Phosphonic Acid Analogs of Fluorophenylalanines as Inhibitors of Human and Porcine Aminopeptidases N: Validation of the Importance of the Substitution of the Aromatic Ring
Abstract
1. Introduction
2. Materials and Methods
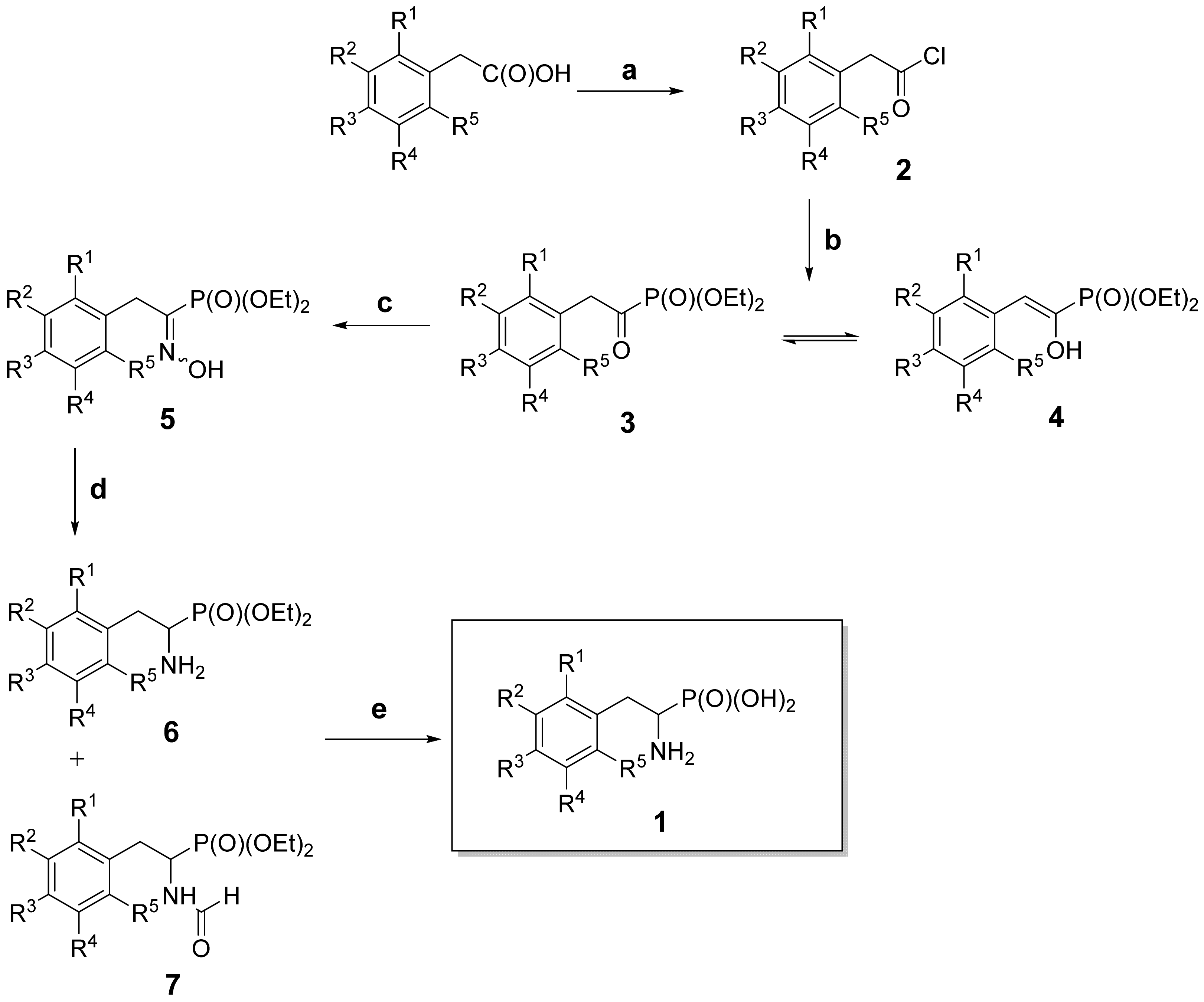
2.1. Chemistry
2.2. Synthesis of the Fluorinated 1-Amino-2-phenylethylphosphonic Acids (1)–General Procedure
2.2.1. 1-Amino-2-phenylethylphosphonic Acid (1a) Was Obtained after Deiodination during Reduction of the Oxime Intermediate to Aminophosphonate Ester
2.2.2. 1-Amino-2-(2-fluorophenyl)ethylphosphonic Acid (1b)
2.2.3. 1-Amino-2-(3-fluorophenyl)ethylphosphonic Acid (1c)
2.2.4. 1-Amino-2-(4-fluorophenyl)ethylphosphonic Acid (1d)
2.2.5. 1-Amino-2-(2,4-difluorophenyl)ethylphosphonic Acid (1e)
2.2.6. 1-Amino-2-(2,5-difluorophenyl)ethylphosphonic Acid (1f)
2.2.7. 1-Amino-2-(2,6-difluorophenyl)ethylphosphonic Acid (1g)
2.2.8. 1-Amino-2-(3,4-difluorophenyl)ethylphosphonic Acid (1h)
2.2.9. 1-Amino-2-(3,5-difluorophenyl)ethylphosphonic Acid (1i)
2.2.10. 1-Amino-2-(2,4,5-trifluorophenyl)ethylphosphonic Acid (1j)
2.2.11. 1-Amino-2-(2,4,6-trifluorophenyl)ethylphosphonic Acid (1k)
2.2.12. 1-Amino-2-(3,4,5-trifluorophenyl)ethylphosphonic Acid (1l)
2.2.13. 1-Amino-2-(2,3,4,5,6-pentafluorophenyl)ethylphosphonic Acid (1m)
2.2.14. 1-Amino-2-(3-chloro-2-fluorophenyl)ethylphosphonic Acid (1n)
2.2.15. 1-Amino-2-(4-chloro-2-fluorophenyl)ethylphosphonic Acid (1o)
2.2.16. 1-Amino-2-(5-chloro-2-fluorophenyl)ethylphosphonic Acid (1p)
2.2.17. 1-Amino-2-(2-chloro-6-fluorophenyl)ethylphosphonic Acid (1r)
2.2.18. 1-Amino-2-(4-chloro-3-fluorophenyl)ethylphosphonic Acid (1s)
2.2.19. 1-Amino-2-(3-chloro-4-fluorophenyl)ethylphosphonic Acid (1t)
2.2.20. 1-Amino-2-(2-chloro-4-fluorophenyl)ethylphosphonic Acid (1u)
2.2.21. 1-Amino-2-(2-trifluoromethylphenyl)ethylphosphonic Acid (1v)
2.2.22. 1-Amino-2-(3-trifluoromethylphenyl)ethylphosphonic Acid (1w)
2.2.23. 1-Amino-2-(4-trifluoromethylphenyl)ethylphosphonic Acid (1x)
2.2.24. 1-Amino-2-(5-fluoro-2-trifluoromethylphenyl)ethylphosphonic Acid (1y)
