Pseudouridine Synthase RsuA Captures an Assembly Intermediate That Is Stabilized by Ribosomal Protein S17
Abstract
1. Introduction
2. Materials and Methods
2.1. Overexpression and Purification of Proteins
2.2. Fluorescence Labeling of RsuA
2.3. 16S 5′-Domain RNA Preparation
2.4. Filter Binding Assay
2.5. FRET-Based RsuA Binding Assay
2.6. RsuA Activity Assay
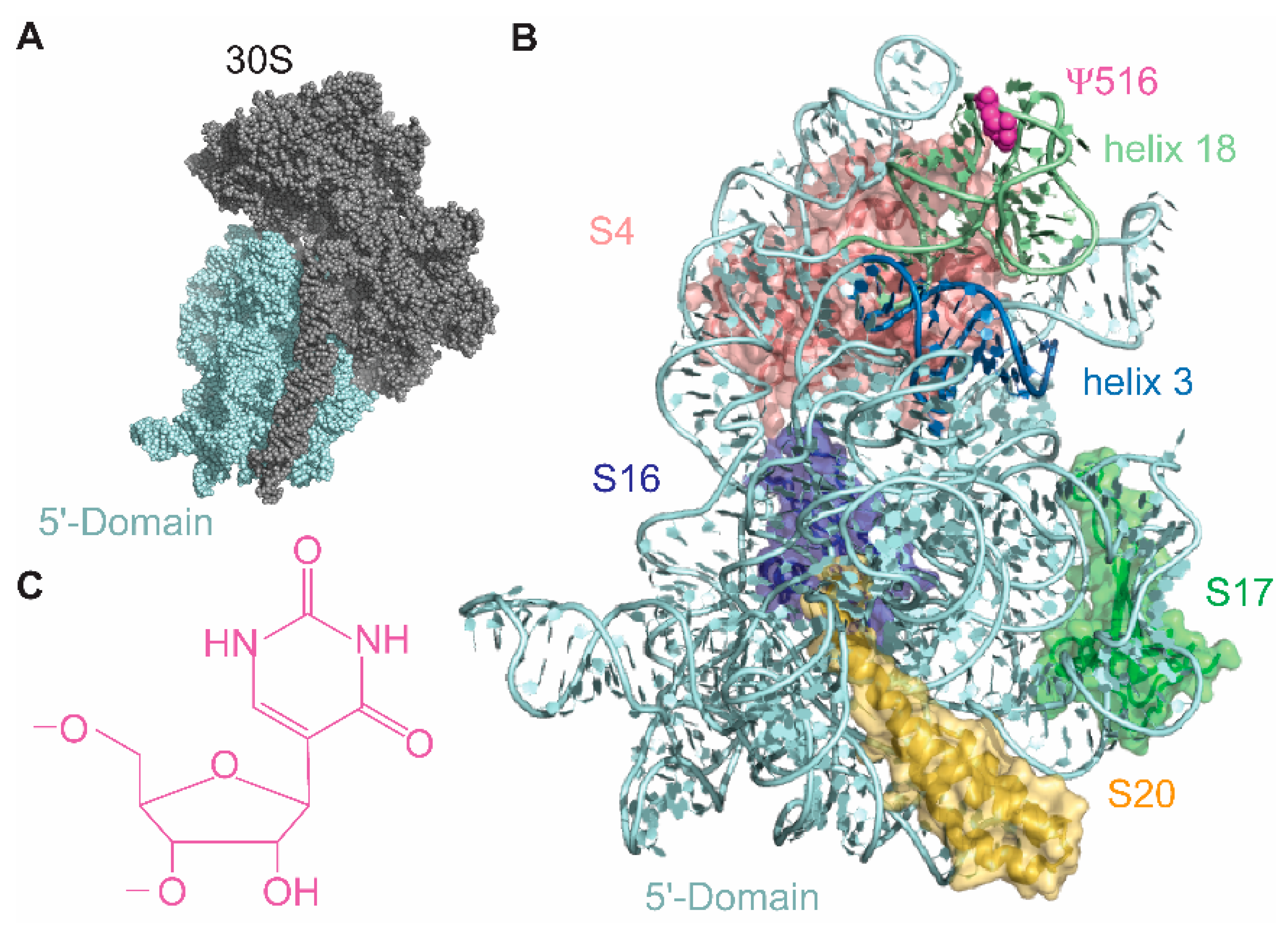
3. Results
3.1. Magnesium Ions Influence RsuA Binding
3.2. RsuA Binds Preferably to 16S Helix 18 Pseudoknot Mutants
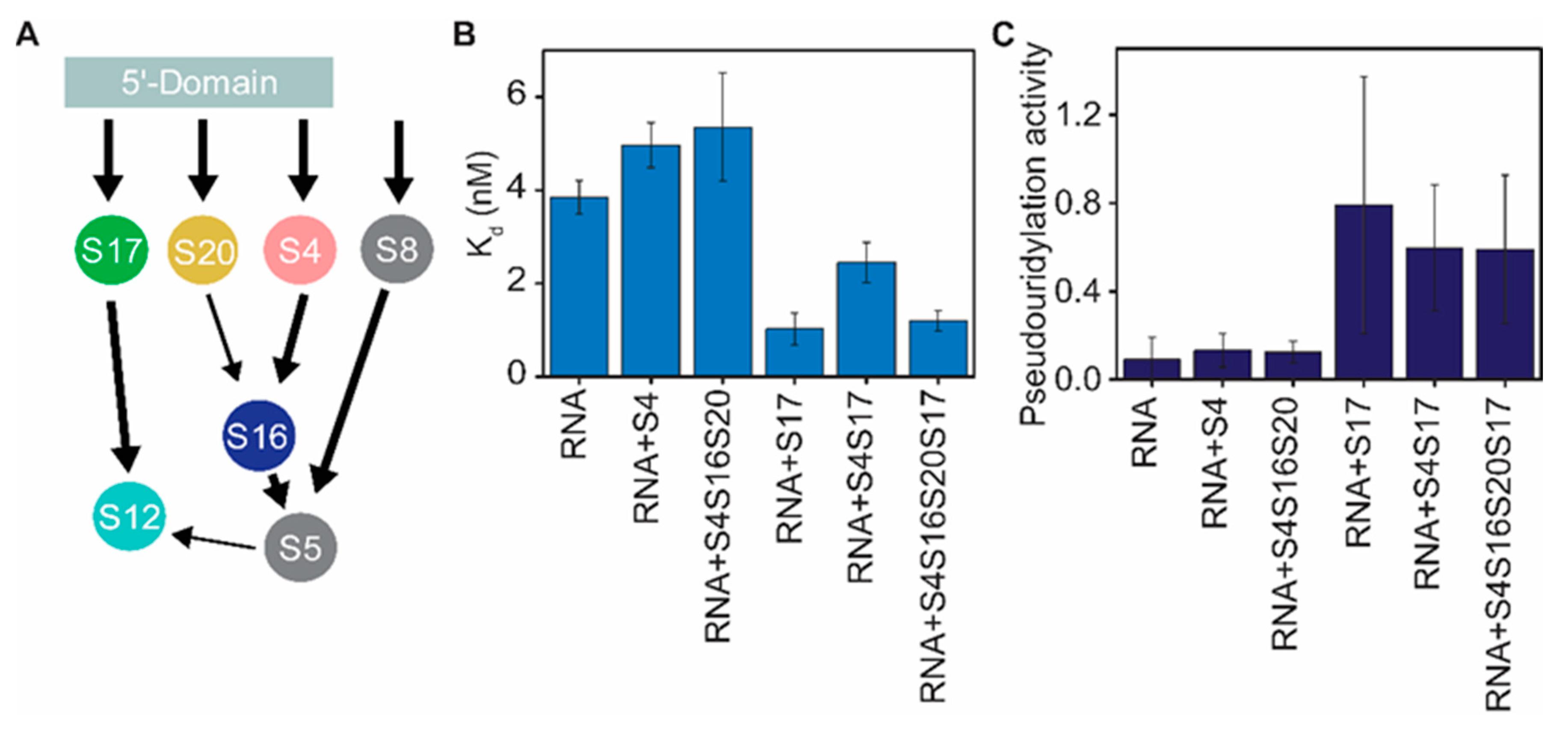
3.3. Binding of RsuA and S17 Is Thermodynamically Cooperative
3.4. RsuA Peripheral Domain Increases the Stability of RsuA-rRNA Complexes
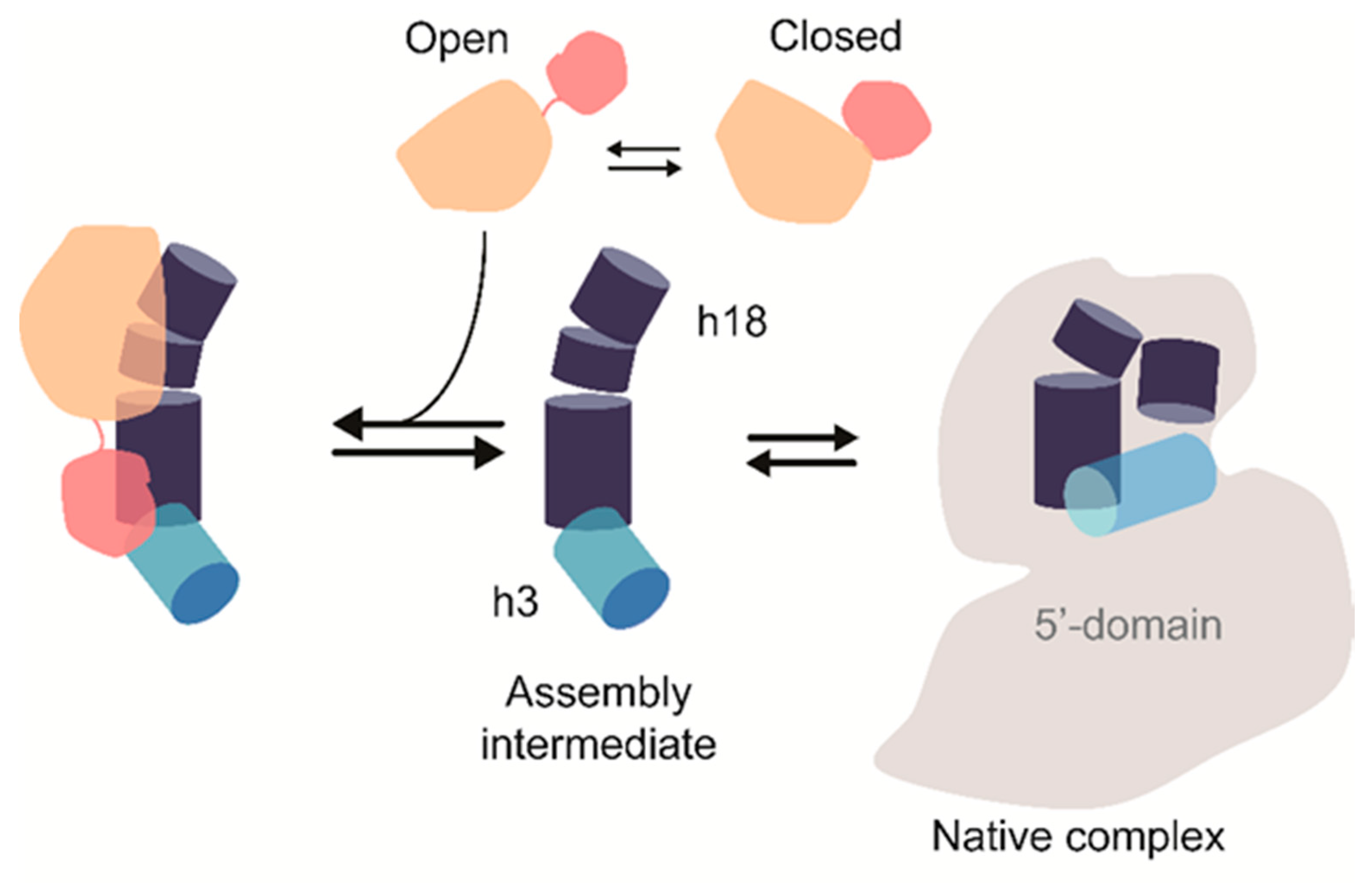
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Held, W.A.; Ballou, B.; Mizushima, S.; Nomura, M. Assembly mapping of 30S ribosomal proteins from Escherichia coli. Further studies. J. Biol. Chem. 1974, 249, 3103–3111. [Google Scholar] [PubMed]
- Held, W.A.; Nomura, M. Escherichia coli 30S ribosomal proteins uniquely required for assembly. J. Biol. Chem. 1975, 250, 3179–3184. [Google Scholar] [PubMed]
- Nowotny, V.; Nierhaus, K.H. Assembly of the 30S subunit from Escherichia coli ribosomes occurs via two assembly domains which are initiated by S4 and S7. Biochemistry 1988, 27, 7051–7055. [Google Scholar] [CrossRef] [PubMed]
- Connolly, K.; Culver, G. Deconstructing ribosome construction. Trends Biochem. Sci. 2009, 34, 256–263. [Google Scholar] [CrossRef]
- Wrzesinski, J.; Bakin, A.; Nurse, K.; Lane, B.G.; Ofengand, J. Purification, cloning, and properties of the 16S RNA pseudouridine 516 synthase from Escherichia coli. Biochemistry 1995, 34, 8904–8913. [Google Scholar] [CrossRef]
