li, X.; Wang, H.; et al. PEST domain mutations in Notch receptors comprise an oncogenic driver segment in triple-negative breast cancer sensitive to a γ-secretase inhibitor. Clin. Cancer Res. 2015, 21, 1487–1496. [Google Scholar] [CrossRef]
- Jung, J.G.; Stoeck, A.; Guan, B.; Wu, R.C.; Zhu, H.; Blackshaw, S.; Shih, I.E.M.; Wang, T.L. Notch3 interactome analysis identified WWP2 as a negative regulator of Notch3 signaling in ovarian cancer. PLoS Genet. 2014, 10, e1004751. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.Y.; Liang, Y.K.; Dou, X.W.; Chen, C.F.; Wei, X.L.; Zeng, D.; Bai, J.W.; Guo, Y.X.; Lin, F.F.; Huang, W.H.; et al. Notch3 inhibits epithelial-mesenchymal transition in breast cancer via a novel mechanism, upregulation of GATA-3 expression. Oncogenesis 2018, 7, 59. [Google Scholar] [CrossRef] [PubMed]
- Sourander, P.; Wålinder, J. Hereditary multi-infarct dementia. Morphological and clinical studies of a new disease. Acta Neuropathol. 1977, 39, 247–254. [Google Scholar] [CrossRef]
- Joutel, A.; Vahedi, K.; Corpechot, C.; Troesch, A.; Chabriat, H.; Vayssière, C.; Cruaud, C.; Maciazek, J.; Weissenbach, J.; Bousser, M.G.; et al. Strong clustering and stereotyped nature of Notch3 mutations in CADASIL patients. Lancet 1997, 350, 1511–1515. [Google Scholar] [CrossRef]
- Dichgans, M.; Mayer, M.; Uttner, I.; Brüning, R.; Müller-Höcker, J.; Rungger, G.; Ebke, M.; Klockgether, T.; Gasser, T. The phenotypic spectrum of CADASIL: Clinical findings in 102 cases. Ann. Neurol. 1998, 44, 731–739. [Google Scholar] [CrossRef] [PubMed]
- Louvi, A.; Arboleda-Velasquez, J.F.; Artavanis-Tsakonas, S. CADASIL: A critical look at a Notch disease. Dev. Neurosci. 2006, 28, 5–12. [Google Scholar] [CrossRef] [PubMed]
- Joutel, A.; Andreux, F.; Gaulis, S.; Domenga, V.; Cecillon, M.; Battail, N.; Piga, N.; Chapon, F.; Godfrain, C.; Tournier-Lasserve, E. The ectodomain of the Notch3 receptor accumulates within the cerebrovasculature of CADASIL patients. J. Clin. Invest. 2000, 5, 597–605. [Google Scholar] [CrossRef]
- Joutel, A.; Favrole, P.; Labauge, P.; Chabriat, H.; Lescoat, C.; Andreux, F.; Domenga, V.; Cécillon, M.; Vahedi, K.; Ducros, A.; et al. Skin biopsy immunostaining with a Notch3 monoclonal antibody for CADASIL diagnosis. Lancet 2001, 358, 2049–2051. [Google Scholar] [CrossRef]
- Monet-Leprêtre, M.; Haddad, I.; Baron-Menguy, C.; Fouillot-Panchal, M.; Riani, M.; Domenga-Denier, V.; Dussaule, C.; Cognat, E.; Vinh, J.; Joutel, A. Abnormal recruitment of extracellular matrix proteins by excess Notch3 ECD: A new pathomechanism in CADASIL. Brain 2013, 136, 1830–1845. [Google Scholar] [CrossRef]
- Yamamoto, Y.; Craggs, L.J.; Watanabe, A.; Booth, T.; Attems, J.; Low, R.W.; Oakley, A.E.; Kalaria, R.N. Brain microvascular accumulation and distribution of the NOTCH3 ectodomain and granular osmiophilic material in CADASIL. J. Neuropathol. Exp. Neurol. 2013, 72, 416–431. [Google Scholar] [CrossRef]
