Identification and Evolution of the WUSCHEL-Related Homeobox Protein Family in Bambusoideae
Abstract
:1. Introduction
2. Methods
2.1. Identification of the WOX Gene Family in Bambusoideae Genomes
2.2. Phylogenetic Tree Construction and Conserved Motif Analysis of WOXs
2.3. Selective Pressure Analysis of Orthologous and Paralogous Genes
2.4. cis-Acting Element Analysis of the P. edulis WOX Promoter
3. Results
3.1. Identification of the WOXs of Bambusoideae
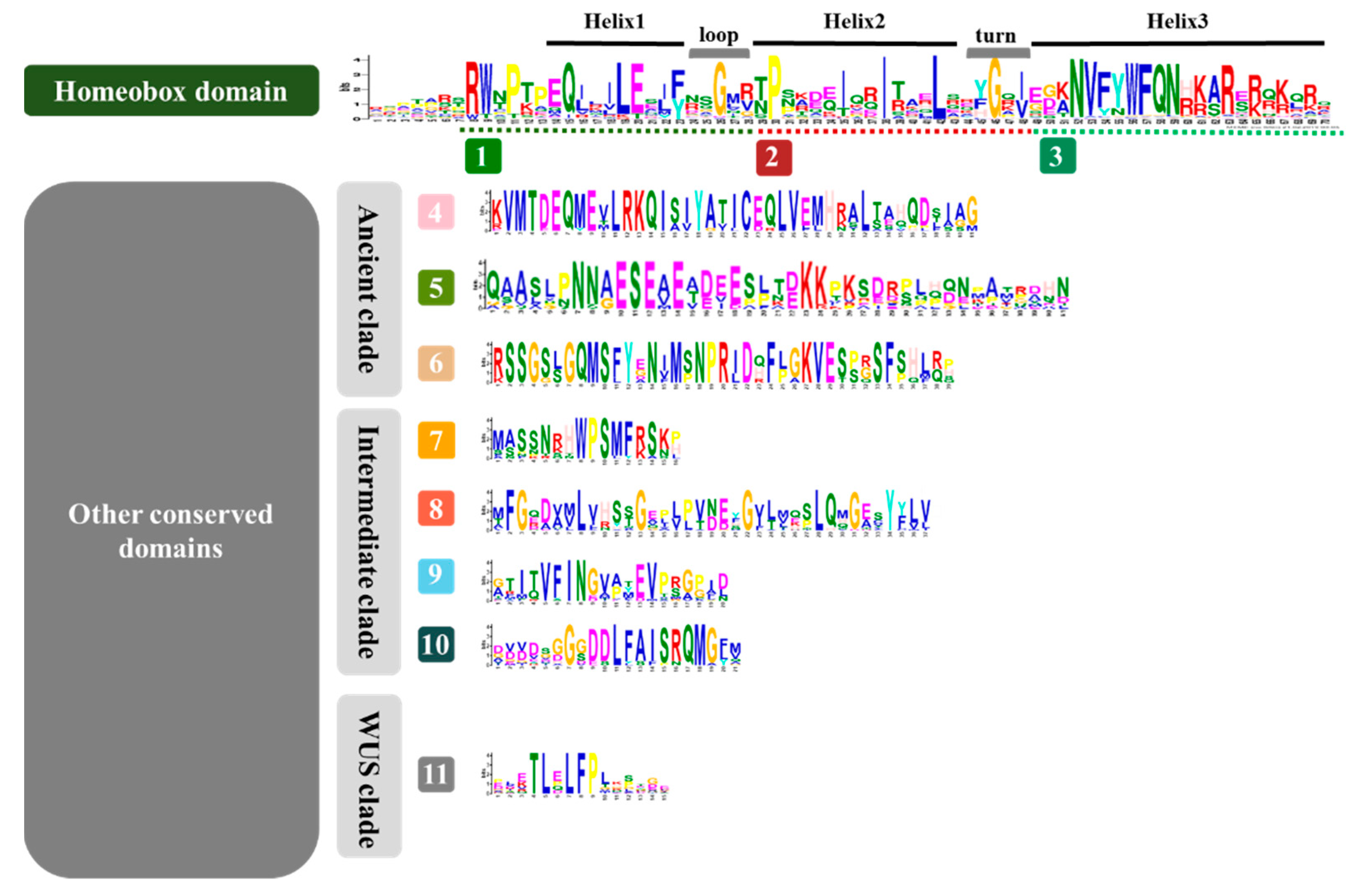
3.2. Identification of Conserved Domains of the WOX Family
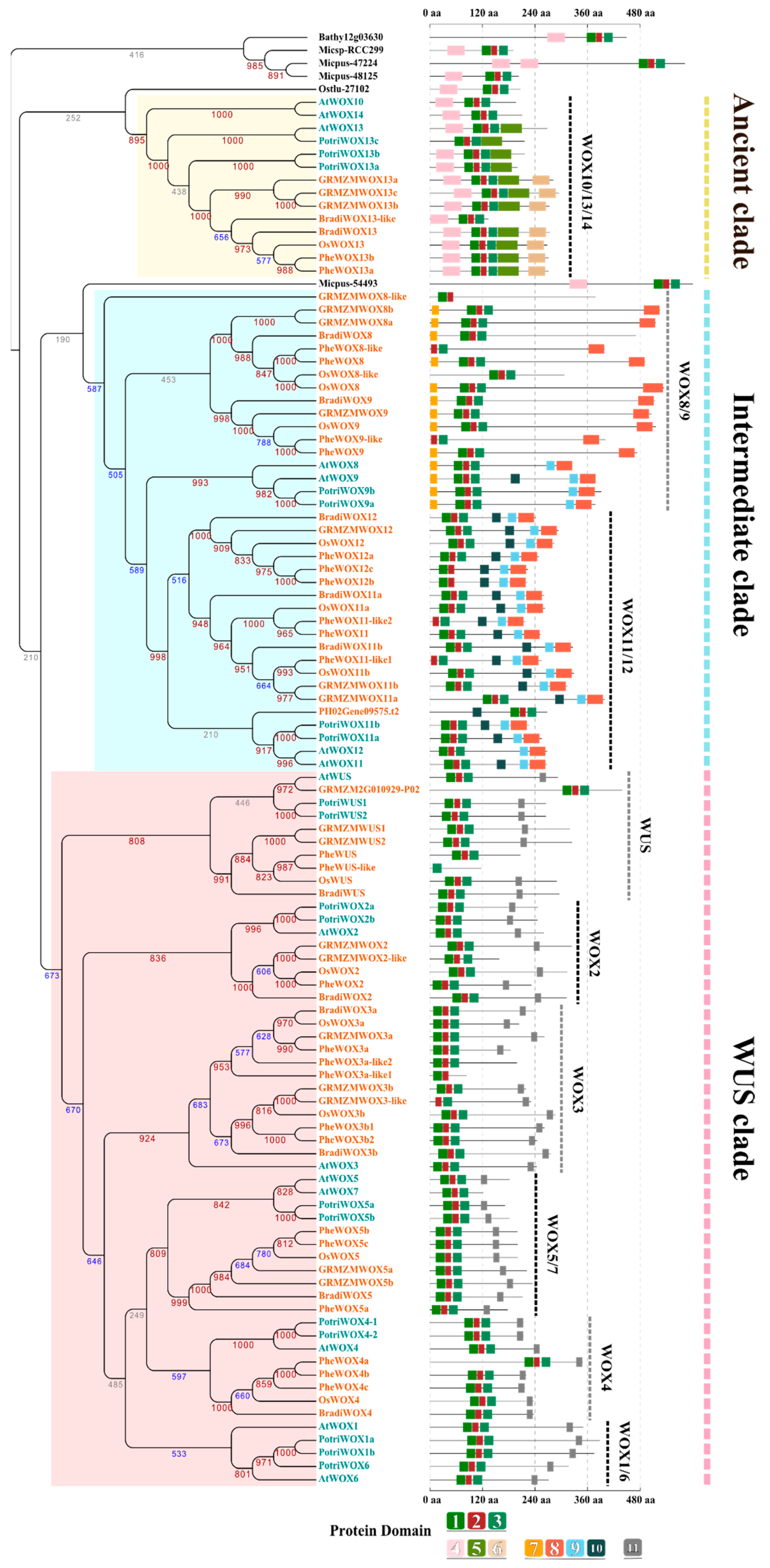
3.3. The Evolution of the WOX Family
3.4. Specific Domain Evolution of WOXs from Different Plants
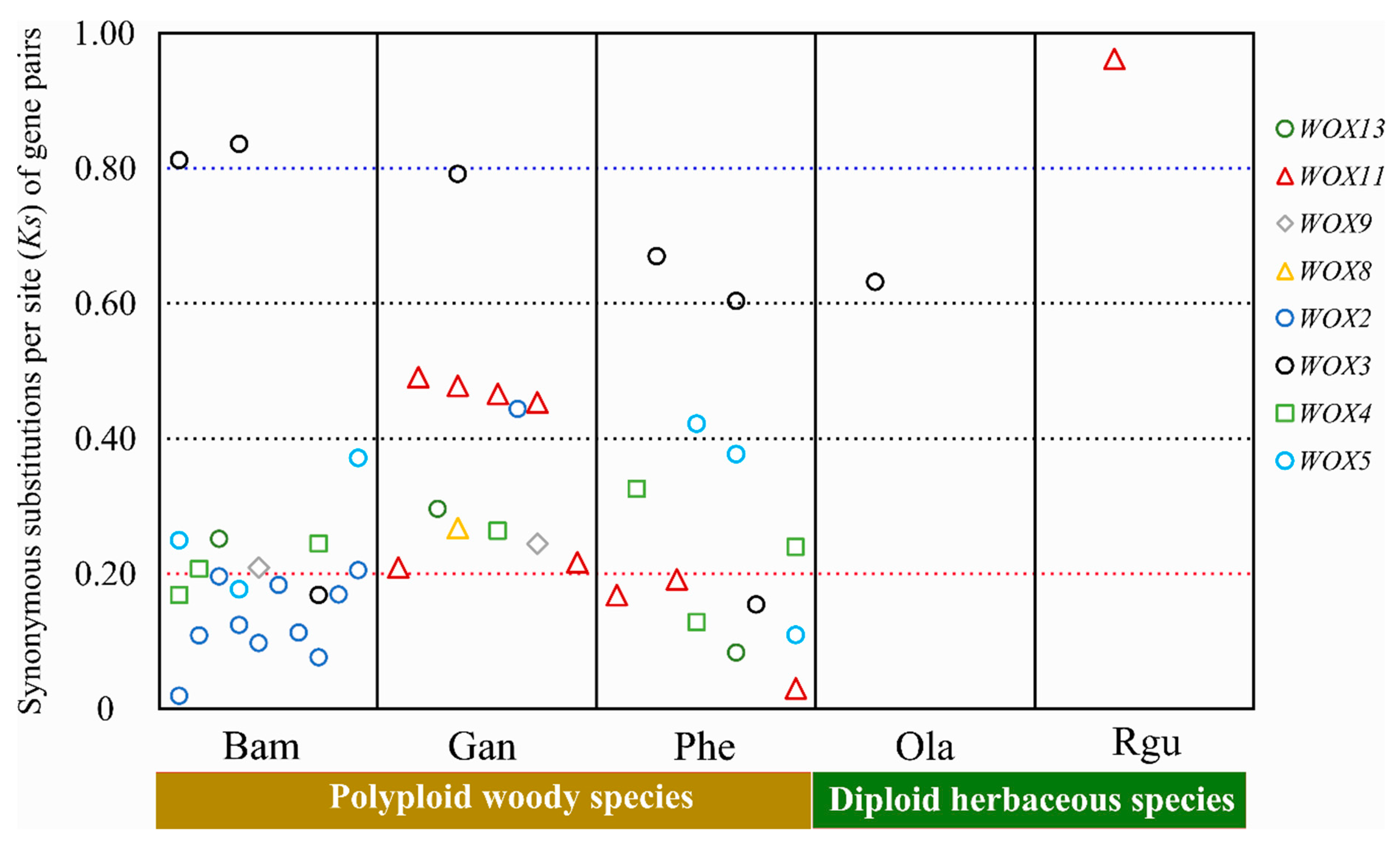
3.5. Evolution of WOXs in Bambusoideae with Different Ploidies
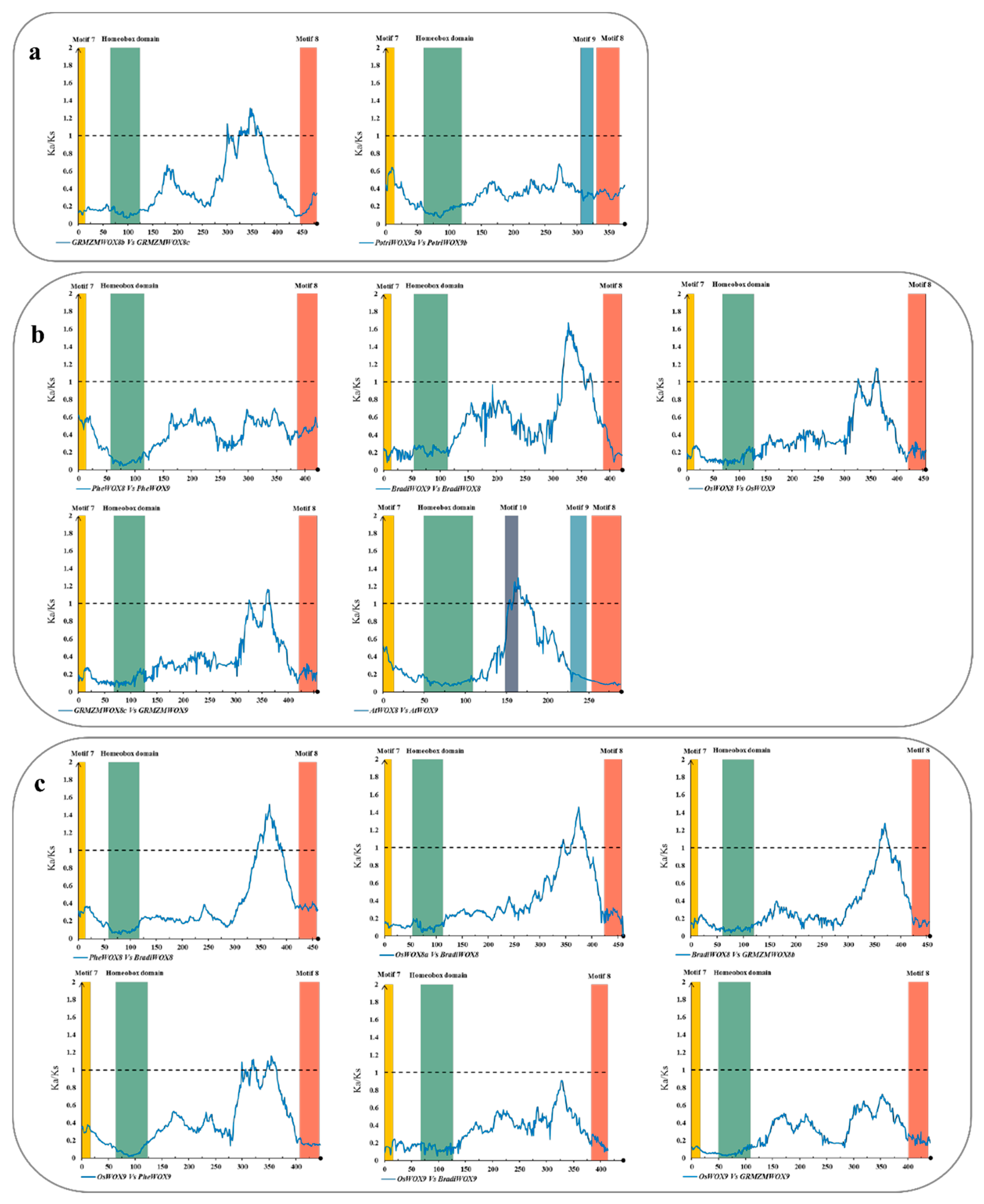
3.6. The Selective Constraints of Homologous Genes within and between Species
3.7. The Expansion of WOXs in Bambusoideae
3.8. The Similar cis-Element Positions of the Orthologous WOX Promoters in Bambusoideae
4. Discussion
4.1. Conserved Domains were Retained during Species Differentiation
4.2. The Evolution of the WUS Clade was Likely Selected by its Conserved and Essential Function in Plants
4.3. The Conserved cis-Element Distribution of Bambusoideae WOXs May Relate to Their Functions during Development
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Guo, Z.H.; Ma, P.F.; Yang, G.Q.; Hu, J.Y.; Liu, Y.L.; Xia, E.H.; Zhong, M.C.; Zhao, L.; Sun, G.L.; Xu, Y.X.; et al. Genome sequences provide insights into the reticulate origin and unique traits of woody bamboos. Mol. Plant 2019, 12, 1353–1365. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Lu, Y.; Li, L.; Zhao, Q.; Feng, Q.I.; Gao, Z.; Lu, H.; Hu, T.; Yao, N.; Liu, K.; et al. The draft genome of the fast-growing non-timber forest species moso bamboo (Phyllostachys heterocycla). Nat. Genet. 2013, 45, 456. [Google Scholar] [PubMed] [Green Version]
- Janzen, D.H. Why bamboos wait so long to flower. Annu. Rev. Ecol. Syst. 1976, 7, 347–391. [Google Scholar] [CrossRef]
- Wei, Q.; Jiao, C.; Guo, L.; Ding, Y.; Cao, J.; Feng, J.; Dong, X.; Mao, L.; Sun, H.; Yu, F. Exploring key cellular processes and candidate genes regulating the primary thickening growth of M oso underground shoots. New Phytol. 2017, 214, 81–96. [Google Scholar] [CrossRef]