- Conrad, J.; Niu, L.; Rudd, K.; Lane, B.G.; Ofengand, J. 16S ribosomal RNA pseudouridine synthase RsuA of Escherichia coli: Deletion, mutation of the conserved Asp102 residue, and sequence comparison among all other pseudouridine synthases. RNA 1999, 5, 751–763. [Google Scholar] [CrossRef]
- Bakin, A.; Kowalak, J.A.; McCloskey, J.A.; Ofengand, J. The single pseudouridine residue in Escherichia coli 16S RNA is located at position 516. Nucleic Acids Res. 1994, 22, 3681–3684. [Google Scholar] [CrossRef]
- Sivaraman, J.; Sauvé, V.; Larocque, R.; Stura, E.A.; Schrag, J.D.; Cygler, M.; Matte, A. Structure of the 16S rRNA pseudouridine synthase RsuA bound to uracil and UMP. Nat. Struct. Biol. 2002, 9, 353–358. [Google Scholar] [CrossRef]
- Davies, C.; Gerstner, R.B.; Draper, D.E.; Ramakrishnan, V.; White, S.W. The crystal structure of ribosomal protein S4 reveals a two-domain molecule with an extensive RNA-binding surface: One domain shows structural homology to the ETS DNA-binding motif. EMBO J. 1998, 17, 4545–4558. [Google Scholar] [CrossRef]
- Aravind, L.; Koonin, E. V Novel Predicted RNA-Binding Domains Associated with the Translation Machinery. J. Mol. Evol. 1999, 48, 291–302. [Google Scholar] [CrossRef]
- Leppik, M.; Liiv, A.; Remme, J. Random pseuoduridylation in vivo reveals critical region of Escherichia coli 23S rRNA for ribosome assembly. Nucleic Acids Res. 2017, 45, 6098–6108. [Google Scholar] [CrossRef] [PubMed]
- Corollo, D.; Blair-Johnson, M.; Conrad, J.; Fiedler, T.; Sun, D.; Wang, L.; Ofengand, J.; Fenna, R. Crystallization and characterization of a fragment of pseudouridine synthase RluC from Escherichia coli. Acta Crystallogr. Sect. D Biol. Crystallogr. 1999, 55, 302–304. [Google Scholar] [CrossRef] [PubMed]