2.2.25. 1-Amino-2-(4-fluoro-3-trifluoromethylphenyl)ethylphosphonic Acid (1z)
2.2.26. (1-Amino-3,3,3-trifluoropropyl)phosphonic acid (8)
2.3. Bioassays
2.3.1. Enzymatic Studies
2.3.2. Molecular Modelling
2.3.3. Crystallography
3. Results and Discussion
3.1. Chemistry
3.2. Enzymatic Studies
3.3. Molecular Docking
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Hooper, N.M.; Lendeckel, U. (Eds.) Aminopeptidases in Biology and Disease; Springer US: Boston, MA, USA, 2004; Volume 2, ISBN 978-1-4613-4698-2. [Google Scholar] [CrossRef]
- Agrawal, N.; Brown, M.A. Genetic associations and functional characterization of M1 aminopeptidases and immune-mediated diseases. Genes Immun. 2014, 15, 521–527. [Google Scholar] [CrossRef] [PubMed]
- Nandan, A.; Nampoothiri, K.M. Molecular advances in microbial aminopeptidases. Bioresour. Technol. 2017, 245, 1757–1765. [Google Scholar] [CrossRef] [PubMed]
- Albiston, A.; Ye, S.; Chai, S. Membrane Bound Members of the M1 Family: More Than Aminopeptidases. Protein Pept. Lett. 2004, 11, 491–500. [Google Scholar] [CrossRef] [PubMed]
- Drinkwater, N.; Lee, J.; Yang, W.; Malcolm, T.R.; McGowan, S. M1 aminopeptidases as drug targets: Broad applications or therapeutic niche? FEBS J. 2017, 284, 1473–1488. [Google Scholar] [CrossRef]
- Mucha, A.; Drag, M.; Dalton, J.P.; Kafarski, P. Metallo-aminopeptidase inhibitors. Biochimie 2010, 92, 1509–1529. [Google Scholar] [CrossRef]
- Hitzerd, S.M.; Verbrugge, S.E.; Ossenkoppele, G.; Jansen, G.; Peters, G.J. Positioning of aminopeptidase inhibitors in next generation cancer therapy. Amino Acids 2014, 46, 793–808. [Google Scholar] [CrossRef]
- Bounaadja, L.; Schmitt, M.; Albrecht, S.; Mouray, E.; Tarnus, C.; Florent, I. Selective inhibition of PfA-M1, over PfA-M17, by an amino-benzosuberone derivative blocks malaria parasites development in vitro and in vivo. Malar. J. 2017, 16, 382. [Google Scholar] [CrossRef]
- Luan, Y.; Xu, W. The Structure and Main Functions of Aminopeptidase N. Curr. Med. Chem. 2007, 14, 639–647. [Google Scholar] [CrossRef]