- Li, L.; Cheng, Z.; Ma, Y.; Bai, Q.; Li, X.; Cao, Z.; Wu, Z.; Gao, J. The association of hormone signalling genes, transcription and changes in shoot anatomy during moso bamboo growth. Plant Biotechnol. J. 2018, 16, 72–85. [Google Scholar] [CrossRef]
- Gehring, W.J.; Qian, Y.Q.; Billeter, M.; Furukubo-Tokunaga, K.; Schier, A.F.; Resendez-Perez, D.; Affolter, M.; Otting, G.; Wüthrich, K. Homeodomain-DNA recognition. Cell 1994, 78, 211–223. [Google Scholar] [CrossRef]
- Ariel, F.D.; Manavella, P.A.; Dezar, C.A.; Chan, R.L. The true story of the HD-Zip family. Trends Plant Sci. 2007, 12, 419–426. [Google Scholar] [CrossRef]
- Van der Graaff, E.; Laux, T.; Rensing, S.A. The WUS homeobox-containing (WOX) protein family. Genome Biol. 2009, 10, 248. [Google Scholar] [CrossRef]
- Zhang, X.; Zong, J.; Liu, J.; Yin, J.; Zhang, D. Genome-wide analysis of WOX gene family in rice, sorghum, maize, Arabidopsis and poplar. J. Integr. Plant Biol. 2010, 52, 1016–1026. [Google Scholar] [CrossRef]
- Haecker, A.; Groß-Hardt, R.; Geiges, B.; Sarkar, A.; Breuninger, H.; Herrmann, M.; Laux, T. Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development 2004, 131, 657–668. [Google Scholar] [CrossRef] [Green Version]
- Hirakawa, Y.; Kondo, Y.; Fukuda, H. TDIF peptide signaling regulates vascular stem cell proliferation via the WOX4 homeobox gene in Arabidopsis. Plant Cell 2010, 22, 2618–2629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Jiao, Y.; Jiao, H.; Zhao, H.; Zhu, Y.-X. Two-step functional innovation of the stem-cell factors WUS/WOX5 during plant evolution. Mol. Biol. Evol. 2016, 34, 640–653. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanaka, W.; Hirano, H.Y. Antagonistic action of TILLERS ABSENT1 and FLORAL ORGAN NUMBER2 regulates stem cell maintenance during axillary meristem development in rice. New Phytol. 2020, 225.2, 974–984. [Google Scholar] [CrossRef] [PubMed]
- Brand, U.; Fletcher, J.C.; Hobe, M.; Meyerowitz, E.M.; Simon, R. Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 2000, 289, 617–619. [Google Scholar] [CrossRef] [PubMed]
- Pi, L.; Aichinger, E.; van der Graaff, E.; Llavata-Peris, C.I.; Weijers, D.; Hennig, L.; Groot, E.; Laux, T. Organizer-derived WOX5 signal maintains root columella stem cells through chromatin-mediated repression of CDF4 expression. Dev. Cell 2015, 33, 576–588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, Z.; Shao, G.; Xiong, J.; Jiao, Y.; Wang, J.; Liu, G.; Meng, X.; Liang, Y.; Xiong, G.; Wang, Y. MONOCULM 3, an ortholog of WUSCHEL in rice, is required for tiller bud formation. J. Genet. Genom. 2015, 42, 71–78. [Google Scholar] [CrossRef]
