Subtilisin-Involved Morphology Engineering for Improved Antibiotic Production in Actinomycetes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains, Plasmids, Media and DNA Techniques
2.2. Growth Measurement and AP-3 Yield Determination
2.3. Mycelial Morphology Observation
2.4. Transcriptome Sequencing and Analysis of A. pretiosum ATCC 31280
2.5. Transcription Analysis by Quantitative Real-Time PCR
2.6. Inactivation and Trans-Complementation of Gene APASM_4178
2.7. Overexpression of Subtilisin Inhibitor Genes
2.8. Heterologous Overexpression of the AdpA-Like Protein APASM_1021
2.9. Electrophoretic Mobility Shift Assays
3. Results
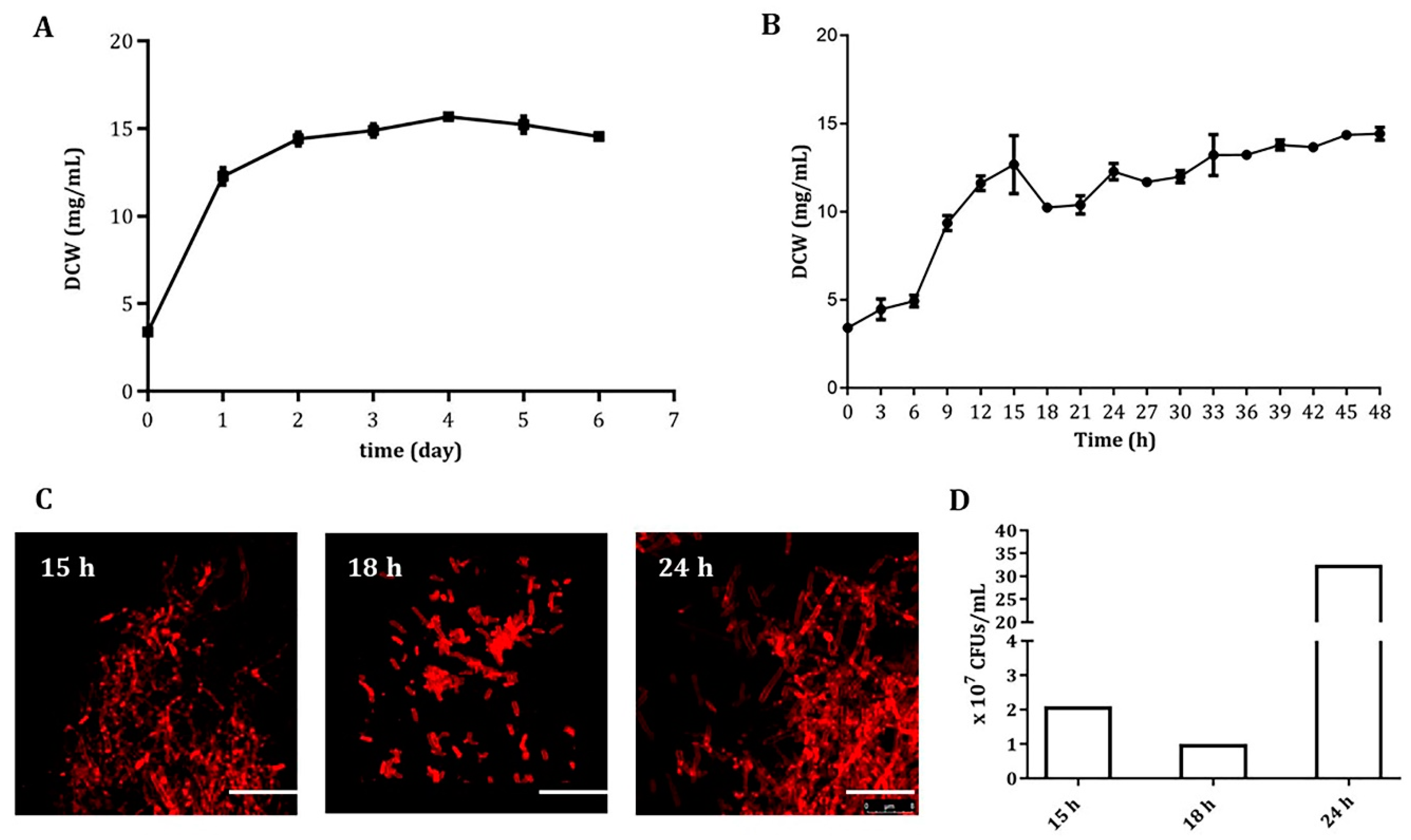
3.1. An Early and Severe Mycelial Fragmentation in Liquid Cultures of A. pretiosum ATCC 31280
3.2. Comparative Transcriptomic Analysis Identified a Subtilisin-Like Protease Gene Responsible for the Mycelial Fragmentation
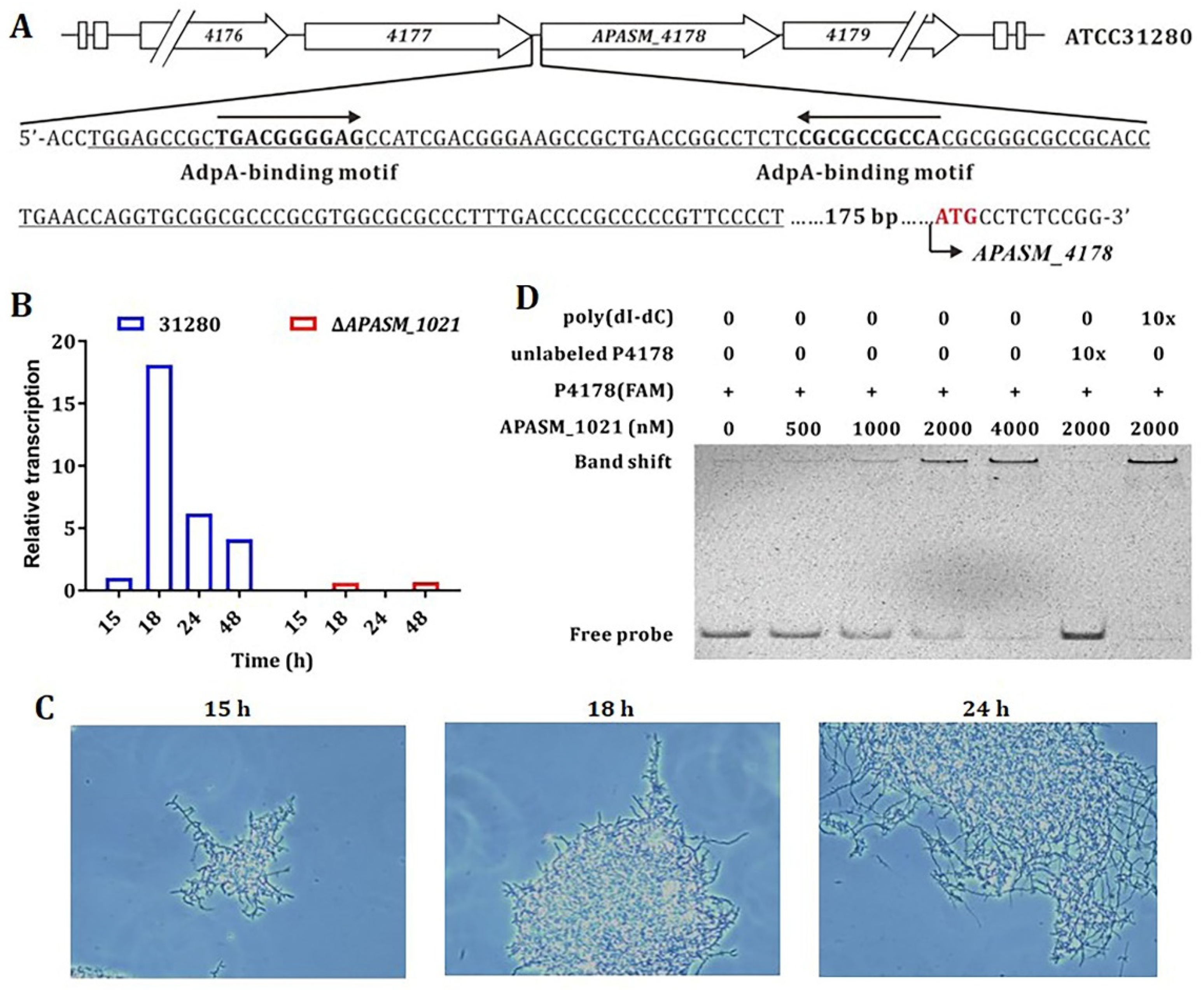
3.3. The Transcription of APASM_4178 and Mycelial Fragmentation is AdpA-Like-Dependent
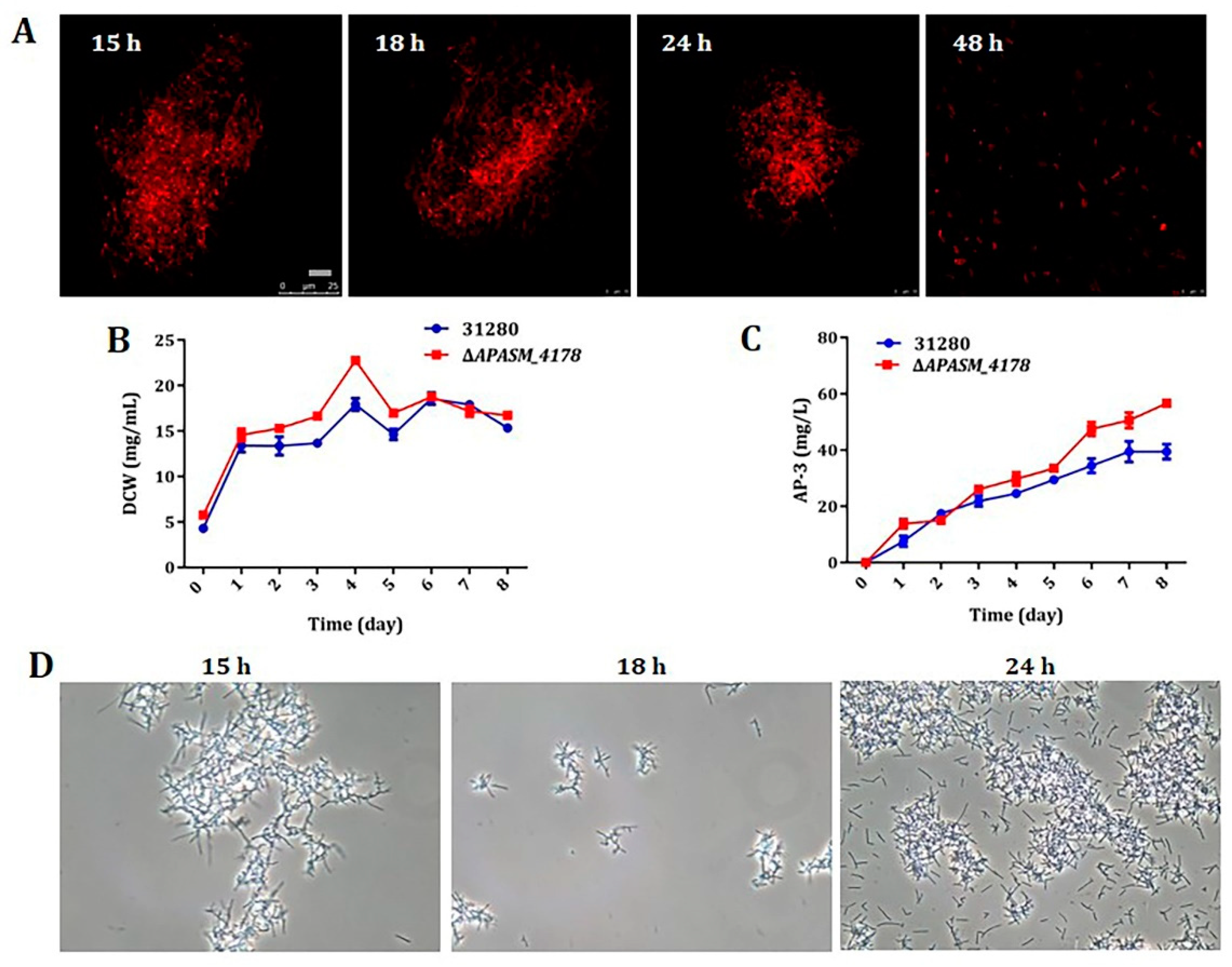
3.4. Overexpression of Subtilisin Inhibitors Alleviated Mycelial Fragmentation and Increased AP-3 Production
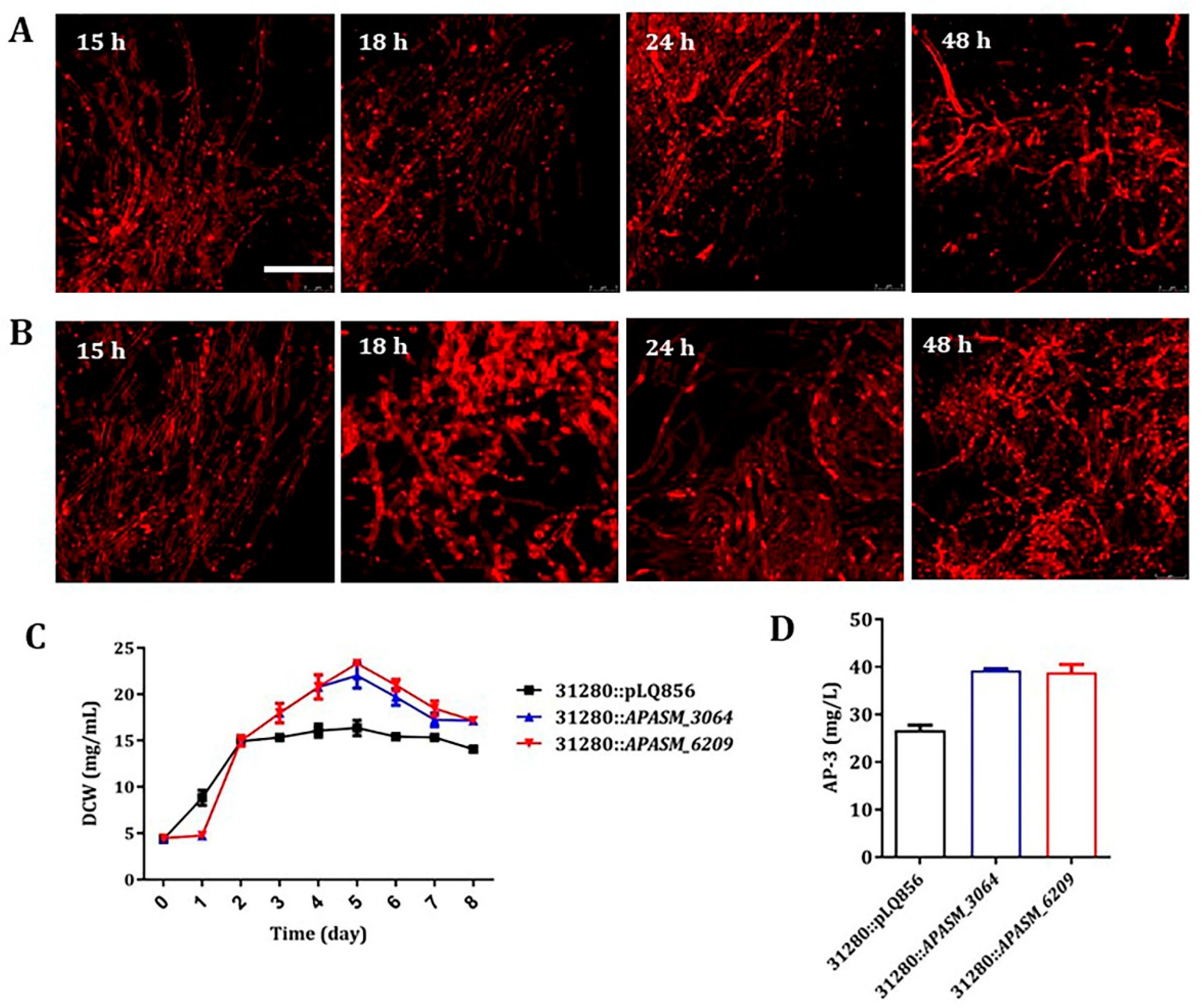
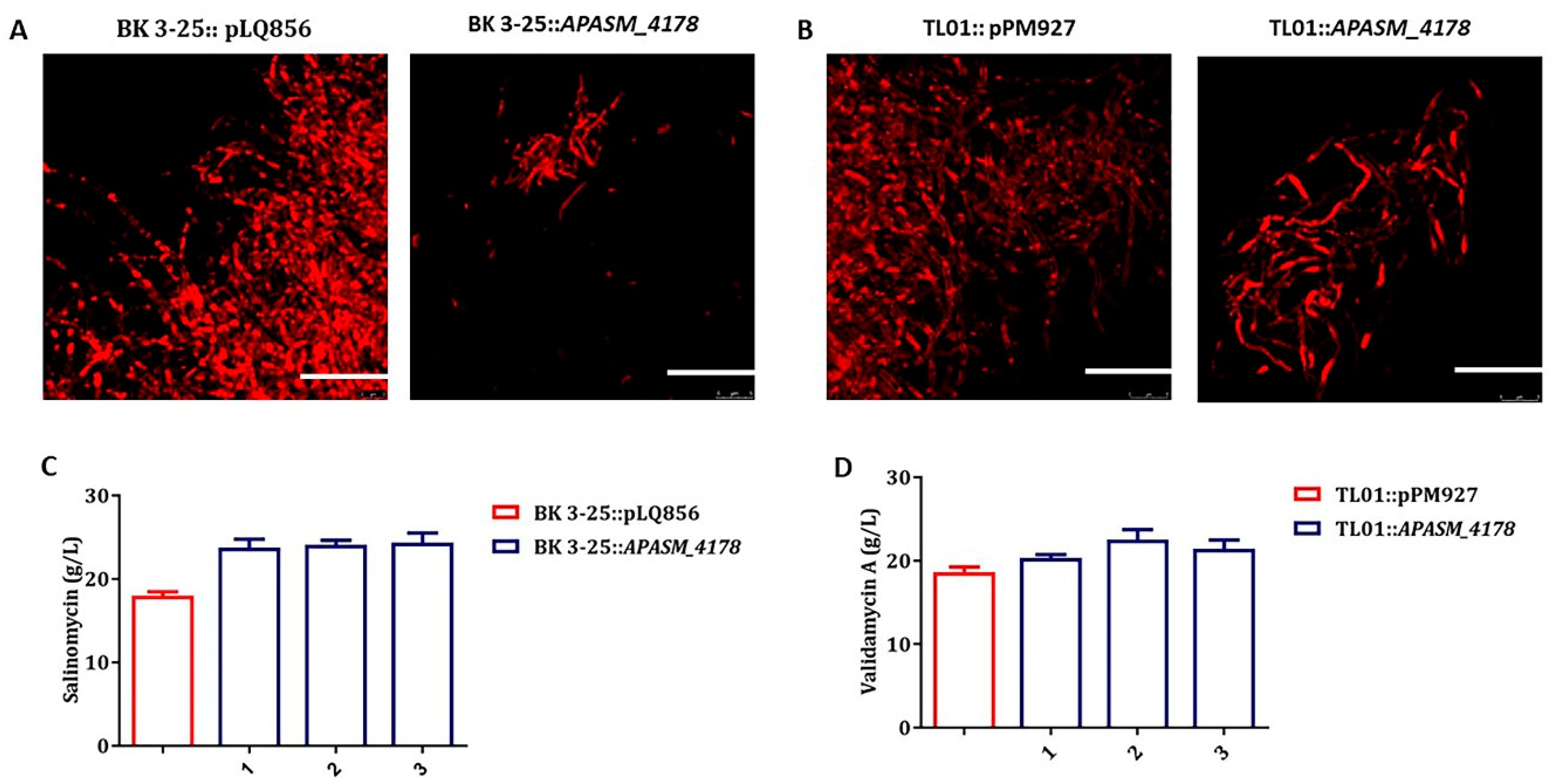
3.5. Overexpression of APASM_4178 Led to Dispersed Mycelia and Improved Antibiotic Yields in Streptomyces Strains
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Flärdh, K.; Buttner, M.J. Streptomyces morphogenetics: Dissecting differentiation in a filamentous bacterium. Nat. Rev. Microbiol. 2009, 7, 36–49. [Google Scholar] [CrossRef] [PubMed]
- Manteca, A.; Alvarez, R.; Salazar, N.; Yagüe, P.; Sanchez, J. Mycelium differentiation and antibiotic production in submerged cultures of Streptomyces coelicolor. Appl. Environ. Microbiol. 2008, 74, 3877–3886. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olmos, E.; Mehmood, N.; Haj Husein, L.; Goergen, J.L.; Fick, M.; Delaunay, S. Effects of bioreactor hydrodynamics on the physiology of Streptomyces. Bioprocess. Biosyst. Eng. 2013, 36, 259–272. [Google Scholar] [CrossRef] [PubMed]