- Chen, L.; Lin, Y.-L.; Peng, G.; Li, F. Structural basis for multifunctional roles of mammalian aminopeptidase N. Proc. Natl. Acad. Sci. USA 2012, 109, 17966–17971. [Google Scholar] [CrossRef]
- Lu, C.; Amin, M.A.; Fox, D.A. CD13/Aminopeptidase N Is a Potential Therapeutic Target for Inflammatory Disorders. J. Immunol. 2020, 204, 3–11. [Google Scholar] [CrossRef]
- Surowiak, P.; Drąg, M.; Materna, V.; Suchocki, S.; Grzywa, R.; Spaczyński, M.; Dietel, M.; Oleksyszyn, J.; Zabel, M.; Lage, H. Expression of aminopeptidase N/CD13 in human ovarian cancers. Int. J. Gynecol. Cancer 2006, 16, 1783–1788. [Google Scholar] [CrossRef] [PubMed]
- Reguera, J.; Santiago, C.; Mudgal, G.; Ordoño, D.; Enjuanes, L.; Casasnovas, J.M. Structural Bases of Coronavirus Attachment to Host Aminopeptidase N and Its Inhibition by Neutralizing Antibodies. PLoS Pathog. 2012, 8, e1002859. [Google Scholar] [CrossRef] [PubMed]
- Kontoyiannis, D.P.; Pasqualini, R.; Arap, W. Aminopeptidase N inhibitors and SARS. Lancet 2003, 361, 1558. [Google Scholar] [CrossRef]
- Wickström, M.; Larsson, R.; Nygren, P.; Gullbo, J. Aminopeptidase N (CD13) as a target for cancer chemotherapy. Cancer Sci. 2011, 102, 501–508. [Google Scholar] [CrossRef]
- Wang, X.; Wang, B.; Zhang, Q. Anti-tumor targeted drug delivery systems mediated by aminopeptidase N/CD13. Acta Pharm. Sin. B 2011, 1, 80–83. [Google Scholar] [CrossRef][Green Version]
- Grzywa, R.; Oleksyszyn, J.; Salvesen, G.S.; Drąg, M. Identification of very potent inhibitor of human aminopeptidase N (CD13). Bioorg. Med. Chem. Lett. 2010, 20, 2497–2499. [Google Scholar] [CrossRef]
- Su, L.; Fang, H.; Xu, W. Aminopeptidase N (EC 3.4.11.2) inhibitors (2006–2010): A patent review. Expert Opin. Ther. Pat. 2011, 21, 1241–1265. [Google Scholar] [CrossRef]
- Albrecht, S.; Al-Lakkis-Wehbe, M.; Orsini, A.; Defoin, A.; Pale, P.; Salomon, E.; Tarnus, C.; Weibel, J.-M. Amino-benzosuberone: A novel warhead for selective inhibition of human aminopeptidase-N/CD13. Bioorg. Med. Chem. 2011, 19, 1434–1449. [Google Scholar] [CrossRef]
