Australian Scorpion Hormurus waigiensis Venom Fractions Show Broad Bioactivity through Modulation of Bio-Impedance and Cytosolic Calcium
Abstract
:1. Introduction
2. Materials and Methods
2.1. Generation of the Recombinant HEK293-RyR1-GCaMP5G Cell Line
2.2. Scorpion Venom Collection
2.3. Scorpion Venom Analysis
2.4. Measurement of rHEK293-RyR1-GCaMP5G Cell Monolayer Bioimpedance
2.5. Calcium Imaging
3. Results
3.1. Scorpion Venom Molecular Characterisation
3.2. Venom-Induced Membrane Impedance Responses
3.3. Venom-Induced Modulation of Cytosolic Calcium
3.4. Comparison of Venom Activity Across Platforms
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ortiz, E.; Gurrola, G.B.; Schwartz, E.F.; Possani, L.D. Scorpion venom components as potential candidates for drug development. Toxicon 2015, 93, 125–135. [Google Scholar] [CrossRef] [PubMed]
- Srairi-Abid, N.; Othman, H.; Aissaoui, D.; BenAissa, R. Anti-tumoral effect of scorpion peptides: Emerging new cellular targets and signaling pathways. Cell Calcium 2019, 80, 160–174. [Google Scholar] [CrossRef] [PubMed]
- Das, B.; Patra, A.; Mukherjee, A.K. Correlation of Venom Toxinome Composition of Indian Red Scorpion (Mesobuthus tamulus) with Clinical Manifestations of Scorpion Stings: Failure of Commercial Antivenom to Immune-Recognize the Abundance of Low Molecular Mass Toxins of This Venom. J. Proteome Res. 2020. [Google Scholar] [CrossRef] [PubMed]
- Ménez, A.; Bontems, F.; Roumestand, C.; Gilquin, B.; Toma, F. Structural basis for functional diversity of animal toxins. Proc. R. Soc. Edinb. Sect. B 1992, 99, 83–103. [Google Scholar] [CrossRef]
- Goudet, C.; Chi, C.W.; Tytgat, J. An overview of toxins and genes from the venom of the Asian scorpion Buthus martensi Karsch. Toxicon 2002, 40, 1239–1258. [Google Scholar] [CrossRef]
- Tytgat, J.; Chandy, K.G.; Garcia, M.L.; Gutman, G.A.; Martin-Eauclaire, M.F.; van der Walt, J.J.; Possani, L.D. A unified nomenclature for short-chain peptides isolated from scorpion venoms: Alpha-KTx molecular subfamilies. Trends Pharmacol. Sci. 1999, 20, 444–447. [Google Scholar] [CrossRef]
