Estrogen Receptors and Ubiquitin Proteasome System: Mutual Regulation
Abstract
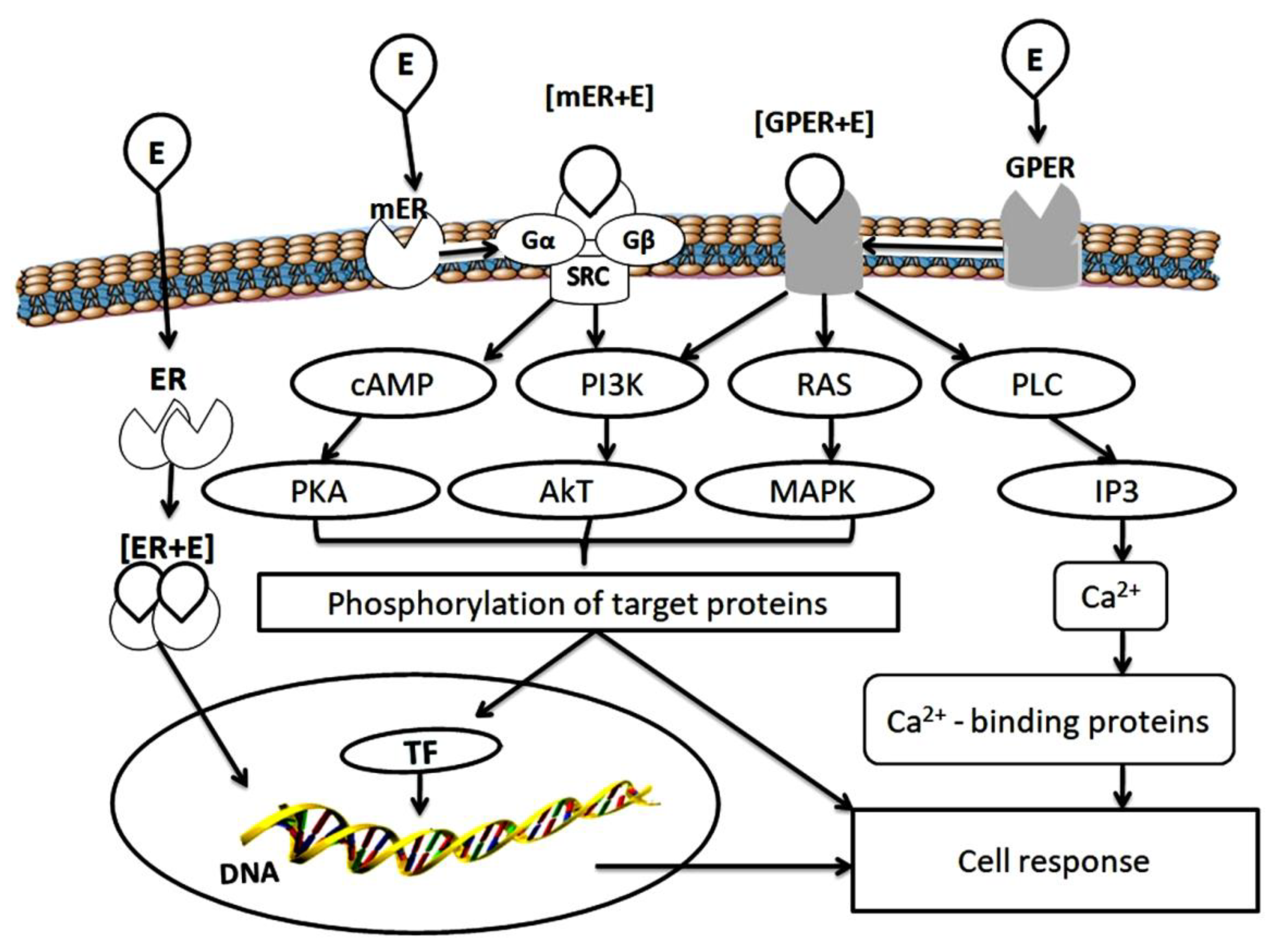
:1. General Introduction
2. Estrogen Receptors
3. Ubiquitin Proteasome System
4. Mutual Regulation of Estrogen Receptors and Ubiquitin Proteasome System
5. Final Remarks
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bondesson, M.; Hao, R.; Lin, C.-Y.; Williams, C.; Gustafsson, J.-A. Estrogen receptor signaling during vertebrate development. Biochim. Biophys. Acta 2015, 1849, 142–151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szmyd, M.; Lloyd, V.; Hallman, K.; Aleck, K.; Mladenovik, V.; McKee, C.; Morse, M.; Bedgood, T.; Dinda, S. The effects of black cohosh on the regulation of estrogen receptor (ERα) and progesterone receptor (PR) in breast cancer cells. Breast Cancer 2018, 10, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koolschijn, P.C.; Peper, J.S.; Crone, E.A. The influence of sex steroids on structural brain maturation in adolescence. PLoS ONE 2014, 9, e83929. [Google Scholar] [CrossRef] [PubMed]
- Jover-Mengual, T.; Castelló-Ruiz, M.; Burguete, M.C.; Jorques, M.; López-Morales, M.A.; Aliena-Valero, A.; Jurado-Rodríguez, A.; Pérez, S.; Centeno, J.M.; Miranda, F.J.; et al. Molecular mechanisms mediating the neuroprotective role of the selective estrogen receptor modulator, bazedoxifene, in acute ischemic stroke: A comparative study with 17β-estradiol. J. Steroid. Biochem. Mol. Biol. 2017, 171, 296–304. [Google Scholar] [CrossRef]
- Andersson, A.; Törnqvist, A.E.; Moverare-Skrtic, S.; Bernardi, A.I.; Farman, H.H.; Chambon, P.; Engdahl, C.; Lagerquist, M.K.; Windahl, S.H.; Carlsten, H.; et al. Roles of activating functions 1 and 2 of estrogen receptor α in lymphopoiesis. J. Endocrinol. 2018, 236, 99–109. [Google Scholar] [CrossRef]
- Lee, C.-H.; Su, S.-C.; Chiang, C.-F.; Chien, C.-Y.; Hsu, C.-C.; Yu, T.-Y.; Huang, S.-M.; Shieh, Y.-S.; Kao, H.-W.; Tsai, C.-S.; et al. Estrogen modulates vascular smooth muscle cell function through downregulation of SIRT1. Oncotarget 2017, 8, 110039–110051. [Google Scholar] [CrossRef] [Green Version]
- Lin, P.-I.; Tai, Y.-T.; Chan, W.-P.; Lin, Y.-L.; Liao, M.-H.; Chen, R.-M. Estrogen/ERα signaling axis participates in osteoblast maturation via upregulating chromosomal and mitochondrial complex gene expressions. Oncotarget 2017, 9, 1169–1186. [Google Scholar] [CrossRef] [Green Version]
- Xia, W.; Yin, J.; Zhang, S.; Guo, C.; Li, Y.; Zhang, Y.; Zhang, A.; Jia, Z.; Chen, H. Parkin modulates ERRα/eNOS signaling pathway in endothelial cells. Cell Physiol. Biochem. 2018, 49, 2022–2034. [Google Scholar] [CrossRef]
- Valéra, M.-C.; Noirrit-Esclassan, E.; Dupuis, M.; Fontaine, C.; Lenfant, F.; Briaux, A.; Cabou, C.; Garcia, C.; Lairez, O.; Foidart, J.-M.; et al. Effect of estetrol, a selective nuclear estrogen receptor modulator, in mouse models of arterial and venous thrombosis. Mol. Cell. Endocrinol. 2018, 477, 132–139. [Google Scholar] [CrossRef]
- Michalek, R.D.; Gerriets, V.A.; Nichols, A.G.; Inoue, M.; Kazmin, D.; Chang, C.-Y.; Dwyer, M.A.; Nelson, E.R.; Pollizzi, K.N.; Ilkayeva, O.; et al. Estrogen-related receptor-α is a metabolic regulator of effector T-cell activation and differentiation. Proc. Natl. Acad. Sci. USA 2011, 108, 18348–18353. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.-C.; Liu, Y.; Xiao, L.-L.; Li, S.-F.; Jiang, J.-H.; Zhao, Y.; Qian, S.-W.; Tang, Q.-Q.; Li, X.-J. Upregulation of miR-125b by estrogen protects against non-alcoholic fatty liver in female mice. J. Hepatol. 2015, 63, 1466–1475. [Google Scholar] [CrossRef] [PubMed]
- Sier, J.H.; Thumser, A.E.; Plant, N.J. Linking physiologically-based pharmacokinetic and genome-scale metabolic networks to understand estradiol biology. BMC Syst. Biol. 2017, 11, 141–156. [Google Scholar] [CrossRef] [PubMed]