- Amin, S.A.; Adhikari, N.; Jha, T. Design of Aminopeptidase N Inhibitors as Anti-cancer Agents. J. Med. Chem. 2018, 61, 6468–6490. [Google Scholar] [CrossRef]
- Węglarz-Tomczak, E.; Talma, M.; Giurg, M.; Westerhoff, H.V.; Janowski, R.; Mucha, A. Neutral metalloaminopeptidases APN and MetAP2 as newly discovered anticancer molecular targets of actinomycin D and its simple analogs. Oncotarget 2018, 9, 29365–29378. [Google Scholar] [CrossRef]
- Lee, J.; Vinh, N.B.; Drinkwater, N.; Yang, W.; Kannan Sivaraman, K.; Schembri, L.S.; Gazdik, M.; Grin, P.M.; Butler, G.S.; Overall, C.M.; et al. Novel Human Aminopeptidase N Inhibitors: Discovery and Optimization of Subsite Binding Interactions. J. Med. Chem. 2019, 62, 7185–7209. [Google Scholar] [CrossRef] [PubMed]
- Talma, M.; Maślanka, M.; Mucha, A. Recent developments in the synthesis and applications of phosphinic peptide analogs. Bioorg. Med. Chem. Lett. 2019, 29, 1031–1042. [Google Scholar] [CrossRef] [PubMed]
- Luan, Y.; Mu, J.; Xu, W. The Review of the Synthesis of Bestatin, an Effective Inhibitor of Aminopeptidase N. Mini. Rev. Org. Chem. 2008, 5, 134–140. [Google Scholar] [CrossRef]
- Lejczak, B.; Kafarski, P.; Zygmunt, J. Inhibition of aminopeptidases by aminophosphonates. Biochemistry 1989, 28, 3549–3555. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Zou, J.; Tian, F.; Shang, Z. Fluorine Bonding—How Does It Work In Protein−Ligand Interactions? J. Chem. Inf. Model. 2009, 49, 2344–2355. [Google Scholar] [CrossRef]
- Zhou, Y.; Wang, J.; Gu, Z.; Wang, S.; Zhu, W.; Aceña, J.L.; Soloshonok, V.A.; Izawa, K.; Liu, H. Next Generation of Fluorine-Containing Pharmaceuticals, Compounds Currently in Phase II–III Clinical Trials of Major Pharmaceutical Companies: New Structural Trends and Therapeutic Areas. Chem. Rev. 2016, 116, 422–518. [Google Scholar] [CrossRef]