- Tanaka, W.; Ohmori, Y.; Ushijima, T.; Matsusaka, H.; Matsushita, T.; Kumamaru, T.; Kawano, S.; Hirano, H.-Y. Axillary meristem formation in rice requires the WUSCHEL ortholog TILLERS ABSENT1. Plant Cell 2015, 27, 1173–1184. [Google Scholar] [CrossRef] [Green Version]
- Ohmori, Y.; Tanaka, W.; Kojima, M.; Sakakibara, H.; Hirano, H.-Y. WUSCHEL-RELATED HOMEOBOX4 is involved in meristem maintenance and is negatively regulated by the CLE gene FCP1 in rice. Plant Cell 2013, 25, 229–241. [Google Scholar] [CrossRef] [Green Version]
- Shimizu, R.; Ji, J.; Kelsey, E.; Ohtsu, K.; Schnable, P.S.; Scanlon, M.J. Tissue specificity and evolution of meristematic WOX3 function. Plant Physiol. 2009, 149, 841–850. [Google Scholar] [CrossRef] [Green Version]
- Yoshikawa, T.; Tanaka, S.-Y.; Masumoto, Y.; Nobori, N.; Ishii, H.; Hibara, K.-I.; Itoh, J.-I.; Tanisaka, T.; Taketa, S. Barley NARROW LEAFED DWARF1 encoding a WUSCHEL-RELATED HOMEOBOX 3 (WOX3) regulates the marginal development of lateral organs. Breed. Sci. 2016, 66, 416–424. [Google Scholar] [CrossRef] [Green Version]
- Ueda, M.; Zhang, Z.; Laux, T. Transcriptional activation of Arabidopsis axis patterning genes WOX8/9 links zygote polarity to embryo development. Dev. Cell 2011, 20, 264–270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lie, C.; Kelsom, C.; Wu, X. WOX2 and STIMPY-LIKE/WOX8 promote cotyledon boundary formation in Arabidopsis. Plant J. 2012, 72, 674–682. [Google Scholar] [CrossRef]
- Liu, J.; Sheng, L.; Xu, Y.; Li, J.; Yang, Z.; Huang, H.; Xu, L. WOX11 and 12 are involved in the first-step cell fate transition during de novo root organogenesis in Arabidopsis. Plant Cell 2014, 26, 1081–1093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, X.; Xu, L. Transcription factors WOX11/12 directly activate WOX5/7 to promote root primordia initiation and organogenesis. Plant Physiol. 2016, 172, 2363–2373. [Google Scholar] [CrossRef] [Green Version]
- Laux, T.; Mayer, K.; Berger, J.; Jurgens, G. The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 1996, 122, 87–96. [Google Scholar] [PubMed]
- Somssich, M.; Je, B.I.; Simon, R.; Jackson, D. CLAVATA-WUSCHEL signaling in the shoot meristem. Development 2016, 143, 3238–3248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, J.; Tian, F.; Yang, D.-C.; Meng, Y.-Q.; Kong, L.; Luo, J.; Gao, G. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017, 45.D1, D1040–D1045. [Google Scholar] [CrossRef] [Green Version]