- Pokharel, A.; Kolla, S.; Matouskova, K.; Vandenberg, L.N. Asymmetric development of the male mouse mammary gland and its response to a prenatal or postnatal estrogen challenge. Reprod. Toxicol. 2018, 82, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Baker, L.; Meldrum, K.K.; Wang, M.-J.; Sankula, R.; Vanam, R.; Raiesdana, A.; Tsai, B.; Hile, K.; Brown, J.W.; Meldrum, D.R. The role of estrogen in cardiovascular disease. J. Surg. Res. 2003, 115, 325–344. [Google Scholar] [CrossRef]
- Clegg, D.; Hevener, A.L.; Moreau, K.L.; Morselli, E.; Criollo, A.; Van Pelt, R.E.; Vieira-Potter, V.J. Sex Hormones and cardiometabolic health: Role of estrogen and estrogen receptors. Endocrinology 2017, 158, 1095–1105. [Google Scholar] [CrossRef] [PubMed]
- Farahmand, M.; Ramezani Tehrani, F.; Khalili, D.; Cheraghi, L.; BahriKhomami, M.; Azizi, F. Association between duration of endogenous estrogen exposure and cardiovascular outcomes: A population—Based cohort study. Life Sci. 2019, 221, 335–340. [Google Scholar] [CrossRef]
- Chen, C.; Gong, X.; Yang, X.; Shang, X.; Du, Q.; Liao, Q.; Xie, R.; Chen, Y.; Xu, J. The roles of estrogen and estrogen receptors in gastrointestinal disease. Oncol. Lett. 2019, 18, 5673–5680. [Google Scholar] [CrossRef] [Green Version]
- Kozasa, K.; Mabuchi, S.; Matsumoto, Y.; Kuroda, H.; Yokoi, E.; Komura, N.; Kawano, M.; Takahashi, R.; Sasano, T.; Shimura, K.; et al. Estrogen stimulates female cancer progression by inducing myeloid-derived suppressive cells: Investigations on pregnant and non-pregnant experimental models. Oncotarget 2019, 10, 1887–1902. [Google Scholar] [CrossRef] [Green Version]
- Milette, S.; Hashimoto, M.; Perrino, S.; Qi, S.; Chen, M.; Ham, B.; Wang, N.; Istomine, R.; Lowy, A.M.; Piccirillo, C.A.; et al. Sexual dimorphism and the role of estrogen in the immune microenvironment of liver metastases. Nat. Commun. 2019, 10, 1–16. [Google Scholar] [CrossRef]
- Powell, E.; Xu, W. Intermolecular interactions identify ligand-selective activity of estrogen receptor alpha/beta dimers. Proc. Natl. Acad. Sci. USA 2008, 105, 19012–19017. [Google Scholar] [CrossRef] [Green Version]
- Yaşar, P.; Ayaz, G.; User, S.D.; Güpür, G.; Muyan, M. Molecular mechanism of estrogen-estrogenreceptor signaling. Reprod. Med. Biol. 2016, 16, 4–20. [Google Scholar] [CrossRef]
- Madak-Erdogan, Z.; Charn, T.H.; Jiang, Y.; Liu, E.T.; Katzenellenbogen, J.A.; Katzenellenbogen, B.S. Integrative genomics of gene and metabolic regulation by estrogen receptors alpha and beta and their coregulators. Mol. Syst. Biol. 2013, 9, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.C.; Weng, Y.J.; Shibu, M.A.; Han, C.K.; Chen, Y.S.; Shen, C.Y.; Lin, Y.M.; Viswanadha, V.P.; Liang, H.Y.; Huang, C.Y. Estrogen and/or estrogen receptor α inhibits BNIP3-induced apoptosis and autophagy in H9c2 cardiomyoblast cells. Int. J. Mol. Sci. 2018, 19, 1298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Z.; Yu, W.; Liu, B.; Yang, M.; Tao, H. Estrogen receptor β induces autophagy of osteosarcoma through the mTOR signaling pathway. J. Orthop. Surg. Res. 2020, 15, 50. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, S.; Eng, V.; Taylor, J.; Lubahn, D.B.; Korach, K.S.; Pfaff, D.W. Roles of estrogen receptor-α gene expression in reproduction-related behaviors in female mice. Endocrinology 1998, 139, 5070–5081. [Google Scholar] [CrossRef]
- Nakata, M.; Ågmo, A.; Sagoshi, S.; Ogawa, S. The role of estrogen receptor β (ERβ) in the establishment of hierarchical social relationships in male mice. Front. Behav. Neurosci. 2018, 12, 245. [Google Scholar] [CrossRef] [Green Version]
- Levin, E.R. Integration of the extranuclear and nuclear actions of estrogen. Mol. Endocrinol. 2005, 19, 1951–1959. [Google Scholar] [CrossRef] [Green Version]
- Hamilton, K.J.; Arao, Y.; Korach, K.S. Estrogen hormone physiology: Reproductive findings from estrogen receptor mutant mice. Reprod. Biol. 2014, 14, 3–8. [Google Scholar] [CrossRef] [Green Version]
- Grober, O.M.; Mutarelli, M.; Giurato, G.; Ravo, M.; Cicatiello, L.; De Filippo, M.R.; Ferraro, L.; Nassa, G.; Papa, M.F.; Paris, O.; et al. Global analysis of estrogen receptor beta binding to breast cancer cell genome reveals an extensive interplay with estrogen receptor alpha for target gene regulation. BMC Genomics 2011, 2, 36. [Google Scholar] [CrossRef]