- Matte, A.; Louie, G.; Sivaraman, J.; Cygler, M.; Burley, S. Structure of the pseudouridine synthase RsuA from Haemophilus influenza. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2005, 61, 350–354. [Google Scholar] [CrossRef] [PubMed]
- Gutgsell, N.; Englund, N.; Niu, L.; Kaya, Y.; Lane, B.G.; Ofengand, J. Deletion of the Escherichia coli pseudouridine synthase gene truB blocks formation of pseudouridine 55 in tRNA in vivo, does not affect exponential growth, but confers a strong selective disadvantage in competition with wild-type cells. RNA 2000, 6, 1870–1881. [Google Scholar] [CrossRef] [PubMed]
- Gutgsell, N.S.; Del Campo, M.; Raychaudhuri, S.; Ofengand, J. A second function for pseudouridine synthases: A point mutant of RluD unable to form pseudouridines 1911, 1915, and 1917 in Escherichia coli 23S ribosomal RNA restores normal growth to an RluD-minus strain. RNA 2001, 7, 990–998. [Google Scholar] [CrossRef] [PubMed]
- Kitagawa, M.; Ara, T.; Arifuzzaman, M.; Ioka-Nakamichi, T.; Inamoto, E.; Toyonaga, H.; Mori, H. Complete set of ORF clones of Escherichia coli ASKA library (A Complete Set of E. coli K -12 ORF A rchive): Unique Resources for Biological Research. DNA Res. 2005, 12, 291–299. [Google Scholar] [CrossRef]
- Culver, G.M.; Noller, H.F. Efficient reconstitution of functional Escherichia coli 30S ribosomal subunits from a complete set of recombinant small subunit ribosomal proteins. RNA 1999, 5, 832–843. [Google Scholar] [CrossRef]
- Kim, H.; Abeysirigunawarden, S.C.; Chen, K.; Mayerle, M.; Ragunathan, K.; Luthey-Schulten, Z.; Ha, T.; Woodson, S.A. Protein-guided RNA dynamics during early ribosome assembly. Nature 2014, 506, 334–338. [Google Scholar] [CrossRef]
- Abeysirigunawardena, S.C.; Woodson, S.A. Differential effects of ribosomal proteins and Mg2+ ions on a conformational switch during 30S ribosome 5′-domain assembly. RNA 2015, 21, 1859–1865. [Google Scholar] [CrossRef]