- Wanat, W.; Talma, M.; Pawełczak, M.; Kafarski, P. Phosphonic Acid Analogues of Phenylglycine as Inhibitors of Aminopeptidases: Comparison of Porcine Aminopeptidase N, Bovine Leucine Aminopeptidase, Tomato Acidic Leucine Aminopeptidase and Aminopeptidase from Barley Seeds. Pharmaceuticals 2019, 12, 139. [Google Scholar] [CrossRef]
- Ziora, Z.; Kafarski, P. Amidoalkylation of Phosphorus Trichloride with Acetamide and Alkyl Oxocycloalkanecarboxylates. Phosphorus. Sulfur. Silicon Relat. Elem. 2009, 184, 1047–1053. [Google Scholar] [CrossRef]
- Soroka, M. The synthesis of 1-aminoalkylphosphonic acids. A revised mechanism of the reaction of phosphorus trichloride, amides and aldehydes or ketones in acetic acid (Oleksyszyn reaction). Liebigs Ann. der Chemie 1990, 1990, 331–334. [Google Scholar] [CrossRef]
- Vassiliou, S.; Węglarz-Tomczak, E.; Berlicki, Ł.; Pawełczak, M.; Nocek, B.; Mulligan, R.; Joachimiak, A.; Mucha, A. Structure-Guided, Single-Point Modifications in the Phosphinic Dipeptide Structure Yield Highly Potent and Selective Inhibitors of Neutral Aminopeptidases. J. Med. Chem. 2014, 57, 8140–8151. [Google Scholar] [CrossRef]
- Węglarz-Tomczak, E.; Berlicki, Ł.; Pawełczak, M.; Nocek, B.; Joachimiak, A.; Mucha, A. A structural insight into the P1 S1 binding mode of diaminoethylphosphonic and phosphinic acids, selective inhibitors of alanine aminopeptidases. Eur. J. Med. Chem. 2016, 117, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Wong, A.H.M.; Zhou, D.; Rini, J.M. The X-ray Crystal Structure of Human Aminopeptidase N Reveals a Novel Dimer and the Basis for Peptide Processing. J. Biol. Chem. 2012, 287, 36804–36813. [Google Scholar] [CrossRef] [PubMed]
- Schrödinger LLC. Schrödinger Release 2018-4: LigPrep; Schrödinger LLC: New York, NY, USA, 2018. [Google Scholar]
- Schrödinger LLC. Schrödinger Release 2018-4: Schrödinger Suite 2018-2 Induced Fit Docking Protocol; Glide; Schrödinger LLC: New York, NY, USA, 2018. [Google Scholar]
- Schrödinger LLC. Schrödinger Release 2018-4: Prime; Schrödinger LLC: New York, NY, USA, 2018. [Google Scholar]
- CrysAlis CCD. CrysAlis RED; Oxford Diffraction Ltd.: Abingdon, UK, 2002.