- Chippaux, J.P.; Goyffon, M. Epidemiology of scorpionism: A global appraisal. Acta Trop. 2008, 107, 71–79. [Google Scholar] [CrossRef]
- Martin-Eauclaire, M.F.; Bougis, P.E. Potassium Channels Blockers from the Venom of Androctonus mauretanicus mauretanicus. J. Toxicol. 2012, 2012, 103608. [Google Scholar] [CrossRef] [Green Version]
- Nicholson, G.M.; Graudins, A.; Wilson, H.I.; Little, M.; Broady, K.W. Arachnid toxinology in Australia: From clinical toxicology to potential applications. Toxicon 2006, 48, 872–898. [Google Scholar] [CrossRef] [Green Version]
- Quintero-Hernandez, V.; Ramirez-Carreto, S.; Romero-Gutierrez, M.T.; Valdez-Velazquez, L.L.; Becerril, B.; Possani, L.D.; Ortiz, E. Transcriptome analysis of scorpion species belonging to the Vaejovis genus. PLoS ONE 2015, 10, e0117188. [Google Scholar] [CrossRef] [Green Version]
- Santibanez-Lopez, C.E.; Possani, L.D. Overview of the Knottin scorpion toxin-like peptides in scorpion venoms: Insights on their classification and evolution. Toxicon 2015, 107, 317–326. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.C.; Corzo, G.; Hahin, R. Scorpion venom peptides without disulfide bridges. IUBMB Life 2005, 57, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Gordon, D.; Karbat, I.; Ilan, N.; Cohen, L.; Kahn, R.; Gilles, N.; Dong, K.; Stuhmer, W.; Tytgat, J.; Gurevitz, M. The differential preference of scorpion alpha-toxins for insect or mammalian sodium channels: Implications for improved insect control. Toxicon 2007, 49, 452–472. [Google Scholar] [CrossRef] [PubMed]
- Pedraza Escalona, M.; Possani, L.D. Scorpion beta-toxins and voltage-gated sodium channels: Interactions and effects. Front. Biosci. 2013, 18, 572–587. [Google Scholar] [CrossRef]
- Pucca, M.B.; Cerni, F.A.; Pinheiro Junior, E.L.; Bordon Kde, C.; Amorim, F.G.; Cordeiro, F.A.; Longhim, H.T.; Cremonez, C.M.; Oliveira, G.H.; Arantes, E.C. Tityus serrulatus venom--A lethal cocktail. Toxicon 2015, 108, 272–284. [Google Scholar] [CrossRef]