- Zhang, Y.; Klein, K.; Sugathan, A.; Nassery, N.; Dombkowski, A.; Zanger, U.M.; Waxman, D.J. Transcriptional profiling of human liver identifies sex-biased genes associated with polygenic dyslipidemia and coronary artery disease. PLoS ONE 2011, 6, e23506. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Lu, Y.; Li, Z.; Zhou, Y.; Gu, Y.; Pang, X.; Wu, J.; Gobin, R.; Yu, J. Oestrogen receptor α regulates the odonto/osteogenic differentiation of stem cells from apical papilla via ERK and JNK MAPK pathways. Cell Prolif. 2018, 51, e12485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tecalco-Cruz, A.C.; Pérez-Alvarado, I.A.; Ramírez-Jarquín, J.O.; Rocha-Zavaleta, L. Nucleo-cytoplasmic transport of estrogen receptor alpha in breast cancer cells. Cell Signal. 2017, 34, 121–132. [Google Scholar] [CrossRef]
- Mohammed, H.; Russell, I.A.; Stark, R.; Rueda, O.M.; Hickey, T.E.; Tarulli, G.A.; Serandour, A.A.; Birrell, S.N.; Bruna, A.; Saadi, A.; et al. Corrigendum: Progesterone receptor modulates ERα action in breast cancer. Nature 2015, 526, 144. [Google Scholar] [CrossRef] [Green Version]
- Rettberg, J.R.; Yao, J.; Brinton, R.D. Estrogen: A master regulator of bioenergetic systems in the brain and body. Front. Neuroendocrinol. 2014, 35, 8–30. [Google Scholar] [CrossRef] [Green Version]
- Simpkins, J.W.; Yang, S.H.; Sarkar, S.N.; Pearce, V. Estrogen actions on mitochondria-physiological and pathological implications. Mol. Cell. Endocrinol. 2008, 290, 51–59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levin, E.R.; Hammes, S.R. Nuclear receptors outside the nucleus: Extranuclear signalling by steroid receptors. Nat. Rev. Mol. Cell. Biol. 2016, 17, 783–797. [Google Scholar] [CrossRef] [Green Version]
- Ranganathan, P.; Nadig, N.; Nambiar, S. Non-canonical Estrogen Signaling in Endocrine Resistance. Front. Endocrinol. 2019, 10, 1–8. [Google Scholar] [CrossRef]
- Maggiolini, M.; Picard, D. The unfolding stories of GPR30, a new membrane-bound estrogen receptor. J. Endocrinol. 2010, 204, 105–114. [Google Scholar] [CrossRef] [Green Version]
- Thomas, P.; Pang, Y.; Filardo, E.J.; Dong, J. Identity of an estrogen membrane receptor coupled to a G protein in human breast cancer cells. Endocrinology 2005, 146, 624–632. [Google Scholar] [CrossRef] [Green Version]
- Prossnitz, E.R.; Barton, M. Estrogen biology: New insights into GPER function and clinical opportunities. Mol. Cell. Endocrinol. 2014, 389, 71–83. [Google Scholar] [CrossRef] [Green Version]
- Trenti, A.; Tedesco, S.; Boscaro, C.; Trevisi, L.; Bolego, C.; Cignarella, A. Estrogen, Angiogenesis, Immunity and Cell Metabolism: Solving the Puzzle. Int. J. Mol. Sci. 2018, 19, 859. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pandey, D.P.; Lappano, R.; Albanito, L.; Madeo, A.; Maggiolini, M.; Picard, D. Estrogenic GPR30 signalling induces proliferation and migration of breast cancer cells through CTGF. EMBO J. 2009, 28, 523–532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanchez, A.M.; Flamini, M.I.; Baldacci, C.; Goglia, L.; Genazzani, A.R.; Simoncini, T. Estrogen receptor-alpha promotes breast cancer cell motility and invasion via focal adhesion kinase and N-WASP. Mol. Endocrinol. 2010, 24, 2114–2125. [Google Scholar] [CrossRef] [PubMed]
- Sharma, G.; Prossnitz, E.R. G-Protein-Coupled Estrogen Receptor (GPER) and Sex-Specific Metabolic Homeostasis. Adv. Exp. Med. Biol. 2017, 1043, 427–453. [Google Scholar] [CrossRef] [PubMed]
- Johnson, A.B.; O’Malley, B.W. Steroid receptor coactivators 1, 2, and 3: Critical regulators of nuclear receptor activity and steroid receptor modulator (SRM)-based cancer therapy. Mol. Cell. Endocrinol. 2011, 348, 430–439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taylor, S.E.; Martin-Hirsch, P.L.; Martin, F.L. Oestrogen receptor splice variants in the pathogenesis of disease. Cancer Lett. 2010, 288, 133–148. [Google Scholar] [CrossRef]