- Zhao, H.; Gao, Z.; Wang, L.; Wang, J.; Wang, S.; Fei, B.; Chen, C.; Shi, C.; Liu, X.; Zhang, H. Chromosome-level reference genome and alternative splicing atlas of moso bamboo (Phyllostachys edulis). GigaScience 2018, 7, giy115. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R. CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2016, 45, D200–D203. [Google Scholar] [CrossRef]
- Chen, C.; Xia, R.; Chen, H.; He, Y. TBtools, a Toolkit for Biologists integrating various biological data handling tools with a user-friendly interface. BioRxiv 2018. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, Z.; Zhang, H.; Gao, S.; Lercher, M.J.; Chen, W.-H.; Hu, S. Evolview v2: An online visualization and management tool for customized and annotated phylogenetic trees. Nucleic Acids Res. 2016, 44, W236–W241. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, H.; Zhou, W.; Landweber, L.F. SWAKK: A web server for detecting positive selection in proteins using a sliding window substitution rate analysis. Nucleic Acids Res. 2016, 34 (Suppl_2), W382–W384. [Google Scholar] [CrossRef] [Green Version]
- Lynch, M.; Conery, J.S. The evolutionary fate and consequences of duplicate genes. Science 2000, 290, 1151–1155. [Google Scholar] [CrossRef] [Green Version]
- Tian, F.; Yang, D.-C.; Meng, Y.-Q.; Jin, J.; Gao, G. PlantRegMap: Charting functional regulatory maps in plants. Nucleic Acids Res. 2020, 48, D1104–D1113. [Google Scholar] [CrossRef]
- Fernández-Mazuecos, M.; Glover, B.J. The evo-devo of plant speciation. Nat. Ecol. Evol. 2017, 1, 0110. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Wu, R.; Qin, G.; Chen, Z.; Gu, H.; Qu, L.J. Over-expression of WOX1 Leads to Defects in Meristem Development and Polyamine Homeostasis in Arabidopsis F. J. Integr. Plant Biol. 2011, 53, 493–506. [Google Scholar] [CrossRef]
- Deveaux, Y.; Toffano-Nioche, C.; Claisse, G.; Thareau, V.; Morin, H.; Laufs, P.; Moreau, H.; Kreis, M.; Lecharny, A. Genes of the most conserved WOX clade in plants affect root and flower development in Arabidopsis. BMC Evol. Biol. 2008, 8, 291. [Google Scholar] [CrossRef] [Green Version]
- Lian, G.; Ding, Z.; Wang, Q.; Zhang, D.; Xu, J. Origins and evolution of WUSCHEL-related homeobox protein family in plant kingdom. Sci. World J. 2014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, C.-C.; Li, F.-W.; Kramer, E.M. Large-scale phylogenomic analysis suggests three ancient superclades of the WUSCHEL-RELATED HOMEOBOX transcription factor family in plants. PLoS ONE 2019, 1. [Google Scholar] [CrossRef] [PubMed]