- Misra, V.K.; Draper, D.E. On the role of magnesium ions in RNA stability. Biopolymers 1998, 48, 113–135. [Google Scholar] [CrossRef]
- Pan, J.; Thirumalai, D.; Woodson, S.A. Magnesium-dependent folding of self-splicing RNA: Exploring the link between cooperativity, thermodynamics, and kinetics. Proc. Natl. Acad. Sci. USA 1999, 96, 6149–6154. [Google Scholar] [CrossRef] [PubMed]
- Leipply, D.; Draper, D.E. Dependence of RNA Tertiary Structural Stability on Mg2+ Concentration: Interpretation of the Hill Equation and Coefficient. Biochemistry 2010, 49, 1843–1853. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gluick, T.C.; Gerstner, R.B.; Draper, D.E. Effects of Mg2+, K+, and H+ on an equilibrium between alternative conformations of an RNA pseudoknot11Edited by B. Honig. J. Mol. Biol. 1997, 270, 451–463. [Google Scholar] [CrossRef] [PubMed]
- Draper, D.E. A guide to ions and RNA structure. RNA 2004, 10, 335–343. [Google Scholar] [CrossRef]
- Adilakshmi, T.; Ramaswamy, P.; Woodson, S.A. Protein-independent folding pathway of the 16S rRNA 5′ domain. J. Mol. Biol. 2005, 351, 508–519. [Google Scholar] [CrossRef]
- Vartikar, J.V.; Draper, D.E. S4-16 S ribosomal RNA complex: Binding constant measurements and specific recognition of a 460-nucleotide region. J. Mol. Biol. 1989, 209, 221–234. [Google Scholar] [CrossRef]
- Siibak, T.; Remme, J. Subribosomal particle analysis reveals the stages of bacterial ribosome assembly at which rRNA nucleotides are modified. RNA 2010, 16, 2023–2032. [Google Scholar] [CrossRef]
- Sergeeva, O.V.; Prokhorova, I.V.; Ordabaev, Y.; Tsvetkov, P.O.; Sergiev, P.V.; Bogdanov, A.A.; Makarov, A.A.; Dontsova, O.A. Properties of small rRNA methyltransferase RsmD: Mutational and kinetic study. RNA 2012, 18, 1178–1185. [Google Scholar] [CrossRef][Green Version]