- Vecht-Lifshitz, S.E.; Sasson, Y.; Braun, S. Nikkomycin production in pellets of Streptomyces tendae. J. Appl. Bacteriol. 1992, 72, 195–200. [Google Scholar] [CrossRef]
- Wardell, J.N.; Stocks, S.M.; Thomas, C.R.; Bushell, M.E. Decreasing the hyphal branching rate of Saccharopolyspora erythraea NRRL 2338 leads to increased resistance to breakage and increased antibiotic production. Biotechnol. Bioeng. 2002, 78, 141–146. [Google Scholar] [CrossRef]
- Park, Y.; Tamura, S.; Koike, Y.; Toriyama, M.; Okabe, M. Mycelial pellet intrastructure visualization and viability prediction in a culture of Streptomyces fradiae using confocal scanning laser microscopy. J. Ferment. Bioeng. 1997, 84, 483–486. [Google Scholar] [CrossRef]
- Jonsbu, E.; McIntyre, M.; Nielsen, J. The influence of carbon sources and morphology on nystatin production by Streptomyces noursei. J. Biotechnol. 2002, 95, 133–144. [Google Scholar] [CrossRef]
- Kumar, P.; Dubey, K.K. Mycelium transformation of Streptomyces toxytricini into pellet: Role of culture conditions and kinetics. Bioresour. Technol. 2017, 228, 339–347. [Google Scholar] [CrossRef]
- Ren, X.-D.; Chen, X.-S.; Tang, L.; Zeng, X.; Wang, L.; Mao, Z.-G. Physiological mechanism of the overproduction of ε-poly-L-lysine by acidic pH shock in fed-batch fermentation. Bioprocess. Biosyst. Eng. 2015, 38, 2085–2094. [Google Scholar] [CrossRef]
- Yen, H.-W.; Hsiao, H.-P. Effects of dissolved oxygen level on rapamycin production by pellet-form of Streptomyces hygroscopicus. J. Biosci. Bioeng. 2013, 116, 366–370. [Google Scholar] [CrossRef]
- van Veluw, G.J.; Petrus, M.L.; Gubbens, J.; de Graaf, R.; de Jong, I.P.; van Wezel, G.P.; Wosten, H.A.; Claessen, D. Analysis of two distinct mycelial populations in liquid-grown Streptomyces cultures using a flow cytometry-based proteomics approach. Appl. Microbiol. Biotechnol. 2012, 96, 1301–1312. [Google Scholar] [CrossRef] [PubMed]