- Nardmann, J.; Reisewitz, P.; Werr, W. Discrete shoot and root stem cell-promoting WUS/WOX5 functions are an evolutionary innovation of angiosperms. Mol. Biol. Evol. 2009, 26, 1745–1755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nardmann, J.; Werr, W. Symplesiomorphies in the WUSCHEL clade suggest that the last common ancestor of seed plants contained at least four independent stem cell niches. New Phytol. 2013, 199, 1081–1092. [Google Scholar] [CrossRef]
- Liu, L.; White, M.J.; MacRae, T.H. Transcription factors and their genes in higher plants: Functional domains, evolution and regulation. Eur. J. Biochem. 1999, 262, 247–257. [Google Scholar] [CrossRef]
- Lin, H.; Niu, L.; McHale, N.A.; Ohme-Takagi, M.; Mysore, K.S.; Tadege, M. Evolutionarily conserved repressive activity of WOX proteins mediates leaf blade outgrowth and floral organ development in plants. Proc. Natl. Acad. Sci. USA 2013, 110, 366–371. [Google Scholar] [CrossRef] [Green Version]
- Ryu, C.H.; Lee, S.; Cho, L.H.; Kim, S.L.; Lee, Y.S.; Choi, S.C.; Jeong, H.J.; Yi, J.; Park, S.J.; Han, C.D. OsMADS50 and OsMADS56 function antagonistically in regulating long day (LD)-dependent flowering in rice. Plant Cell Environ. 2009, 32, 1412–1427. [Google Scholar] [CrossRef]
- Harrison, C.J.; Morris, J.L. The origin and early evolution of vascular plant shoots and leaves. Philos. Trans. R. Soc. B Biol. Sci. 2018, 373, 20160496. [Google Scholar] [CrossRef]
- Nardmann, J.; Werr, W. The shoot stem cell niche in angiosperms: Expression patterns of WUS orthologues in rice and maize imply major modifications in the course of mono-and dicot evolution. Mol. Biol. Evol. 2006, 23, 2492–2504. [Google Scholar] [CrossRef] [Green Version]
- Nardmann, J.; Werr, W. The invention of WUS-like stem cell-promoting functions in plants predates leptosporangiate ferns. Plant Mol. Biol. 2012, 78, 123–134. [Google Scholar] [CrossRef]