- Gilchrist, J.; Bosmans, F. Animal toxins can alter the function of Nav1.8 and Nav1.9. Toxins 2012, 4, 620–632. [Google Scholar] [CrossRef]
- Gordon, D.; Gurevitz, M. The selectivity of scorpion alpha-toxins for sodium channel subtypes is determined by subtle variations at the interacting surface. Toxicon 2003, 41, 125–128. [Google Scholar] [CrossRef]
- Jimenez-Vargas, J.M.; Restano-Cassulini, R.; Possani, L.D. Toxin modulators and blockers of hERG K(+) channels. Toxicon 2012, 60, 492–501. [Google Scholar] [CrossRef]
- Bartok, A.; Panyi, G.; Varga, Z. Potassium channel blocking peptide toxins from scorpion venom. In Scorpion Venoms; Gopalakrishnakone, P., Possani, L.D., Schwartz, E., Rodríguez de la Vega, C.R., Eds.; Springer Netherlands: Dordrecht, The Netherlands, 2015; pp. 493–527. [Google Scholar]
- Yu, M.; Liu, S.L.; Sun, P.B.; Pan, H.; Tian, C.L.; Zhang, L.H. Peptide toxins and small-molecule blockers of BK channels. Acta Pharmacol. Sin. 2016, 37, 56–66. [Google Scholar] [CrossRef] [Green Version]
- Wanke, E.; Restano-Cassulini, R. Toxins interacting with ether-a-go-go-related gene voltage-dependent potassium channels. Toxicon 2007, 49, 239–248. [Google Scholar] [CrossRef]
- Norton, R.S.; McDonough, S.I. Peptides targeting voltage-gated calcium channels. Curr. Pharm. Des. 2008, 14, 2480–2491. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Franco, J.; Fill, M. Approaching ryanodine receptor therapeutics from the calcin angle. J. Gen. Physiol. 2016, 147, 369–373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dardevet, L.; Rani, D.; Aziz, T.A.; Bazin, I.; Sabatier, J.M.; Fadl, M.; Brambilla, E.; De Waard, M. Chlorotoxin: A helpful natural scorpion peptide to diagnose glioma and fight tumor invasion. Toxins 2015, 7, 1079–1101. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, E.F.; Capes, E.M.; Diego-Garcia, E.; Zamudio, F.Z.; Fuentes, O.; Possani, L.D.; Valdivia, H.H. Characterization of hadrucalcin, a peptide from Hadrurus gertschi scorpion venom with pharmacological activity on ryanodine receptors. Br. J. Pharmacol. 2009, 157, 392–403. [Google Scholar] [CrossRef] [Green Version]