- van Dissel, D.; Claessen, D.; van Wezel, G.P. Chapter One—Morphogenesis of Streptomyces in submerged cultures. Adv. Appl. Microbiol. 2014, 89, 1–45. [Google Scholar] [PubMed]
- Flärdh, K. Essential role of DivIVA in polar growth and morphogenesis in Streptomyces coelicolor A3(2). Mol. Microbiol. 2003, 49, 1523–1536. [Google Scholar] [CrossRef] [PubMed]
- Holmes, N.A.; Walshaw, J.; Leggett, R.M.; Thibessard, A.; Dalton, K.A.; Gillespie, M.D.; Hemmings, A.M.; Gust, B.; Kelemen, G.H. Coiled-coil protein Scy is a key component of a multiprotein assembly controlling polarized growth in Streptomyces. Proc. Natl. Acad. Sci. USA 2013, 110, E397–E406. [Google Scholar] [CrossRef] [Green Version]
- Kawamoto, S.; Watanabe, H.; Hesketh, A.; Ensign, J.C.; Ochi, K. Expression analysis of the ssgA gene product, associated with sporulation and cell division in Streptomyces griseus. Microbiology 1997, 143, 1077–1086. [Google Scholar] [CrossRef] [Green Version]
- Xu, H.; Chater, K.F.; Deng, Z.; Tao, M. A cellulose synthase-like protein involved in hyphal tip growth and morphological differentiation in Streptomyces. J. Bacteriol. 2008, 190, 4971–4978. [Google Scholar] [CrossRef] [Green Version]
- Liman, R.; Facey, P.D.; van Keulen, G.; Dyson, P.J.; Del Sol, R. A laterally acquired galactose oxidase-like gene is required for aerial development during osmotic stress in Streptomyces coelicolor. PLoS ONE 2013, 8, e54112. [Google Scholar] [CrossRef] [Green Version]
- Kwak, J.; McCue, L.A.; Trczianka, K.; Kendrick, K.E. Identification and characterization of a developmentally regulated protein, EshA, required for sporogenic hyphal branches in Streptomyces griseus. J. Bacteriol. 2001, 183, 3004–3015. [Google Scholar] [CrossRef] [Green Version]
- Koebsch, I.; Overbeck, J.; Piepmeyer, S.; Meschke, H.; Schrempf, H. A molecular key for building hyphae aggregates: The role of the newly identified Streptomyces protein HyaS. Microb. Biotechnol. 2009, 2, 343–360. [Google Scholar] [CrossRef] [Green Version]