- Gray, F.; Polivka, M.; Viswanathan, A.; Baudrimont, M.; Bousser, M.G.; Chabriat, H. Apoptosis in cerebral autosomal-dominant arteriopathy with subcortical infarcts and leukoencephalopathy. J. Neuropathol. Exp. Neurol. 2007, 66, 597–607. [Google Scholar] [CrossRef] [PubMed]
- Panahi, M.; Yousefi Mesri, N.; Samuelsson, E.B.; Coupland, K.G.; Forsell, C.; Graff, C.; Tikka, S.; Winblad, B.; Viitanen, M.; Karlström, H.; et al. Differences in proliferation rate between CADASIL and control vascular smooth muscle cells are related to increased TGFβ expression. J. Cell Mol. Med. 2018, 22, 3016–3024. [Google Scholar] [CrossRef] [PubMed]
- Rajani, R.M.; Ratelade, J.; Domenga-Denier, V.; Hase, Y.; Kalimo, H.; Kalaria, R.N.; Joutel, A. Blood brain barrier leakage is not a consistent feature of white matter lesions in CADASIL. Acta Neuropathol. Commun. 2019, 7, 187. [Google Scholar] [CrossRef] [PubMed]
- Rutten, J.W.; Dauwerse, H.G.; Gravesteijn, G.; van Belzen, M.J.; van der Grond, J.; Polke, J.M.; Bernal-Quiros, M.; Lesnik Oberstein, S.A. Archetypal NOTCH3 mutations frequent in public exome: Implications for CADASIL. Ann. Clin. Transl. Neurol. 2016, 3, 844–853. [Google Scholar] [CrossRef]
- Rutten, J.W.; Van Eijsden, B.J.; Duering, M.; Jouvent, E.; Opherk, C.; Pantoni, L.; Federico, A.; Dichgans, M.; Markus, H.S.; Chabriat, H.; et al. The effect of NOTCH3 pathogenic variant position on CADASIL disease severity: NOTCH3 EGFr 1-6 pathogenic variant are associated with a more severe phenotype and lower survival compared with EGFr 7-34 pathogenic variant. Genet. Med. 2019, 21, 676–682. [Google Scholar] [CrossRef]
- Kelley, M.R.; Kidd, S.; Deutsch, W.A.; Young, M.W. Mutations altering the structure of epidermal growth factor-like coding sequences at the Drosophila Notch locus. Cell 1987, 51, 539–548. [Google Scholar] [CrossRef]
- Bardot, B.; Mok, L.P.; Thayer, T.; Ahimou, F.; Wesley, C. The Notch amino terminus regulates protein levels and Delta-induced clustering of Drosophila Notch receptors. Exp. Cell Res. 2005, 304, 202–223. [Google Scholar] [CrossRef]
- Arboleda-Velasquez, J.F.; Lopera, F.; Lopez, E.; Frosch, M.P.; Sepulveda-Falla, D.; Gutierrez, J.E.; Vargas, S.; Medina, M.; Martinez De Arrieta, C.; Lebo, R.V.; et al. C455R Notch3 mutation in a Colombian CADASIL kindred with early onset of stroke. Neurology 2002, 59, 277–279. [Google Scholar] [CrossRef]
- Monet-Leprêtre, M.; Bardot, B.; Lemaire, B.; Domenga, V.; Godin, O.; Dichgans, M.; Tournier-Lasserve, E.; Cohen-Tannoudji, M.; Chabriat, H.; Joutel, A. Distinct phenotypic and functional features of CADASIL mutations in the Notch3 ligand binding domain. Brain 2009, 132, 1601–1612. [Google Scholar] [CrossRef]