- Sheldrick, G.M. A short history of SHELX. Acta Crystallogr. Sect. A Found. Crystallogr. 2008, 64, 112–122. [Google Scholar] [CrossRef] [PubMed]
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. Sect. C Struct. Chem. 2015, 71, 3–8. [Google Scholar] [CrossRef]
- Macrae, C.F.; Bruno, I.J.; Chisholm, J.A.; Edgington, P.R.; McCabe, P.; Pidcock, E.; Rodriguez-Monge, L.; Taylor, R.; van de Streek, J.; Wood, P.A. Mercury CSD 2.0—New features for the visualization and investigation of crystal structures. J. Appl. Crystallogr. 2008, 41, 466–470. [Google Scholar] [CrossRef]
- Kwiczak-Yiğitbaşı, J.; Pirat, J.-L.; Virieux, D.; Volle, J.-N.; Janiak, A.; Hoffmann, M.; Pluskota-Karwatka, D. Fluorinated phosphonate analogues of phenylalanine: Synthesis, X-ray and DFT studies. Arab. J. Chem. 2020, 13, 2384–2399. [Google Scholar] [CrossRef]
- Kowalik, J.; Kupczyk-Subotkowska, L.; Mastalerz, P. Preparation of Dialkyl 1-Aminoalkanephosphonates by Reduction of Dialkyl 1-Hydroxyiminoalkanephosphonates with Zinc in Formic Acid. Synthesis (Stuttg.) 1981, 1981, 57–58. [Google Scholar] [CrossRef]
- Maier, L.; Diel, P.J. Organic phosphorus compounds 97. 1 Synthesis and properties of 1-amino-2-aryl- and 2-pyridyl-ethylphosphonic acids and derivatives. Phosphorus. Sulfur. Silicon Relat. Elem. 1991, 62, 15–27. [Google Scholar] [CrossRef]
- Drąg, M.; Grembecka, J.; Pawełczak, M.; Kafarski, P. α-Aminoalkylphosphonates as a tool in experimental optimisation of P1 side chain shape of potential inhibitors in S1 pocket of leucine- and neutral aminopeptidases. Eur. J. Med. Chem. 2005, 40, 764–771. [Google Scholar] [CrossRef]
- Tam, C.C.; Mattocks, K.L.; Tishler, M. Enol-keto tautomerism of -ketophosphonates. Proc. Natl. Acad. Sci. USA 1981, 78, 3301–3304. [Google Scholar] [CrossRef]
- Abiraj, K.; Gowda, D.C. Zinc/ammonium formate: A new facile system for the rapid and selective reduction of oximes to amines. J. Chem. Res. 2003, 332–333. [Google Scholar] [CrossRef]
- Jnaneshwara, G.K.; Sudalai, A.; Deshpande, V.H. Palladium-catalysed Transfer Hydrogenation of Azobenzenes and Oximes using Ammonium Formate. J. Chem. Res. 1998, 3, 160–161. [Google Scholar] [CrossRef]
- Abiraj, K.; Gowda, D.C. Magnesium-Catalyzed Proficient Reduction of Oximes to Amines Using Ammonium Formate. Synth. Commun. 2004, 34, 599–605. [Google Scholar] [CrossRef]
- Baláž, M.; Kudličková, Z.; Vilková, M.; Imrich, J.; Balážová, Ľ.; Daneu, N. Mechanochemical Synthesis and Isomerization of N-Substituted Indole-3-carboxaldehyde Oximes. Molecules 2019, 24, 3347. [Google Scholar] [CrossRef]
- Liu, G.-B.; Zhao, H.-Y.; Yang, B.; Thiemann, T. Zinc dust-mediated reductive degradation of decabromodiphenyl ether. Green Chem. Lett. Rev. 2010, 3, 1–6. [Google Scholar] [CrossRef]
- Tashiro, M.; Fukata, G. Studies on selective preparation of aromatic compounds. XII. The selective reductive dehalogenation of some halophenols with zinc powder in basic and acidic media. J. Org. Chem. 1977, 42, 835–838. [Google Scholar] [CrossRef]