- Housley, D.M.; Housley, G.D.; Liddell, M.J.; Jennings, E.A. Scorpion toxin peptide action at the ion channel subunit level. Neuropharmacology 2017, 127, 46–78. [Google Scholar] [CrossRef] [PubMed]
- Oliveira-Mendes, B.B.R.; Horta, C.C.R.; do Carmo, A.O.; Biscoto, G.L.; Sales-Medina, D.F.; Leal, H.G.; Brandao-Dias, P.F.P.; Miranda, S.E.M.; Aguiar, C.J.; Cardoso, V.N.; et al. CPP-Ts: A new intracellular calcium channel modulator and a promising tool for drug delivery in cancer cells. Sci. Rep. 2018, 8, 14739. [Google Scholar] [CrossRef] [PubMed]
- Berridge, M.J. Capacitative calcium entry. Biochem. J. 1995, 312, 1–11. [Google Scholar] [CrossRef]
- Luo, D.; Sun, H.; Xiao, R.P.; Han, Q. Caffeine induced Ca2+ release and capacitative Ca2+ entry in human embryonic kidney (HEK293) cells. Eur. J. Pharmacol. 2005, 509, 109–115. [Google Scholar] [CrossRef]
- Morton-Jones, R.T.; Cannell, M.B.; Housley, G.D. Ca2+ entry via AMPA-type glutamate receptors triggers Ca2+-induced Ca2+ release from ryanodine receptors in rat spiral ganglion neurons. Cell Calcium 2008, 43, 356–366. [Google Scholar] [CrossRef]
- Franzini-Armstrong, C.; Protasi, F. Ryanodine receptors of striated muscles: A complex channel capable of multiple interactions. Physiol. Rev. 1997, 77, 699–729. [Google Scholar] [CrossRef]
- Bers, D.M. Macromolecular complexes regulating cardiac ryanodine receptor function. J. Mol. Cell. Cardiol. 2004, 37, 417–429. [Google Scholar] [CrossRef] [PubMed]
- Alexander, S.P.; Kelly, E.; Marrion, N.V.; Peters, J.A.; Faccenda, E.; Harding, S.D.; Pawson, A.J.; Sharman, J.L.; Southan, C.; Davies, J.A.; et al. The concise guide to pharmacology 2017/18: Other ion channels. Br. J. Pharmacol. 2017, 174 (Suppl. 1), S195–S207. [Google Scholar] [CrossRef] [Green Version]
- Takeshima, H.; Nishimura, S.; Matsumoto, T.; Ishida, H.; Kangawa, K.; Minamino, N.; Matsuo, H.; Ueda, M.; Hanaoka, M.; Hirose, T.; et al. Primary structure and expression from complementary DNA of skeletal muscle ryanodine receptor. Nature 1989, 339, 439–445. [Google Scholar] [CrossRef] [PubMed]