- van Dissel, D.; Willemse, J.; Zacchetti, B.; Claessen, D.; Pier, G.B.; van Wezel, G.P. Production of poly-β-1,6-N-acetylglucosamine by MatAB is required for hyphal aggregation and hydrophilic surface adhesion by Streptomyces. Microb Cell 2018, 5, 269–279. [Google Scholar] [CrossRef]
- Nguyen, H.T.; Pham, V.T.T.; Nguyen, C.T.; Pokhrel, A.R.; Kim, T.-S.; Kim, D.; Na, K.; Yamaguchi, T.; Sohng, J.K. Exploration of cryptic organic photosensitive compound as Zincphyrin IV in Streptomyces venezuelae ATCC 15439. Appl. Microbiol. Biotechnol. 2020, 104, 713–724. [Google Scholar] [CrossRef] [PubMed]
- van Wezel, G.P.; Krabben, P.; Traag, B.A.; Keijser, B.J.F.; Kerste, R.; Vijgenboom, E.; Heijnen, J.J.; Kraal, B. Unlocking Streptomyces spp. for use as sustainable industrial production platforms by morphological engineering. Appl. Environ. Microbiol. 2006, 72, 5283–5288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kupchan, S.M.; Komoda, Y.; Court, W.A.; Thomas, G.J.; Smith, R.M.; Karim, A.; Gilmore, C.J.; Haltiwanger, R.C.; Bryan, R.F. Maytansine, a novel antileukemic ansa macrolide from Maytenus ovatus. J. Am. Chem. Soc. 1972, 94, 1354–1356. [Google Scholar] [CrossRef] [PubMed]
- Higashide, E.; Asai, M.; Ootsu, K.; Tanida, S.; Kozai, Y.; Hasegawa, T.; Kishi, T.; Sugino, Y.; Yoneda, M. Ansamitocin, a group of novel maytansinoid antibiotics with antitumour properties from Nocardia. Nature 1977, 270, 721–722. [Google Scholar] [CrossRef] [PubMed]
- Land, M.; Lapidus, A.; Mayilraj, S.; Chen, F.; Copeland, A.; Del Rio, T.G.; Nolan, M.; Lucas, S.; Tice, H.; Cheng, J.-F.; et al. Complete genome sequence of Actinosynnema mirum type strain (101T). Stand. Genom. Sci. 2009, 1, 46–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Snipes, C.E.; Duebelbeis, D.O.; Olson, M.; Hahn, D.R.; Dent, W.H.; Gilbert, J.R.; Werk, T.L.; Davis, G.E.; Lee-Lu, R.; Graupner, P.R. The ansacarbamitocins: Polar ansamitocin derivatives. J. Nat. Prod. 2007, 70, 1578–1581. [Google Scholar] [CrossRef] [PubMed]