- Klinge, C.M. miRNAs and estrogen action. Trends Endocrinol. Metab. 2012, 23, 223–233. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Wu, H.-T.; Zhu, N.; Shi, Y.-N.; Liu, Z.; Ao, B.-X.; Liao, D.-F.; Zheng, X.-L.; Qin, L. Steroid receptor RNA activator: Biologic function and role in disease. Clin. Chim. Acta 2016, 459, 137–146. [Google Scholar] [CrossRef]
- Le Romancer, M.; Poulard, C.; Cohen, P.; Sentis, S.; Renoir, J.M.; Corbo, L. Cracking the estrogen receptor’s posttranslational code in breast tumors. Endocr. Rev. 2011, 32, 597–622. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Ye, Z.-W.; Chen, W.; Manevich, Y.; Mehrotra, S.; Ball, L.E.; Janssen-Heininger, Y.M.; Tew, K.D.; Townsend, D.M. S-Glutathionylation of estrogen receptor alpha affects dendritic cell function. J. Biol. Chem. 2018, 293, 4366–4380. [Google Scholar] [CrossRef] [Green Version]
- Pickart, C.M.; Cohen, R. Proteasomes and their kin: Proteases in the machine age. Nat. Rev. Mol. Cell. Biol. 2004, 5, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K. The proteasome: Overiew of structure and function. Proc. Jpn. Acad. Ser. B 2009, 85, 12–36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sadre-Bazzaz, K.; Whitby, F.G.; Robinson, H.; Formosa, T. Structure of a Blm10 complex reveals common mechanisms for proteasome binding and gate opening. Mol. Cell. 2010, 37, 728–735. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, T.; Grune, T. Structure of the proteasome. Prog. Mol. Biol. Transl. Sci. 2012, 109, 1–39. [Google Scholar] [CrossRef] [PubMed]
- Raynes, R.; Pomatto, L.C.; Davies, K.J. Degradation of oxidized proteins by the proteasome: Distinguishing between the 20S, 26S, and immunoproteasome proteolytic pathways. Mol. Aspects Med. 2016, 50, 41–55. [Google Scholar] [CrossRef] [Green Version]
- Ciechanover, A.; Heller, H.; Katz-Etzion, R.; Hershko, A. Activation of the heat-stable polypeptide of the ATP-dependent proteolytic system. Proc. Natl. Acad. Sci. USA 1981, 78, 761–765. [Google Scholar] [CrossRef] [Green Version]
- Hershko, A.; Heller, H.; Elias, S.; Ciechanover, A. Components of ubiquitin-protein ligase system. Resolution, affinity purification, and role in protein breakdown. J. Biol. Chem. 1983, 258, 8206–8214. [Google Scholar]
- Hershko, A.; Ciechanover, A. The ubiquitin system. Annu. Rev. Biochem. 1998, 67, 425–479. [Google Scholar] [CrossRef]
- Reiss, Y.; Heller, H.; Hershko, A. Binding sites of ubiquitin-protein ligase. Binding of ubiquitin-protein conjugates and of ubiquitin-carrier protein. J. Biol. Chem. 1989, 264, 10378–10383. [Google Scholar]
- Dohmen, R.J.; Madura, K.; Bartel, B.; Varshavsky, A. The N-end rule is mediated by the UBC2 (RAD6) ubiquitin-conjugating enzyme. Proc. Natl. Acad. Sci. USA 1991, 88, 7351–7355. [Google Scholar] [CrossRef] [Green Version]
- Rechsteiner, M.; Rogers, S.W. PEST sequences and regulation by proteolysis. Trends Biochem. Sci. 1996, 21, 267–271. [Google Scholar] [CrossRef]
- King, R.W.; Glotzer, M.; Kirschner, M.W. Mutagenic analysis of the destruction signal of mitotic cyclins and structural characterization of ubiquitinated intermediates. Mol. Biol. Cell 1996, 7, 1343–1357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Varshavsky, A. The N-end rule: Functions, mysteries, uses. Proc. Natl. Acad. Sci. USA 1996, 93, 12142–12149. [Google Scholar] [CrossRef] [Green Version]
- Kim, T.K.; Maniatis, T. Regulation of interferon-gamma-activated STAT1 by the ubiquitin-proteasome pathway. Science 1996, 273, 1717–1719. [Google Scholar] [CrossRef]
- Storey, A.; Thomas, M.; Kalita, A.; Harwood, C.; Gardiol, D.; Mantovani, F.; Breuer, J.; Leigh, I.M.; Matlashewski, G.; Banks, L. Role of a p53 polymorphism in the development of human papillomavirus-associated cancer. Nature 1998, 393, 229–234. [Google Scholar] [CrossRef]
- Chau, V.; Tobias, J.W.; Bachmair, A.; Marriott, D.; Ecker, D.J.; Gonda, D.K.; Varshavsky, A. Multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science 1989, 243, 1576–1583. [Google Scholar] [CrossRef]