- Nakai, J.; Imagawa, T.; Hakamat, Y.; Shigekawa, M.; Takeshima, H.; Numa, S. Primary structure and functional expression from cDNA of the cardiac ryanodine receptor/calcium release channel. FEBS Lett. 1990, 271, 169–177. [Google Scholar] [CrossRef] [Green Version]
- Hakamata, Y.; Nakai, J.; Takeshima, H.; Imoto, K. Primary structure and distribution of a novel ryanodine receptor/calcium release channel from rabbit brain. FEBS Lett. 1992, 312, 229–235. [Google Scholar] [CrossRef] [Green Version]
- Fill, M.; Copello, J.A. Ryanodine receptor calcium release channels. Physiol. Rev. 2002, 82, 893–922. [Google Scholar] [CrossRef] [Green Version]
- Ali, S.Z.; Taguchi, A.; Rosenberg, H. Malignant hyperthermia. Best Pract. Res. Clin. Anaesthesiol. 2003, 17, 519–533. [Google Scholar] [CrossRef]
- Priori, S.G.; Napolitano, C.; Tiso, N.; Memmi, M.; Vignati, G.; Bloise, R.; Sorrentino, V.; Danieli, G.A. Mutations in the cardiac ryanodine receptor gene (hRyR2) underlie catecholaminergic polymorphic ventricular tachycardia. Circulation 2001, 103, 196–200. [Google Scholar] [CrossRef] [Green Version]
- Quintero-Hernandez, V.; Jimenez-Vargas, J.M.; Gurrola, G.B.; Valdivia, H.H.; Possani, L.D. Scorpion venom components that affect ion-channels function. Toxicon 2013, 76, 328–342. [Google Scholar] [CrossRef] [Green Version]
- Xiao, L.; Gurrola, G.B.; Zhang, J.; Valdivia, C.R.; SanMartin, M.; Zamudio, F.Z.; Zhang, L.; Possani, L.D.; Valdivia, H.H. Structure-function relationships of peptides forming the calcin family of ryanodine receptor ligands. J. Gen. Physiol. 2016, 147, 375–394. [Google Scholar] [CrossRef] [Green Version]