- Nardmann, J.; Ji, J.; Werr, W.; Scanlon, M.J. The maize duplicate genes narrow sheath1 and narrow sheath2 encode a conserved homeobox gene function in a lateral domain of shoot apical meristems. Development 2004, 131, 2827–2839. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burki, F.; Inagaki, Y.; Bråte, J.; Archibald, J.M.; Keeling, P.J.; Cavalier-Smith, T.; Sakaguchi, M.; Hashimoto, T.; Horak, A.; Kumar, S. Large-scale phylogenomic analyses reveal that two enigmatic protist lineages, Telonemia and Centroheliozoa, are related to photosynthetic chromalveolates. Genome Biol. Evol. 2009, 1, 231–238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Hamyat, M.; Liu, C.; Ahmad, S.; Gao, X.; Guo, C.; Wang, Y.; Guo, Y. Identification and characterization of the WOX family genes in five Solanaceae species reveal their conserved roles in peptide signaling. Genes 2018, 9, 260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palovaara, J.; Hakman, I. WOX2 and polar auxin transport during spruce embryo pattern formation. Plant Signal. Behav. 2009, 4, 153–155. [Google Scholar] [CrossRef] [Green Version]
- Ji, J.; Shimizu, R.; Sinha, N.; Scanlon, M.J. Analyses of WOX4 transgenics provide further evidence for the evolution of the WOX gene family during the regulation of diverse stem cell functions. Plant Signal. Behav. 2010, 5, 916–920. [Google Scholar] [CrossRef] [Green Version]
- Schoof, H.; Lenhard, M.; Haecker, A.; Mayer, K.F.; Jürgens, G.; Laux, T. The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 2000, 100, 635–644. [Google Scholar] [CrossRef] [Green Version]
- Sarkar, A.K.; Luijten, M.; Miyashima, S.; Lenhard, M.; Hashimoto, T.; Nakajima, K.; Scheres, B.; Heidstra, R.; Laux, T. Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 2007, 446, 811. [Google Scholar] [CrossRef]
- Ikeda, M.; Mitsuda, N.; Ohme-Takagi, M. Arabidopsis WUSCHEL is a bifunctional transcription factor that acts as a repressor in stem cell regulation and as an activator in floral patterning. Plant Cell 2009, 21, 3493–3505. [Google Scholar] [CrossRef] [Green Version]
- Lenhard, M.; Bohnert, A.; Jürgens, G.; Laux, T. Termination of stem cell maintenance in Arabidopsis floral meristems by interactions between WUSCHEL and AGAMOUS. Cell 2001, 105, 805–814. [Google Scholar] [CrossRef] [Green Version]
- Vandenbussche, M.; Horstman, A.; Zethof, J.; Koes, R.; Rijpkema, A.S.; Gerats, T. Differential recruitment of WOX transcription factors for lateral development and organ fusion in Petunia and Arabidopsis. Plant Cell 2009, 21, 2269–2283. [Google Scholar] [CrossRef] [Green Version]