- Dammer, E.B.; Na, C.H.; Xu, P.; Seyfried, N.T.; Duong, D.M.; Cheng, D.; Gearing, M.; Rees, H.; Lah, J.J.; Levey, A.I.; et al. Polyubiquitin linkage profiles in three models of proteolytic stress suggest the etiology of Alzheimer disease. J. Biol. Chem. 2011, 286, 10457–10465. [Google Scholar] [CrossRef] [Green Version]
- Ciechanover, A.; Stanhill, A. The complexity of recognition of ubiquitinated substrates by the 26S proteasome. Biochim. Biophys. Acta 2014, 1843, 86–96. [Google Scholar] [CrossRef] [Green Version]
- Spence, J.; Sadis, S.; Haas, A.L.; Finley, D. A ubiquitin mutant with specific defects in DNA repair and multiubiquitination. Mol. Cell. Biol. 1995, 15, 1265–1273. [Google Scholar] [CrossRef] [Green Version]
- Ohtake, F.; Tsuchiya, H.; Saeki, Y.; Tanaka, K. K63 ubiquitylation triggers proteasomal degradation by seeding branched ubiquitin chains. Proc. Natl. Acad. Sci. USA 2018, 115, E1401–E1408. [Google Scholar] [CrossRef] [Green Version]
- Meyer, H.-J.; Rape, M. Enhanced protein degradation by branched ubiquitin chains. Cell 2014, 157, 910–921. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yau, R.G.; Doerner, K.; Castellanos, E.R.; Haakonsen, D.L.; Werner, A.; Wang, N.; Yang, X.W.; Martinez-Martin, N.; Matsumoto, M.L.; Dixit, V.M.; et al. Assembly and Function of Heterotypic Ubiquitin Chains in Cell-Cycle and Protein Quality Control. Cell 2017, 171, 918–933. [Google Scholar] [CrossRef] [Green Version]
- Glickman, M.H.; Rubin, D.M.; Fried, V.A.; Finley, D. The regulatory particle of the Saccharomyces cerevisiae proteasome. Mol. Cell. Biol. 1998, 18, 3149–3162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanna, J.; Finley, D. A proteasome for all occasions. FEBS Lett. 2007, 581, 2854–2861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verma, R.; Aravind, L.; Oania, R.; McDonald, W.H.; Yates, J.R., 3rd; Koonin, E.V.; Deshaies, R.J. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science 2002, 298, 611–615. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.; Li, P.; Song, L.; Jeffrey, P.D.; Chenova, T.A.; Wilkinson, K.D.; Cohen, R.E.; Shi, Y. Structure and mechanisms of the proteasome-associated deubiquitinating enzyme USP14. EMBO J. 2005, 24, 3747–3756. [Google Scholar] [CrossRef]
- Hamazaki, J.; Iemura, S.; Natsume, T.; Yashiroda, H.; Tanaka, K.; Murata, S. A novel proteasome interacting protein recruits the deubiquitinating enzyme UCH37 to 26S proteasomes. EMBO J. 2006, 25, 4524–4536. [Google Scholar] [CrossRef] [Green Version]
- Yao, T.; Song, L.; Xu, W.; DeMartino, G.N.; Florens, L.; Swanson, S.K.; Washburn, M.P.; Conaway, R.C.; Conaway, J.W.; Cohen, R.E. Proteasome recruitment and activation of the Uch37 deubiquitinating enzyme by Adrm1. Nat. Cell Biol. 2006, 8, 994–1002. [Google Scholar] [CrossRef]
- Schreiner, P.; Chen, X.; Husnjak, K.; Randles, L.; Zhang, N.; Elsasser, S.; Finley, D.; Dikic, I.; Walters, K.J.; Groll, M. Ubiquitin docking at the proteasome through a novel pleckstrin-homology domain interaction. Nature 2008, 453, 548–552. [Google Scholar] [CrossRef] [Green Version]
- Husnjak, K.; Elsasser, S.; Zhang, N.; Chen, X.; Randles, L.; Shi, Y.; Hofmann, K.; Walters, K.J.; Finley, D.; Dikic, I. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature 2008, 453, 481–488. [Google Scholar] [CrossRef] [Green Version]
- Braun, B.C.; Glickman, M.; Kraft, R.; Dahlmann, B.; Kloetzel, P.M.; Finley, D.; Schmidt, M. The base of the proteasome regulatory particle exhibits chaperone-like activity. Nat. Cell Biol. 1999, 1, 221–226. [Google Scholar] [CrossRef] [PubMed]
- Strickland, E.; Hakala, K.; Thomas, P.J.; DeMartino, G.N. Recognition of misfolding proteins by PA700, the regulatory subcomplex of the 26 S proteasome. J. Biol. Chem. 2000, 275, 5565–5572. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, K.; Kasahara, M. The MHC class I ligand-generating system: Roles of immunoproteasomes and the interferon-gamma-inducible proteasome activator PA28. Immunol. Rev. 1998, 163, 161–176. [Google Scholar] [CrossRef]