- Tscherne, J.S.; Nurse, K.; Popienick, P.; Michel, H.; Sochacki, M.; Ofengand, J. Purification, Cloning, and Characterization of the 16S RNA m5C967 Methyltransferase from Escherichia coli. Biochemistry 1999, 38, 1884–1892. [Google Scholar] [CrossRef]
- Powers, T.; Noller, H.F. Hydroxyl radical footprinting of ribosomal proteins on 16S rRNA. RNA 1995, 1, 194–209. [Google Scholar]
- Brodersen, D.E.; Clemons, W.M.; Carter, A.P.; Wimberly, B.T.; Ramakrishnan, V. Crystal structure of the 30 S ribosomal subunit from Thermus thermophilus: Structure of the proteins and their interactions with 16 S RNA. J. Mol. Biol. 2002, 316, 725–768. [Google Scholar] [CrossRef] [PubMed]
- Stern, S.; Wilson, R.C.; Noller, H.F. Localization of the binding site for protein S4 on 16S ribosomal RNA by chemical and enzymatic probing and primer extension. J. Mol. Biol. 1986, 192, 101–110. [Google Scholar] [CrossRef]
- Bellur, D.L.; Woodson, S.A. A minimized rRNA-binding site for ribosomal protein S4 and its implications for 30S assembly. Nucleic Acids Res. 2009, 37, 1886–1896. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ramaswamy, P.; Woodson, S.A. Global stabilization of rRNA structure by ribosomal proteins S4, S17, and S20. J. Mol. Biol. 2009, 392, 666–677. [Google Scholar] [CrossRef]
- Bakin, A.; Ofengand, J. Four newly located pseudouridylate residues in Escherichia coli 23S ribosomal RNA are all at the peptidyltransferase center: Analysis by the application of a new sequencing technique. Biochemistry 1993, 32, 9754–9762. [Google Scholar] [CrossRef]
- Ofengand, J.; Del Campo, M.; Kaya, Y. Mapping Pseudouridines in RNA Molecules. Methods 2001, 25, 365–373. [Google Scholar] [CrossRef]