- Morgan, B.P.; Scholz, J.M.; Ballinger, M.D.; Zipkin, I.D.; Bartlett, P.A. Differential Binding Energy: A Detailed Evaluation of the Influence of Hydrogen-Bonding and Hydrophobic Groups on the Inhibition of Thermolysin by Phosphorus-Containing Inhibitors. J. Am. Chem. Soc. 1991, 113, 297–307. [Google Scholar] [CrossRef]





| Cmpd. | R1 | R2 | R3 | R4 | R5 | pAPN Ki (µM) | hAPN Ki (µM) |
|---|---|---|---|---|---|---|---|
| 1a | H | H | H | H | H | 73.3 ± 7.6 | 15.5 ± 1.7 |
| 1b | F | H | H | H | H | 256 ± 19 | 18.1 ± 3.7 |
| 1c | H | F | H | H | H | 26.4 ± 2.0 | 3.39 ± 0.71 |
| 1d | H | H | F | H | H | 41.4 ± 2.5 | 4.86 ± 1.18 |
| 1e | F | H | F | H | H | 95.6 ± 9.6 | 13.5 ± 2.7 |
| 1f | F | H | H | F | H | 86.3 ± 8.2 | 25.9 ± 5.0 |
| 1g | F | H | H | H | F | 725 ± 121 | 327 ± 98 |
| 1h | H | F | F | H | H | 15.7 ± 2.5 | 1.71 ± 0.52 |
| 1i | H | F | H | F | H | 8.09 ± 1.1 | 0.529 ± 0.088 |
| 1j | F | H | F | F | H | 78.5 ± 3.9 | 15.0 ± 2.2 |
| 1k | F | H | F | H | F | 969 ± 158 | 225 ± 47 |
| 1l | H | F | F | F | H | 7.10 ± 0.93 | 0.793 ± 0.138 |
| 1m | F | F | F | F | F | 808 ± 132 | 139 ± 71 |
| 1n | F | Cl | H | H | H | 85.1 ± 9.1 | 18.5 ± 7.1 |
| 1o | F | H | Cl | H | H | 160 ± 11 | 17.8 ± 2.09 |
| 1p | F | H | H | Cl | H | 47.2 ± 1.4 | 4.30 ± 0.44 |
| 1r | F | H | H | H | Cl | 2557 ± 729 | 360 ± 125 |
| 1s | H | F | Cl | H | H | 14.8 ± 0.9 | 1.01 ± 0.16 |
| 1t | H | H | F | Cl | H | 6.93 ± 0.78 | 0.542 ± 0.152 |
| 1u | H | H | F | H | Cl | 106 ± 12 | 29.1 ± 10.3 |
| 1v | CF3 | H | H | H | H | 2335 ± 617 | Inactive a |
| 1w | H | CF3 | H | H | H | 66.0 ± 7.3 | 2.53 ± 0.4 |
| 1x | H | H | CF3 | H | H | 257 ± 27 | 24.9 ± 3.0 |
| 1y | CF3 | H | H | F | H | Inactive a | Inactive a |
| 1z | H | CF3 | F | H | H | 67.7 ± 5.6 | 3.55 ± 0.43 |
| 8 | 755 ± 230 | 84.7 ± 15.8 | |||||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wanat, W.; Talma, M.; Dziuk, B.; Pirat, J.-L.; Kafarski, P. Phosphonic Acid Analogs of Fluorophenylalanines as Inhibitors of Human and Porcine Aminopeptidases N: Validation of the Importance of the Substitution of the Aromatic Ring. Biomolecules 2020, 10, 579. https://doi.org/10.3390/biom10040579
Wanat W, Talma M, Dziuk B, Pirat J-L, Kafarski P. Phosphonic Acid Analogs of Fluorophenylalanines as Inhibitors of Human and Porcine Aminopeptidases N: Validation of the Importance of the Substitution of the Aromatic Ring. Biomolecules. 2020; 10(4):579. https://doi.org/10.3390/biom10040579
Chicago/Turabian StyleWanat, Weronika, Michał Talma, Błażej Dziuk, Jean-Luc Pirat, and Paweł Kafarski. 2020. "Phosphonic Acid Analogs of Fluorophenylalanines as Inhibitors of Human and Porcine Aminopeptidases N: Validation of the Importance of the Substitution of the Aromatic Ring" Biomolecules 10, no. 4: 579. https://doi.org/10.3390/biom10040579
APA StyleWanat, W., Talma, M., Dziuk, B., Pirat, J.-L., & Kafarski, P. (2020). Phosphonic Acid Analogs of Fluorophenylalanines as Inhibitors of Human and Porcine Aminopeptidases N: Validation of the Importance of the Substitution of the Aromatic Ring. Biomolecules, 10(4), 579. https://doi.org/10.3390/biom10040579