- Li, X.-M.; Li, X.-M.; Lu, C.-H. Abscisic acid-type sesquiterpenes and ansamycins from Amycolatopsis alba DSM 44262. J. Asian Nat. Prod. Res. 2017, 19, 946–953. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.G.; Liu, Q.; Wang, H.F.; Park, D.J.; Guo, J.W.; Kim, C.J.; Zhang, Y.M.; Li, W.J. Nocardiopsis ansamitocini sp. nov., a new producer of ansamitocin P-3 of the genus Nocardiopsis. Int. J. Syst. Evol. Microbiol. 2016, 66, 230–235. [Google Scholar] [CrossRef]
- Mecklenburg, L. A brief introduction to antibody–drug conjugates for toxicologic pathologists. Toxicol. Pathol. 2018, 46, 746–752. [Google Scholar] [CrossRef] [Green Version]
- Lambert, J.M.; Chari, R.V.J. Ado-trastuzumab Emtansine (T-DM1): An antibody–drug conjugate (ADC) for HER2-positive breast cancer. J. Med. Chem. 2014, 57, 6949–6964. [Google Scholar] [CrossRef]
- Yu, T.-W.; Bai, L.; Clade, D.; Hoffmann, D.; Toelzer, S.; Trinh, K.Q.; Xu, J.; Moss, S.J.; Leistner, E.; Floss, H.G. The biosynthetic gene cluster of the maytansinoid antitumor agent ansamitocin from Actinosynnema pretiosum. Proc. Natl. Acad. Sci. USA 2002, 99, 7968–7973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, Q.; Shen, Y.; Bai, L. Biosynthesis of 3,5-AHBA-derived natural products. Nat. Prod. Rep. 2012, 29, 243–263. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Fan, Y.; Nambou, K.; Hu, F.; Imanaka, T.; Wei, L.; Hua, Q. Improvement of ansamitocin P-3 production by Actinosynnema mirum with fructose as the sole carbon source. Appl. Biochem. Biotechnol. 2015, 175, 2845–2856. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Gao, Y.; Zhou, J.; Wei, L.; Chen, J.; Hua, Q. Process optimization with alternative carbon sources and modulation of secondary metabolism for enhanced ansamitocin P-3 production in Actinosynnema pretiosum. J. Biotechnol. 2014, 192, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Bandi, S.; Kim, Y.; Chang, Y.K.; Shang, G.D.; Yu, T.W.; Floss, H.G. Construction of asm2 deletion mutant of Actinosynnema pretiosum and medium optimization for ansamitocin P-3 production using statistical approach. J. Microbiol. Biotechnol. 2006, 16, 1338–1346. [Google Scholar]