- Strehl, B.; Seifert, U.; Kruger, E.; Heink, S.; Kuckelkorn, U.; Kloetzel, P.M. Interferon-gamma, the functional plasticity of the ubiquitin-proteasome system, and MHC class I antigen processing. Immunol. Rev. 2005, 207, 19–30. [Google Scholar] [CrossRef] [PubMed]
- Unno, M.; Mizushima, T.; Morimoto, Y.; Tomisugi, Y.; Tanaka, K.; Yasuoka, N.; Tsukihara, T. The structure of the mammalian 20S proteasome at 2.75 A resolution. Structure 2002, 10, 609–618. [Google Scholar] [CrossRef] [Green Version]
- Kisselev, A.F.; Akopian, T.N.; Woo, K.M.; Goldberg, A.L. The sizes of peptides generated from protein by mammalian 26 and 20 S proteasomes. J. Biol. Chem. 1999, 274, 3363–3371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Astakhova, T.M.; Bozhok, G.A.; Alabedal’karim, N.M.; Karpova, Y.D.; Lyupina, Y.V.; Ushakova, E.M.; Legach, E.I.; Bondarenko, T.P.; Sharova, N.P. Proteasome expression in ovarian heterotopic allografts of Wistar and August rats under induction of donor specific tolerance. Russ. J. Dev. Biol. 2019, 50, 261–267. [Google Scholar] [CrossRef]
- Spirina, L.V.; Yunusova, N.V.; Kondakova, I.V. Association of growth factors, HIF-1 and NF-kappa B expression with proteasomes in endometrial cancer. Mol. Biol. Rep. 2012, 39, 8655–8662. [Google Scholar] [CrossRef]
- Heldin, J.; O’Callaghan, P.; Hernández Vera, R.; Fuchs, P.F.; Gerwins, P.; Kreuger, J. FGD5 sustains vascular endothelial growth factor A (VEGFA) signaling through inhibition of proteasome-mediated VEGF receptor 2 degradation. Cell Signal. 2017, 40, 125–132. [Google Scholar] [CrossRef]
- Zhou, W.; Slingerland, J.M. Links between oestrogen receptor activation and proteolysis: Relevance to hormone-regulated cancer therapy. Nat. Rev. Cancer 2014, 14, 26–38. [Google Scholar] [CrossRef]
- Iizuka, M.; Susa, T.; Tamamori Adachi, M.; Okinaga, H.; Okazaki, T. Intrinsic ubiquitin E3 ligase activity of histone acetyltransferase Hbo1 for estrogen receptor α. Proc. Jpn. Acad. Ser. B. Phys. Biol. Sci. 2017, 93, 498–510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Helzer, K.T.; Hooper, C.; Miyamoto, S.; Alarid, E.T. Ubiquitylation of nuclear receptors: New linkages and therapeutic implications. J. Mol. Endocrinol. 2015, 54, R151–R167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, M.; Park, A.; Nephew, K.P. CHIP (carboxyl terminus of Hsc70-interacting protein) promotes basal and geldanamycin-induced degradation of estrogen receptor-alpha. Mol. Endocrinol. 2005, 19, 2901–2914. [Google Scholar] [CrossRef] [Green Version]
- Duong, V.; Boulle, N.; Daujat, S.; Chauvet, J.; Bonnet, S.; Neel, H.; Cavaillès, V. Differential regulation of estrogen receptor alpha turnover and transactivation by Mdm2 and stress-inducing agents. Cancer Res. 2007, 67, 5513–5521. [Google Scholar] [CrossRef] [Green Version]
- Sanchez, M.; Picard, N.; Sauvé, K.; Tremblay, A. Coordinate regulation of estrogen receptor β degradation by Mdm2 and CREB-binding protein in response to growth signals. Oncogene 2013, 32, 117–126. [Google Scholar] [CrossRef]
- Rajbhandari, P.; Schalper, K.A.; Solodin, N.M.; Ellison-Zelski, S.J.; Ping Lu, K.; Rimm, D.L.; Alarid, E.T. Pin1 modulates ERα levels in breast cancer through inhibition of phosphorylation-dependent ubiquitination and degradation. Oncogene 2014, 33, 1438–1447. [Google Scholar] [CrossRef] [Green Version]
- Sun, J.; Zhou, W.; Kaliappan, K.; Nawaz, Z.; Slingerland, J.M. ERα phosphorylation at Y537 by Src triggers E6-AP-ERα binding, ERα ubiquitylation, promoter occupancy, and target gene expression. Mol. Endocrinol. 2012, 26, 1567–1577. [Google Scholar] [CrossRef] [Green Version]
- Bhatt, S.; Xiao, Z.; Meng, Z.; Katzenellenbogen, B.S. Phosphorylation by p38 mitogen-activated protein kinase promotes estrogen receptor α turnover and functional activity via the SCF (Skp2) proteasomal complex. Mol. Cell. Biol. 2012, 32, 1928–1943. [Google Scholar] [CrossRef] [Green Version]