- Stahl, Y.; Wink, R.H.; Ingram, G.C.; Simon, R. A signaling module controlling the stem cell niche in Arabidopsis root meristems. Curr. Biol. 2009, 19, 909–914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yadav, R.K.; Perales, M.; Gruel, J.; Girke, T.; Jönsson, H.; Reddy, G.V. WUSCHEL protein movement mediates stem cell homeostasis in the Arabidopsis shoot apex. Genes Dev. 2011, 25, 2025–2030. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, S.H.; Yoo, S.C.; Zhang, H.; Pandeya, D.; Koh, H.J.; Hwang, J.Y.; Kim, G.T.; Paek, N.C. The rice narrow leaf2 and narrow leaf3 loci encode WUSCHEL-related homeobox 3 A (O s WOX 3 A) and function in leaf, spikelet, tiller and lateral root development. New Phytol. 2013, 198, 1071–1084. [Google Scholar] [CrossRef] [PubMed]
- Tadege, M.; Lin, H.; Bedair, M.; Berbel, A.; Wen, J.; Rojas, C.M.; Niu, L.; Tang, Y.; Sumner, L.; Ratet, P. STENOFOLIA regulates blade outgrowth and leaf vascular patterning in Medicago truncatula and Nicotiana sylvestris. Plant Cell 2011, 23, 2125–2142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McHale, N.A.; Marcotrigiano, M. LAM1 is required for dorsoventrality and lateral growth of the leaf blade in Nicotiana. Development 1998, 125, 4235–4243. [Google Scholar] [PubMed]
- Hecker, A.; Brand, L.H.; Peter, S.; Simoncello, N.; Kilian, J.; Harter, K.; Gaudin, V.; Wanke, D. The Arabidopsis GAGA-binding factor basic pentacysteine6 recruits the polycomb-repressive complex1 component like heterochromatin protein1 to GAGA DNA motifs. Plant Physiol. 2015, 168, 1013–1024. [Google Scholar] [CrossRef]
- Meister, R.J.; Williams, L.A.; Monfared, M.M.; Gallagher, T.L.; Kraft, E.A.; Nelson, C.G.; Gasser, C.S. Definition and interactions of a positive regulatory element of the Arabidopsis INNER NO OUTER promoter. Plant J. 2004, 37, 426–438. [Google Scholar] [CrossRef]
- Guo, Y.; Qin, G.; Gu, H.; Qu, L.-J. Dof5. 6/HCA2, a Dof transcription factor gene, regulates interfascicular cambium formation and vascular tissue development in Arabidopsis. Plant Cell 2009, 21, 3518–3534. [Google Scholar] [CrossRef] [Green Version]
- Kang, H.G.; Foley, R.C.; Oñate-Sánchez, L.; Lin, C.; Singh, K.B. Target genes for OBP3, a Dof transcription factor, include novel basic helix-loop-helix domain proteins inducible by salicylic acid. Plant J. 2003, 35, 362–372. [Google Scholar] [CrossRef]
- Kang, H.G.; Singh, K.B. Characterization of salicylic acid-responsive, Arabidopsis Dof domain proteins: Overexpression of OBP3 leads to growth defects. Plant J. 2000, 21, 329–339. [Google Scholar] [CrossRef]