- Zhao, M.; Fan, Y.; Wei, L.; Hu, F.; Hua, Q. Effects of the methylmalonyl-CoA metabolic pathway on ansamitocin production in Actinosynnema pretiosum. Appl. Biochem. Biotechnol. 2017, 181, 1167–1178. [Google Scholar] [CrossRef]
- Fan, Y.; Hu, F.; Wei, L.; Bai, L.; Hua, Q. Effects of modulation of pentose-phosphate pathway on biosynthesis of ansamitocins in Actinosynnema pretiosum. J. Biotechnol. 2016, 230, 3–10. [Google Scholar] [CrossRef]
- Du, Z.-Q.; Zhong, J.-J. Rational approach to improve ansamitocin P-3 production by integrating pathway engineering and substrate feeding in Actinosynnema pretiosum. Biotechnol. Bioeng. 2018, 115, 2456–2466. [Google Scholar] [CrossRef]
- Ning, X.; Wang, X.; Wu, Y.; Kang, Q.; Bai, L. Identification and engineering of post-PKS modification bottlenecks for ansamitocin P-3 titer improvement in Actinosynnema pretiosum subsp. pretiosum ATCC 31280. Biotechnol. J. 2017, 12, 1700484. [Google Scholar] [CrossRef]
- Kieser, T.; Bibb, M.J.; Buttner, M.J.; Chater, K.F.; Hopwood, D.A. Practical Streptomyces Genetics; John Innes Foundation: Norwich, UK, 2000. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Hellman, L.M.; Fried, M.G. Electrophoretic mobility shift assay (EMSA) for detecting protein-nucleic acid interactions. Nat. Protoc. 2007, 2, 1849–1861. [Google Scholar] [CrossRef] [PubMed]
- Qu, S.; Kang, Q.; Wu, H.; Wang, L.; Bai, L. Positive and negative regulation of GlnR in validamycin A biosynthesis by binding to different loci in promoter region. Appl. Microbiol. Biotechnol. 2015, 99, 4771–4783. [Google Scholar] [CrossRef] [PubMed]