- Joutel, A.; Monet, M.; Domenga, V.; Riant, F.; Tournier-Lasserve, E. Pathogenic mutations associated with cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy differently affect Jagged1 binding and Notch3 activity via the RBP/JK signaling Pathway. Am. J. Hum. Genet. 2004, 74, 338–347. [Google Scholar] [CrossRef]
- Monet, M.; Domenga, V.; Lemaire, B.; Souilhol, C.; Langa, F.; Babinet, C.; Gridley, T.; Tournier-Lasserve, E.; Cohen-Tannoudji, M.; Joutel, A. The archetypal R90C CADASIL-NOTCH3 mutation retains NOTCH3 function in vivo. Hum. Mol. Genet. 2007, 16, 982–992. [Google Scholar] [CrossRef] [PubMed]
- Kast, J.; Hanecker, P.; Beaufort, N.; Giese, A.; Joutel, A.; Dichgans, M.; Opherk, C.; Haffner, C. Sequestration of latent TGF-β binding protein 1 into CADASIL-related Notch3-ECD deposits. Acta Neuropathol. Commun. 2014, 2, 96. [Google Scholar] [CrossRef] [PubMed]
- Capone, C.; Cognat, E.; Ghezali, L.; Baron-Menguy, C.; Aubin, D.; Mesnard, L.; Stöhr, H.; Domenga-Denier, V.; Nelson, M.T.; Joutel, A. Reducing Timp3 or vitronectin ameliorates disease manifestations in CADASIL mice. Ann. Neurol. 2016, 79, 387–403. [Google Scholar] [CrossRef] [PubMed]
- Dotti, M.T.; Federico, A.; Mazzei, R.; Bianchi, S.; Scali, O.; Conforti, F.L.; Sprovieri, T.; Guidetti, D.; Aguglia, U.; Consoli, D. The spectrum of Notch3 mutations in 28 Italian CADASIL families. J. Neurol. Neurosurg. Psychiatry 2005, 76, 736–738. [Google Scholar] [CrossRef] [PubMed]
- Moccia, M.; Mosca, L.; Erro, R.; Cervasio, M.; Allocca, R.; Vitale, C.; Leonardi, A.; Caranci, F.; Del Basso-De Caro, M.L.; Barone, P.; et al. Hypomorphic NOTCH3 mutation in an Italian family with CADASIL features. Neurobiol. Aging 2015, 36, 547. [Google Scholar] [CrossRef] [PubMed]
- Rutten, J.W.; Boon, E.M.; Liem, M.K.; Dauwerse, J.G.; Pont, M.J.; Vollebregt, E.; Maat-Kievit, A.J.; Ginjaar, H.B.; Lakeman, P.; van Duinen, S.G.; et al. Hypomorphic NOTCH3 alleles do not cause CADASIL in humans. Hum. Mutat. 2013, 34, 1486–1489. [Google Scholar] [CrossRef]
- Lesnik Oberstein, S.A.; van Duinen, S.G.; van den Boom, R.; Maat-Schieman, M.L.; van Buchem, M.A.; van Houwelingen, H.C.; Hegeman-Kleinn, I.M.; Ferrari, M.D.; Breuning, M.H.; Haan, J. Evaluation of diagnostic NOTCH3 immunostaining in CADASIL. Acta Neuropathol. 2003, 106, 107–111. [Google Scholar] [CrossRef]
- Pippucci, T.; Maresca, A.; Magini, P.; Cenacchi, G.; Donadio, V.; Palombo, F.; Papa, V.; Incensi, A.; Gasparre, G.; Valentino, M.L.; et al. Homozygous NOTCH3 null mutation and impaired NOTCH3 signaling in recessive early-onset arteriopathy and cavitating leukoencephalopathy. EMBO Mol. Med. 2015, 7, 848–858. [Google Scholar] [CrossRef]
- Henshall, T.L.; Keller, A.; He, L.; Johansson, B.R.; Wallgard, E.; Raschperger, E.; Mäe, M.A.; Jin, S.; Betsholtz, C.; Lendahl, U. Notch3 is necessary for blood vessel integrity in the central nervous system. Arterioscler. Thromb. Vasc. Biol. 2015, 35, 409–420. [Google Scholar] [CrossRef]