- Bakin, A.; Ofengand, J. Mapping of the 13 pseudouridine residues in Saccharomyces cerevisiae small subunit ribosomal RNA to nucleotide resolution. Nucleic Acids Res. 1995, 23, 3290–3294. [Google Scholar] [CrossRef]
- Gilham, P.T.; Ho, N.W.Y. Reaction of pseudouridine and inosine with N-cyclohexyl-N’-β-(4-methylmorpholinium) ethylcarbodiimide. Biochemistry 1971, 10, 3651–3657. [Google Scholar] [CrossRef]
- Ho, N.W.Y.; Gilham, P.T. The Reversible Chemical Modification of Uracil, Thymine, and Guanine Nucleotides and the Modification of the Action of Ribonuclease on Ribonucleic Acid*. Biochemistry 1967, 6, 3632–3639. [Google Scholar] [CrossRef]
- Wimberly, B.; Brodersen, D.; Clemons, W.; Morgan-Warren, R.; Carter, A.; Vonrhein, C.; Hartsch, T.; Ramakrishnan, V. Structure of the 30S Ribosomal Subunit. Nature 2000, 407, 327–339. [Google Scholar] [CrossRef]
- Kim, H.D.; Nienhaus, G.U.; Ha, T.; Orr, J.W.; Williamson, J.R.; Chu, S. Mg2+-dependent conformational change of RNA studied by fluorescence correlation and FRET on immobilized single molecules. Proc. Natl. Acad. Sci. USA 2002, 99, 4284–4289. [Google Scholar] [CrossRef] [PubMed]
- Adilakshmi, T.; Bellur, D.L.; Woodson, S.A. Concurrent nucleation of 16S folding and induced fit in 30S ribosome assembly. Nature 2008, 455, 1268–1272. [Google Scholar] [CrossRef] [PubMed]
- Williamson, J.R. Induced fit in RNA–protein recognition. Nat. Struct. Biol. 2000, 7, 834–837. [Google Scholar] [CrossRef] [PubMed]
- Leulliot, N.; Varani, G. Current Topics in RNA−Protein Recognition: Control of Specificity and Biological Function through Induced Fit and Conformational Capture. Biochemistry 2001, 40, 7947–7956. [Google Scholar] [CrossRef]
- Boehr, D.D.; Nussinov, R.; Wright, P.E. The role of dynamic conformational ensembles in biomolecular recognition. Nat. Chem. Biol. 2009, 5, 789–796. [Google Scholar] [CrossRef]