- Vujaklija, D.; Horinouchi, S.; Beppu, T. Detection of an A-factor-responsive protein that binds to the upstream activation sequence of strR, a regulatory gene for streptomycin biosynthesis in Streptomyces griseus. J. Bacteriol. 1993, 175, 2652–2661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rabyk, M.; Yushchuk, O.; Rokytskyy, I.; Anisimova, M.; Ostash, B. Genomic insights into evolution of AdpA family master regulators of morphological differentiation and secondary metabolism in Streptomyces. J. Mol. Evol. 2018, 86, 204–215. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, H.; Tomono, A.; Ohnishi, Y.; Horinouchi, S. DNA-binding specificity of AdpA, a transcriptional activator in the A-factor regulatory cascade in Streptomyces griseus. Mol. Microbiol. 2004, 53, 555–572. [Google Scholar] [CrossRef] [PubMed]
- Horinouchi, S.; Beppu, T. Hormonal control by A-factor of morphological development and secondary metabolism in Streptomyces. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2007, 83, 277–295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Satow, Y.; Mitsui, Y.; Iitaka, Y.; Murao, S.; Sato, S. Crystallization and preliminary X-ray investigation of a new alkaline protease inhibitor and its complex with subtilisin BPN′. J. Mol. Biol. 1973, 75, 745-IN22. [Google Scholar] [CrossRef]
- Kim, D.W.; Hesketh, A.; Kim, E.S.; Song, J.Y.; Lee, D.H.; Kim, I.S.; Chater, K.F.; Lee, K.J. Complex extracellular interactions of proteases and a protease inhibitor influence multicellular development of Streptomyces coelicolor. Mol. Microbiol. 2008, 70, 1180–1193. [Google Scholar] [CrossRef]
- Zhang, X.; Lu, C.; Bai, L. Mechanism of salinomycin overproduction in Streptomyces albus as revealed by comparative functional genomics. Appl. Microbiol. Biotechnol. 2017, 101, 4635–4644. [Google Scholar] [CrossRef]
- Tan, G.-Y.; Bai, L.; Zhong, J.-J. Exogenous 1,4-butyrolactone stimulates A-factor-like cascade and validamycin biosynthesis in Streptomyces hygroscopicus 5008. Biotechnol. Bioeng. 2013, 110, 2984–2993. [Google Scholar] [CrossRef]