- Tripathy, A.; Resch, W.; Xu, L.; Valdivia, H.H.; Meissner, G. Imperatoxin A induces subconductance states in Ca2+ release channels (ryanodine receptors) of cardiac and skeletal muscle. J. Gen. Physiol. 1998, 111, 679–690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gurrola, G.B.; Capes, E.M.; Zamudio, F.Z.; Possani, L.D.; Valdivia, H.H. Imperatoxin A, a Cell-Penetrating Peptide from Scorpion Venom, as a Probe of Ca2+-Release Channels/Ryanodine Receptors. Pharmaceuticals 2010, 3, 1093–1107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du Plessis, L.H.; Elgar, D.; du Plessis, J.L. Southern African scorpion toxins: An overview. Toxicon 2008, 51, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Boisseau, S.; Mabrouk, K.; Ram, N.; Garmy, N.; Collin, V.; Tadmouri, A.; Mikati, M.; Sabatier, J.M.; Ronjat, M.; Fantini, J.; et al. Cell penetration properties of maurocalcine, a natural venom peptide active on the intracellular ryanodine receptor. Biochim. Biophys. Acta 2006, 1758, 308–319. [Google Scholar] [CrossRef] [PubMed]
- Monod, L.; Prendini, L. Evidence for Eurogondwana: The roles of dispersal, extinction and vicariance in the evolution and biogeography of Indo-Pacific Hormuridae (Scorpiones: Scorpionoidea). Cladistics 2015, 31, 71–111. [Google Scholar] [CrossRef]
- Smith, J.J.; Vetter, I.; Lewis, R.J.; Peigneur, S.; Tytgat, J.; Lam, A.; Gallant, E.M.; Beard, N.A.; Alewood, P.F.; Dulhunty, A.F. Multiple actions of phi-LITX-Lw1a on ryanodine receptors reveal a functional link between scorpion DDH and ICK toxins. Proc. Natl. Acad. Sci. USA 2013, 110, 8906–8911. [Google Scholar] [CrossRef] [Green Version]
- Querfurth, H.W.; Haughey, N.J.; Greenway, S.C.; Yacono, P.W.; Golan, D.E.; Geiger, J.D. Expression of ryanodine receptors in human embryonic kidney (HEK293) cells. Biochem. J. 1998, 334 Pt 1, 79–86. [Google Scholar] [CrossRef] [Green Version]
- Tong, J.; Du, G.G.; Chen, S.R.; MacLennan, D.H. HEK-293 cells possess a carbachol- and thapsigargin-sensitive intracellular Ca2+ store that is responsive to stop-flow medium changes and insensitive to caffeine and ryanodine. Biochem. J. 1999, 343 Pt 1, 39–44. [Google Scholar] [CrossRef]
- Akerboom, J.; Chen, T.W.; Wardill, T.J.; Tian, L.; Marvin, J.S.; Mutlu, S.; Calderon, N.C.; Esposti, F.; Borghuis, B.G.; Sun, X.R.; et al. Optimization of a GCaMP calcium indicator for neural activity imaging. J. Neurosci. 2012, 32, 13819–13840. [Google Scholar] [CrossRef]