- Powers, G.L.; Ellison-Zelski, S.J.; Casa, A.J.; Lee, A.V.; Alarid, E.T. Proteasome inhibition represses ERα gene expression in ER+ cells—A new link between proteasome activity and estrogen signaling in breast cancer. Oncogene 2010, 29, 1509–1518. [Google Scholar] [CrossRef] [Green Version]
- Amita, M.; Takahashi, T.; Igarashi, H.; Nagase, S. Clomiphene citrate down-regulates estrogenreceptor-α through the ubiquitin-proteasome pathway in a human endometrial cancer cell line. Mol. Cell. Endocrinol. 2016, 428, 142–147. [Google Scholar] [CrossRef]
- Singh, V.; Sharma, V.; Verma, V.; Pandey, D.; Yadav, S.K.; Maikhuri, J.P.; Gupta, G. Apigenin manipulates the ubiquitin-proteasome system to rescue estrogen receptor-β from degradation and induce apoptosis in prostate cancer cells. Eur. J. Nutr. 2015, 54, 1255–1267. [Google Scholar] [CrossRef] [PubMed]
- Wijayaratne, A.L.; McDonnell, D.P. The human estrogen receptor alpha is a ubiquitinated protein whose stability is affected differentially by agonists, antagonists, and selective estrogen receptor modulators. J. Biol. Chem. 2001, 276, 35684–35692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohoka, N.; Morita, Y.; Nagai, K.; Shimokawa, K.; Ujikawa, O.; Fujimori, I.; Ito, M.; Hayase, Y.; Okuhira, K.; Shibata, N.; et al. Derivatization of inhibitor of apoptosis protein (IAP) ligands yields improved inducers of estrogen receptor α degradation. J. Biol. Chem. 2018, 293, 6776–6790. [Google Scholar] [CrossRef] [Green Version]
- Xia, X.; Liao, Y.; Huang, C.; Liu, Y.; He, J.; Shao, Z.; Jiang, L.; Dou, Q.P.; Liu, J.; Huang, H. Deubiquitination and stabilization of estrogen receptor α by ubiquitin-specific protease 7 promotes breast tumorigenesis. Cancer Lett. 2019, 465, 118–128. [Google Scholar] [CrossRef]
- Kos, M.; Reid, G.; Denger, S.; Gannon, F. Minireview: Genomic organization of the human ERalpha gene promoter region. Mol. Endocrinol. 2001, 15, 2057–2063. [Google Scholar] [CrossRef]
- Powers, G.L.; Rajbhandari, P.; Solodin, N.M.; Bickford, B.; Alarid, E.T. The proteasome inhibitor bortezomib induces an inhibitory chromatin environment at a distal enhancer of the estrogen receptor-α gene. PLoS ONE 2013, 8, e81110. [Google Scholar] [CrossRef]
- Zhang, H.; Sun, L.; Liang, J.; Yu, W.; Zhang, Y.; Wang, Y.; Chen, Y.; Li, R.; Sun, X.; Shang, Y. The catalytic subunit of the proteasome is engaged in the entire process of estrogen receptor-regulated transcription. EMBO J. 2006, 25, 4223–4233. [Google Scholar] [CrossRef]
- Prenzel, T.; Begus-Nahrmann, Y.; Kramer, F.; Hennion, M.; Hsu, C.; Gorsler, T.; Hintermair, C.; Eick, D.; Kremmer, E.; Simons, M.; et al. Estrogen-dependent gene transcription in human breast cancer cells relies upon proteasome-dependent monoubiquitination of histone H2B. Cancer Res. 2011, 71, 5739–5753. [Google Scholar] [CrossRef] [Green Version]
- Cheng, S.B.; Quinn, J.A.; Graeber, C.T.; Filardo, E.J. Down-modulation of the G-protein-coupled estrogen receptor, GPER, from the cell surface occurs via a trans-Golgi-proteasome pathway. J. Biol. Chem. 2011, 286, 22441–22455. [Google Scholar] [CrossRef] [Green Version]
- Portbury, A.L.; Ronnebaum, S.M.; Zungu, M.; Patterson, C.; Willis, M.S. Back to your heart: Ubiquitin proteasome system-regulated signal transduction. J. Mol. Cell. Cardiol. 2012, 52, 526–537. [Google Scholar] [CrossRef] [Green Version]
- Heino, T.J.; Chagin, A.S.; Sävendahl, L. The novel estrogen receptor G-protein-coupled receptor 30 is expressed in human bone. J. Endocrinol. 2008, 197, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Shashova, E.E.; Lyupina, Y.V.; Glushchenko, S.A.; Slonimskaya, E.M.; Savenkova, O.V.; Kulikov, A.M.; Gornostaev, N.G.; Kondakova, I.V.; Sharova, N.P. Proteasome functioning in breast cancer: Connection with clinical-pathological factors. PLoS ONE 2014, 9, e109933. [Google Scholar] [CrossRef] [PubMed]
- Sassin, W. Zu den Grenzen menschlicher Erkenntnis. Beacon J. Stud. Ideol. Ment. Dimens. 2018, 1, 010310202. Available online: http://hdl.handle.net//20.500.12656/thebeacon.1.010310202 (accessed on 1 December 2019).