- Romano, J.M.; Dubos, C.; Prouse, M.B.; Wilkins, O.; Hong, H.; Poole, M.; Kang, K.Y.; Li, E.; Douglas, C.J.; Western, T.L. AtMYB61, an R2R3-MYB transcription factor, functions as a pleiotropic regulator via a small gene network. New Phytol. 2012, 195, 774–786. [Google Scholar] [CrossRef] [PubMed]
- Pastore, J.J.; Limpuangthip, A.; Yamaguchi, N.; Wu, M.-F.; Sang, Y.; Han, S.-K.; Malaspina, L.; Chavdaroff, N.; Yamaguchi, A.; Wagner, D. LATE MERISTEM IDENTITY2 acts together with LEAFY to activate APETALA1. Development 2011, 138, 3189–3198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gregis, V.; Sessa, A.; Dorca-Fornell, C.; Kater, M.M. The Arabidopsis floral meristem identity genes AP1, AGL24 and SVP directly repress class B and C floral homeotic genes. Plant J. 2009, 60, 626–637. [Google Scholar] [CrossRef] [PubMed]
- Costanzo, E.; Trehin, C.; Vandenbussche, M. The role of WOX genes in flower development. Ann. Bot. 2014, 114, 1545–1553. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Taxonomic Group | Species | Ancient Clade | Intermediate Clade | WUSCHEL (WUS) Clade | Other | Total |
|---|---|---|---|---|---|---|
| Dicots | Arabidopsis thaliana | 3 | 4 | 8 | - | 15 |
| Populus trichocarpa | 3 | 4 | 11 | - | 18 | |
| Monocots | Oryza sativa | 1 | 6 | 6 | - | 13 |
| Brachypodium distachyon | 2 | 5 | 6 | - | 13 | |
| Zea mays | 3 | 7 | 10 | - | 20 | |
| Olyra latifolia | 1 | 5 | 6 | - | 12 | |
| Raddia guianensis | 1 | 3 | 5 | - | 9 | |
| Guadua angustifolia | 2 | 9 | 7 | - | 18 | |
| Phyllostachys edulis | 2 | 11 | 14 | - | 27 | |
| Bonia amplexicaulis | 2 | 6 | 18 | - | 26 | |
| Chlorophyta | Micromonas pusilla | - | - | - | 4 | 4 |
| Micromonas sp. | - | - | - | 1 | 1 | |
| Ostreococcus tauri, | - | - | - | 1 | 1 | |
| Bathycoccus prasinos | - | - | - | 1 | 1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Li, J.; Cai, M.; Zheng, H.; Cheng, Z.; Gao, J. Identification and Evolution of the WUSCHEL-Related Homeobox Protein Family in Bambusoideae. Biomolecules 2020, 10, 739. https://doi.org/10.3390/biom10050739
Li X, Li J, Cai M, Zheng H, Cheng Z, Gao J. Identification and Evolution of the WUSCHEL-Related Homeobox Protein Family in Bambusoideae. Biomolecules. 2020; 10(5):739. https://doi.org/10.3390/biom10050739
Chicago/Turabian StyleLi, Xiangyu, Juan Li, Miaomiao Cai, Huifang Zheng, Zhanchao Cheng, and Jian Gao. 2020. "Identification and Evolution of the WUSCHEL-Related Homeobox Protein Family in Bambusoideae" Biomolecules 10, no. 5: 739. https://doi.org/10.3390/biom10050739