- Cognat, E.; Baron-Menguy, C.; Domenga-Denier, V.; Cleophax, S.; Fouillade, C.; Monet-Leprêtre, M.; Dewerchin, M.; Joutel, A. Archetypal Arg169Cys mutation in NOTCH3 does not drive the pathogenesis in cerebral autosomal dominant arteriopathy with subcortical infarcts and leucoencephalopathy via a loss-of-function mechanism. Stroke 2014, 45, 842–849. [Google Scholar] [CrossRef]
- Meng, H.; Zhang, X.; Yu, G.; Lee, S.J.; Chen, Y.E.; Prudovsky, I.; Wang, M.M. Biochemical characterization and cellular effects of CADASIL mutants of NOTCH3. PLoS ONE 2012, 7, e44964. [Google Scholar] [CrossRef] [PubMed]
- Arboleda-Velasquez, J.F.; Manent, J.; Lee, J.H.; Tikka, S.; Ospina, C.; Vanderburg, C.R.; Frosch, M.P.; Rodríguez-Falcón, M.; Villen, J.; Gygi, S.; et al. Hypomorphic Notch 3 alleles link Notch signaling to ischemic cerebral small-vessel disease. Proc. Natl. Acad. Sci. USA 2011, 108, E128–E135. [Google Scholar] [CrossRef] [PubMed]
- Machuca-Parra, A.I.; Bigger-Allen, A.A.; Sanchez, A.V.; Boutabla, A.; Cardona-Vélez, J.; Amarnani, D.; Saint-Geniez, M.; Siebel, C.W.; Kim, L.A.; D’Amore, P.A.; et al. Therapeutic antibody targeting of Notch3 signaling prevents mural cell loss in CADASIL. J. Exp. Med. 2017, 214, 2271–2282. [Google Scholar] [CrossRef] [PubMed]
- Ghezali, L.; Capone, C.; Baron-Menguy, C.; Ratelade, J.; Christensen, S.; Østergaard Pedersen, L.; Domenga-Denier, V.; Pedersen, J.T.; Joutel, A. Notch3 (ECD) immunotherapy improves cerebrovascular responses in CADASIL mice. Ann. Neurol. 2018, 84, 246–259. [Google Scholar] [CrossRef] [PubMed]
- Neves, K.B.; Harvey, A.P.; Moreton, F.; Montezano, A.C.; Rios, F.J.; Alves-Lopes, R.; Nguyen Dinh Cat, A.; Rocchicciolli, P.; Delles, C.; Joutel, A.; et al. ER stress and Rho kinase activation underlie the vasculopathy of CADASIL. JCI Insight 2019, 4, 131344. [Google Scholar] [CrossRef] [PubMed]
- Ling, C.; Liu, Z.; Song, M.; Zhang, W.; Wang, S.; Liu, X.; Ma, S.; Sun, S.; Fu, L.; Chu, Q.; et al. Modeling CADASIL vascular pathologies with patient-derived induced pluripotent stem cells. Protein Cell 2019, 10, 249–271. [Google Scholar] [CrossRef] [PubMed]
- Kelleher, J.; Dickinson, A.; Cain, S.; Hu, Y.; Bates, N.; Harvey, A.; Ren, J.; Zhang, W.; Moreton, F.C.; Muir, K.W.; et al. Patient-Specific iPSC Model of a Genetic Vascular Dementia Syndrome Reveals Failure of Mural Cells to Stabilize Capillary Structures. Stem Cell Rep. 2019, 13, 817–831. [Google Scholar] [CrossRef]
- Rand, M.D.; Grimm, L.M.; Artavanis-Tsakonas, S.; Patriub, V.; Blacklow, S.C.; Sklar, J.; Aster, J.C. Calcium depletion dissociates and activates heterodimeric notch receptors. Mol. Cell Biol. 2000, 20, 1825–1835. [Google Scholar] [CrossRef]
- Takahashi, K.; Adachi, K.; Yoshizaki, K.; Kunimoto, S.; Kalaria, R.N.; Watanabe, A. Mutations in NOTCH3 cause the formation and retention of aggregates in the endoplasmic reticulum, leading to impaired cell proliferation. Hum. Mol. Genet. 2010, 19, 79–89. [Google Scholar] [CrossRef]