- Amitai, S.; Kolodkin-Gal, I.; Hananya-Meltabashi, M.; Sacher, A.; Engelberg-Kulka, H. Escherichia coli MazF Leads to the Simultaneous Selective Synthesis of Both “Death Proteins” and “Survival Proteins”. PLoS Genet. 2009, 5, e1000390. [Google Scholar] [CrossRef]
- Byrgazov, K.; Vesper, O.; Moll, I. Ribosome heterogeneity: Another level of complexity in bacterial translation regulation. Curr. Opin. Microbiol. 2013, 16, 133–139. [Google Scholar] [CrossRef]
- Kaberdina, A.; Szaflarski, W.; Nierhaus, K.; Moll, I. An Unexpected Type of Ribosomes Induced by Kasugamycin: A Look into Ancestral Times of Protein Synthesis? Mol. Cell 2009, 33, 227–236. [Google Scholar] [CrossRef]



© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jayalath, K.; Frisbie, S.; To, M.; Abeysirigunawardena, S. Pseudouridine Synthase RsuA Captures an Assembly Intermediate That Is Stabilized by Ribosomal Protein S17. Biomolecules 2020, 10, 841. https://doi.org/10.3390/biom10060841
Jayalath K, Frisbie S, To M, Abeysirigunawardena S. Pseudouridine Synthase RsuA Captures an Assembly Intermediate That Is Stabilized by Ribosomal Protein S17. Biomolecules. 2020; 10(6):841. https://doi.org/10.3390/biom10060841
Chicago/Turabian StyleJayalath, Kumudie, Sean Frisbie, Minhchau To, and Sanjaya Abeysirigunawardena. 2020. "Pseudouridine Synthase RsuA Captures an Assembly Intermediate That Is Stabilized by Ribosomal Protein S17" Biomolecules 10, no. 6: 841. https://doi.org/10.3390/biom10060841
APA StyleJayalath, K., Frisbie, S., To, M., & Abeysirigunawardena, S. (2020). Pseudouridine Synthase RsuA Captures an Assembly Intermediate That Is Stabilized by Ribosomal Protein S17. Biomolecules, 10(6), 841. https://doi.org/10.3390/biom10060841