- Nagpal, N.; Ahmad, H.M.; Molparia, B.; Kulshreshtha, R. MicroRNA-191, an estrogen responsive microRNA, functions as an oncogenic regulator in human breast cancer. Carcinogenesis 2013, 34, 1889–1899. [Google Scholar] [CrossRef]
- Di Leva, G.; Piovan, C.; Gasparini, P.; Ngankeu, A.; Taccioli, C.; Briskin, D.; Cheung, D.G.; Bolon, B.; Anderlucci, L.; Alder, H.; et al. Estrogen mediated-activation of miR-191/425 cluster modulates tumorigenicity of breast cancer cells depending on estrogen receptor status. PLoS Genet. 2013, 9, e1003311. [Google Scholar] [CrossRef]
- Aue, G.; Du, Y.; Cleveland, S.M.; Smith, S.B.; Dave, U.P.; Liu, D.; Weniger, M.A.; Metais, J.Y.; Jenkins, N.A.; Copeland, N.G.; et al. Sox4 cooperates with PU.1 haploinsufficiency in murine myeloid leukemia. Blood 2011, 118, 4674–4681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, X.W.; Wang, P.; Liu, J.Q.; Zhang, H.; Xi, W.D.; Jia, X.H.; Wang, K.K. Coordinated regulation of the immunoproteasome subunits by PML/RARα and PU.1 in acute promyelocytic leukemia. Oncogene 2014, 33, 2700–2708. [Google Scholar] [CrossRef] [Green Version]
- Astakhova, T.M.; Morozov, A.V.; Erokhov, P.A.; Mikhailovskaya, M.I.; Akopov, S.B.; Chupikova, N.I.; Safarov, R.R.; Sharova, N.P. Combined Effect of Bortezomib and Menadione Sodium Bisulfite on Proteasomes of Tumor Cells: The Dramatic Decrease of Bortezomib Toxicity in a Preclinical Trial. Cancers 2018, 10, 351. [Google Scholar] [CrossRef] [Green Version]
- Donskikh, O.A. Horror Zivilisationis, oder Horror der Subjektivität. Beacon J. Stud. Ideol. Ment. Dimens. 2019, 2, 020110205. Available online: http://hdl.handle.net//20.500.12656/thebeacon.2.020110205 (accessed on 23 January 2020).
- Thaler, S.; Thiede, G.; Hengstler, J.G.; Schad, A.; Schmidt, M.; Sleeman, J.P. The proteasome inhibitor Bortezomib (Velcade) as potential inhibitor of estrogen receptor-positive breast cancer. Int. J. Cancer 2015, 137, 686–697. [Google Scholar] [CrossRef]
- Chai, F.; Liang, Y.; Bi, J.; Chen, L.; Zhang, F.; Cui, Y.; Jiang, J. REGγ regulates ERα degradation via ubiquitin-proteasome pathway in breast cancer. Biochem. Biophys. Res. Commun. 2015, 456, 534–540. [Google Scholar] [CrossRef] [PubMed]
- Shashova, E.E.; Astakhova, T.M.; Plekhanova, A.S.; Bogomyagkova, Y.V.; Lyupina, Y.V.; Sumedi, I.R.; Slonimskaya, E.M.; Erokhov, P.A.; Abramova, E.B.; Rodoman, G.V.; et al. Changes in proteasome chymotrypsin-like activity during the development of human mammary and thyroid carcinomas. Bull. Exp. Biol. Med. 2013, 156, 242–244. [Google Scholar] [CrossRef] [PubMed]
- Kondakova, I.V.; Spirina, L.V.; Shashova, E.E.; Koval, V.D.; Kolomiets, L.A.; Chernyshova, A.L.; Slonimskaya, E.M. Proteasome activity in tumors of the female reproductive system. Russ. J. Bioorg. Chem. 2012, 38, 89–92. [Google Scholar] [CrossRef]
- Kondakova, I.V.; Spirina, L.V.; Koval, V.D.; Shashova, E.E.; Choinzonov, E.L.; Ivanova, E.V.; Kolomiets, L.A.; Chernyshova, A.L.; Slonimskaya, E.M.; Usynin, E.A.; et al. Chymotrypsin-like activity and subunit composition of proteasomes in human cancers. Mol. Biol. 2014, 48, 384–389. [Google Scholar] [CrossRef]
- Maynadier, M.; Basile, I.; Gallud, A.; Gary-Bobo, M.; Garcia, M. Combination treatment with proteasome inhibitors and antiestrogens has a synergistic effect mediated by p21WAF1 in estrogenreceptor-positive breast cancer. Oncol. Rep. 2016, 36, 1127–1134. [Google Scholar] [CrossRef]
- Xia, X.; Liao, Y.; Guo, Z.; Li, Y.; Jiang, L.; Zhang, F.; Huang, C.; Liu, Y.; Wang, X.; Liu, N.; et al. Targeting proteasome-associated deubiquitinases as a novel strategy for the treatment of estrogen receptor-positive breast cancer. Oncogenesis 2018, 7, 1–12. [Google Scholar] [CrossRef] [Green Version]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kondakova, I.V.; Shashova, E.E.; Sidenko, E.A.; Astakhova, T.M.; Zakharova, L.A.; Sharova, N.P. Estrogen Receptors and Ubiquitin Proteasome System: Mutual Regulation. Biomolecules 2020, 10, 500. https://doi.org/10.3390/biom10040500
Kondakova IV, Shashova EE, Sidenko EA, Astakhova TM, Zakharova LA, Sharova NP. Estrogen Receptors and Ubiquitin Proteasome System: Mutual Regulation. Biomolecules. 2020; 10(4):500. https://doi.org/10.3390/biom10040500
Chicago/Turabian StyleKondakova, Irina V., Elena E. Shashova, Evgenia A. Sidenko, Tatiana M. Astakhova, Liudmila A. Zakharova, and Natalia P. Sharova. 2020. "Estrogen Receptors and Ubiquitin Proteasome System: Mutual Regulation" Biomolecules 10, no. 4: 500. https://doi.org/10.3390/biom10040500
APA StyleKondakova, I. V., Shashova, E. E., Sidenko, E. A., Astakhova, T. M., Zakharova, L. A., & Sharova, N. P. (2020). Estrogen Receptors and Ubiquitin Proteasome System: Mutual Regulation. Biomolecules, 10(4), 500. https://doi.org/10.3390/biom10040500