- Koch, L.E. The taxonomy, geographic distribution and evolutionary radiation of australo-papuan scorpions. West. Aust. Mus. 1977, 5 Pt 2, 83–367. [Google Scholar]
- Denelavas, A.; Weibel, F.; Prummer, M.; Imbach, A.; Clerc, R.G.; Apfel, C.M.; Hertel, C. Real-time cellular impedance measurements detect Ca2+ channel-dependent oscillations of morphology in human H295R adrenoma cells. Biochim. Biophys. Acta 2011, 1813, 754–762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodriguez de la Vega, R.C.; Schwartz, E.F.; Possani, L.D. Mining on scorpion venom biodiversity. Toxicon 2010, 56, 1155–1161. [Google Scholar] [CrossRef] [PubMed]
- Chaousis, S.; Smout, M.; Wilson, D.; Loukas, A.; Mulvenna, J.; Seymour, J. Rapid short term and gradual permanent cardiotoxic effects of vertebrate toxins from Chironex fleckeri (Australian box jellyfish) venom. Toxicon 2014, 80, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Chuang, R.S.; Jaffe, H.; Cribbs, L.; Perez-Reyes, E.; Swartz, K.J. Inhibition of T-type voltage-gated calcium channels by a new scorpion toxin. Nat. Neurosci. 1998, 1, 668–674. [Google Scholar] [CrossRef]
- Olamendi-Portugal, T.; Bartok, A.; Zamudio-Zuniga, F.; Balajthy, A.; Becerril, B.; Panyi, G.; Possani, L.D. Isolation, chemical and functional characterization of several new K+-channel blocking peptides from the venom of the scorpion Centruroides tecomanus. Toxicon 2016, 115, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Smith, J.J.; Hill, J.M.; Little, M.J.; Nicholson, G.M.; King, G.F.; Alewood, P.F. Unique scorpion toxin with a putative ancestral fold provides insight into evolution of the inhibitor cystine knot motif. Proc. Natl. Acad. Sci. USA 2011, 108, 10478–10483. [Google Scholar] [CrossRef] [Green Version]
- Gangur, A.N.; Smout, M.; Liddell, M.J.; Seymour, J.E.; Wilson, D.; Northfield, T.D. Changes in predator exposure, but not in diet, induce phenotypic plasticity in scorpion venom. Proc. Biol. Sci. 2017, 284. [Google Scholar] [CrossRef] [Green Version]