- Watanabe-Hosomi, A.; Watanabe, Y.; Tanaka, M.; Nakagawa, M.; Mizuno, T. Transendocytosis is impaired in CADASIL-mutant NOTCH3. Exp. Neurol. 2012, 233, 303–311. [Google Scholar] [CrossRef]
- Dziewulska, D.; Rafalowska, J. Is the increased expression of ubiquitin in CADASIL syndrome a manifestation of aberrant endocytosis in the vascular smooth muscle cells? J. Clin. Neurosci. 2008, 15, 535–540. [Google Scholar] [CrossRef]
- Hanemaaijer, E.S.; Panahi, M.; Swaddiwudhipong, N.; Tikka, S.; Winblad, B.; Viitanen, M.; Piras, A.; Behbahani, H. Autophagy-lysosomal defect in human CADASIL vascular smooth muscle cells. Eur. J. Cell Biol. 2018, 97, 557–567. [Google Scholar] [CrossRef]
- Gravesteijn, G.; Dauwerse, J.G.; Overzier, M.; Brouwer, G.; Hegeman, I.; Mulder, A.A.; Baas, F.; Kruit, M.C.; Terwindt, G.M.; van Duinen, S.G.; et al. Naturally occurring NOTCH3 exon skipping attenuates NOTCH3 protein aggregation and disease severity in CADASIL patients. Hum. Mol. Genet. 2020, in press. [Google Scholar] [CrossRef]
- Li, X.; Zhang, X.; Leathers, R.; Makino, A.; Huang, C.; Parsa, P.; Macias, J.; Yuan, J.X.; Jamieson, S.W.; Thistlethwaite, P.A. Notch3 signaling promotes the development of pulmonary arterial hypertension. Nat. Med. 2009, 15, 1289–1297. [Google Scholar] [CrossRef]
- Wang, W.; Liu, J.; Ma, A.; Miao, R.; Jin, Y.; Zhang, H.; Xu, K.; Wang, C.; Wang, J. mTORC1 is involved in hypoxia-induced pulmonary hypertension through the activation of Notch3. J. Cell Physiol. 2014, 229, 2117–2125. [Google Scholar] [CrossRef]
- Wang, Y.; Dai, S.; Cheng, X.; Prado, E.; Yan, L.; Hu, J.; He, Q.; Lv, Y.; Lv, Y.; Du, L. Notch3 signaling activation in smooth muscle cells promotes extrauterine growth restriction-induced pulmonary hypertension. Nutr. Metab. Cardiovasc. Dis. 2019, 29, 639–651. [Google Scholar] [CrossRef]
- Chida, A.; Shintani, M.; Matsushita, Y.; Sato, H.; Eitoku, T.; Nakayama, T.; Furutani, Y.; Hayama, E.; Kawamura, Y.; Inai, K.; et al. Mutations of NOTCH3 in childhood pulmonary arterial hypertension. Mol. Genet. Genomic Med. 2014, 2, 229–239. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hosseini-Alghaderi, S.; Baron, M. Notch3 in Development, Health and Disease. Biomolecules 2020, 10, 485. https://doi.org/10.3390/biom10030485
Hosseini-Alghaderi S, Baron M. Notch3 in Development, Health and Disease. Biomolecules. 2020; 10(3):485. https://doi.org/10.3390/biom10030485
Chicago/Turabian StyleHosseini-Alghaderi, Samira, and Martin Baron. 2020. "Notch3 in Development, Health and Disease" Biomolecules 10, no. 3: 485. https://doi.org/10.3390/biom10030485
APA StyleHosseini-Alghaderi, S., & Baron, M. (2020). Notch3 in Development, Health and Disease. Biomolecules, 10(3), 485. https://doi.org/10.3390/biom10030485