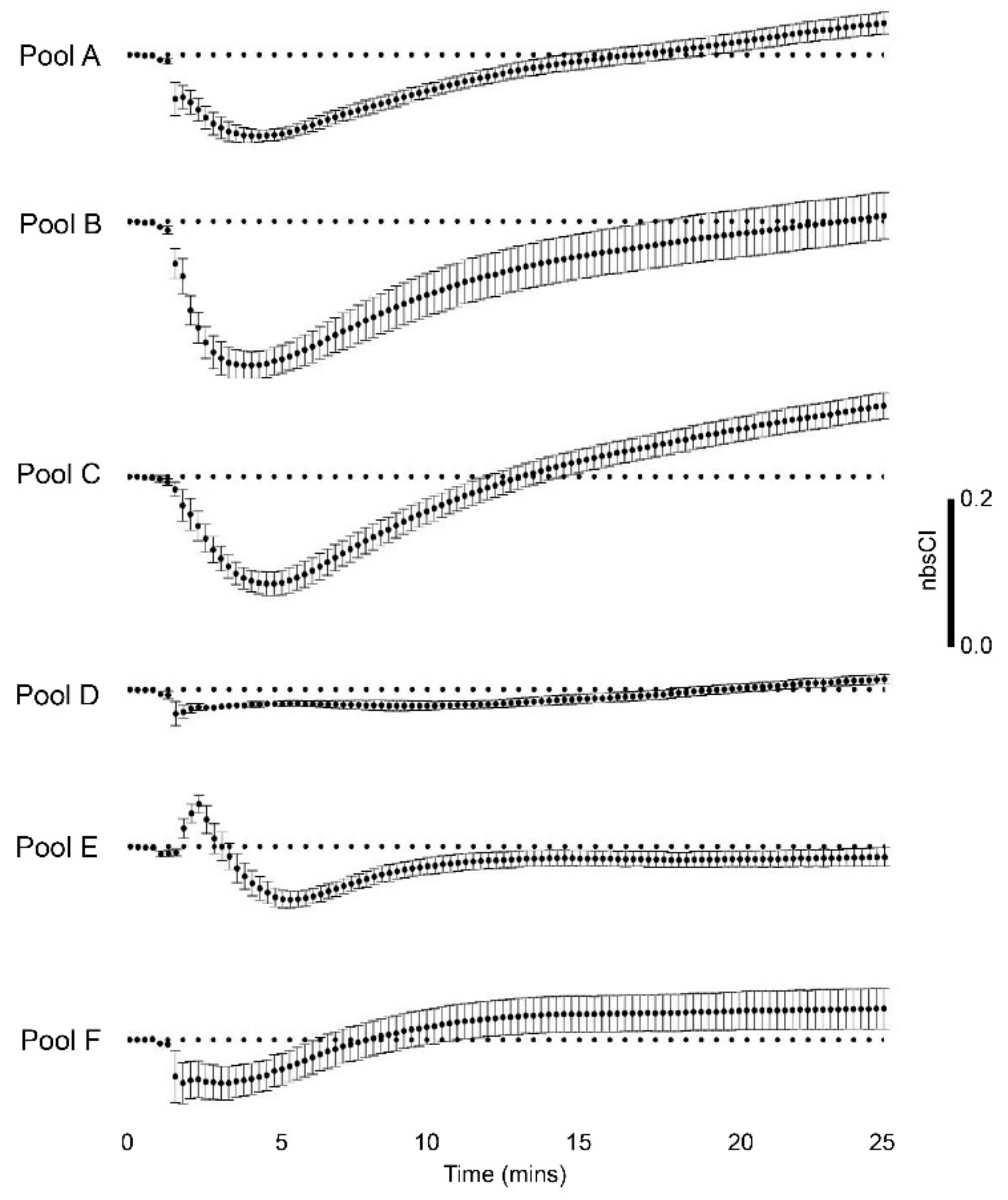
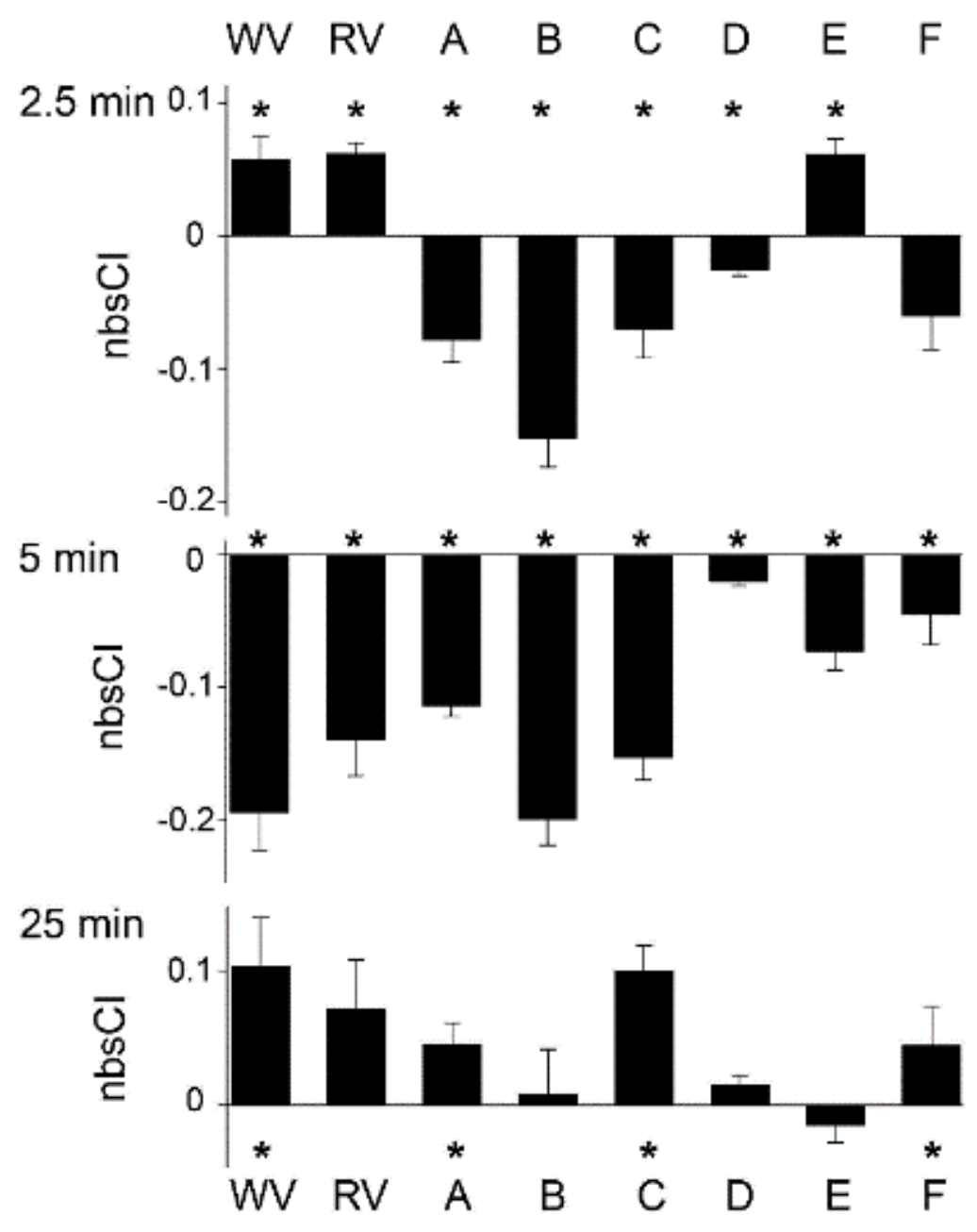

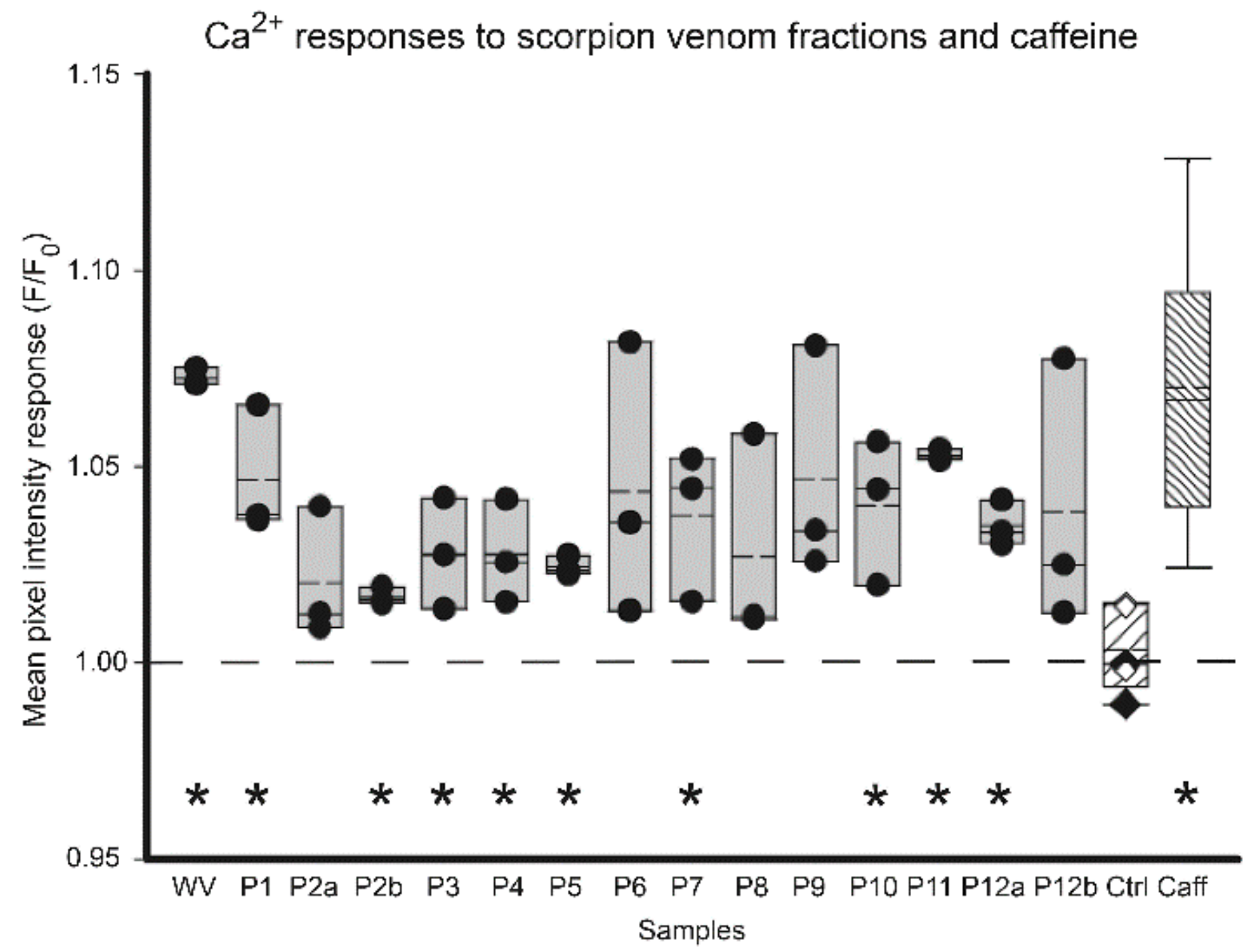
| Rank Order | xCELLigence-Based Bio-Impedance for Venom Pools | Mean Cell Index (Normalised Background-Subtracted) | Toxin-Induced Ca2+ Response for Venom Pool Equivalents | Sum of F/F0 Peak Samples * |
|---|---|---|---|---|
| 1 | Pool B (peak samples 3 ‒4) | ‒0.199 | Pool D (peak samples 8‒10) | 3.114 |
| 2 | Pool C (peak samples 5‒7) | ‒0.153 | Pool C (peak samples 5‒7) | 3.105 |
| 3 | Pool A (peak samples 1‒2a,b) | ‒0.114 | Pool A (peak samples 1‒2a,b) | 3.048 |
| 4 | Pool E (peak sample 11) | ‒0.073 | Pool F (peak samples 12a,b) | 2.074 |
| 5 | Pool F (peak samples 12a,b) | ‒0.045 | Pool B (peak samples 3‒4) | 2.056 |
| 6 | Pool D (peak samples 8‒10) | −0.020 | Pool E (peak sample 11) | 1.053 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Housley, D.M.; Pinyon, J.L.; von Jonquieres, G.; Perera, C.J.; Smout, M.; Liddell, M.J.; Jennings, E.A.; Wilson, D.; Housley, G.D. Australian Scorpion Hormurus waigiensis Venom Fractions Show Broad Bioactivity through Modulation of Bio-Impedance and Cytosolic Calcium. Biomolecules 2020, 10, 617. https://doi.org/10.3390/biom10040617
Housley DM, Pinyon JL, von Jonquieres G, Perera CJ, Smout M, Liddell MJ, Jennings EA, Wilson D, Housley GD. Australian Scorpion Hormurus waigiensis Venom Fractions Show Broad Bioactivity through Modulation of Bio-Impedance and Cytosolic Calcium. Biomolecules. 2020; 10(4):617. https://doi.org/10.3390/biom10040617
Chicago/Turabian StyleHousley, David M., Jeremy L. Pinyon, Georg von Jonquieres, Chamini J. Perera, Michael Smout, Michael J. Liddell, Ernest A. Jennings, David Wilson, and Gary D. Housley. 2020. "Australian Scorpion Hormurus waigiensis Venom Fractions Show Broad Bioactivity through Modulation of Bio-Impedance and Cytosolic Calcium" Biomolecules 10, no. 4: 617. https://doi.org/10.3390/biom10040617