- Watanabe, K.; Okuda, T.; Yokose, K.; Furumai, T.; Maruyama, H.B. Actinosynnema mirum, a new producer of nocardicin antibiotics. J. Antibiot. 1983, 36, 321–324. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.W.; Lee, B.R.; You, S.; Kim, E.J.; Kim, J.N.; Song, E.; Yang, Y.H.; Hwang, D.; Kim, B.G. Transcriptome analysis of wild-type and afsS deletion mutant strains identifies synergistic transcriptional regulator of afsS for a high antibiotic-producing strain of Streptomyces coelicolor A3(2). Appl. Microbiol. Biotechnol. 2018, 102, 3243–3253. [Google Scholar] [CrossRef] [PubMed]
- Fondi, M.; Pinatel, E.; Talà, A.; Damiano, F.; Consolandi, C.; Mattorre, B.; Fico, D.; Testini, M.; De Benedetto, G.E.; Siculella, L.; et al. Time-resolved transcriptomics and constraint-based modeling identify system-level metabolic features and overexpression targets to increase spiramycin production in Streptomyces ambofaciens. Front. Microbiol. 2017, 8, 835. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, M.; Taguchi, S.; Yamada, S.; Kojima, S.; Miura, K.I.; Momose, H. A novel member of the subtilisin-like protease family from Streptomyces albogriseolus. J. Bacteriol. 1997, 179, 430–438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vos, P.; Simons, G.; Siezen, R.J.; de Vos, W.M. Primary structure and organization of the gene for a procaryotic, cell envelope-located serine proteinase. J. Biol. Chem. 1989, 264, 13579–13585. [Google Scholar] [PubMed]
- Hirano, S.; Kato, J.Y.; Ohnishi, Y.; Horinouchi, S. Control of the Streptomyces subtilisin inhibitor gene by AdpA in the A-factor regulatory cascade in Streptomyces griseus. J. Bacteriol. 2006, 188, 6207–6216. [Google Scholar] [CrossRef] [Green Version]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, Y.; Kang, Q.; Zhang, L.-L.; Bai, L. Subtilisin-Involved Morphology Engineering for Improved Antibiotic Production in Actinomycetes. Biomolecules 2020, 10, 851. https://doi.org/10.3390/biom10060851
Wu Y, Kang Q, Zhang L-L, Bai L. Subtilisin-Involved Morphology Engineering for Improved Antibiotic Production in Actinomycetes. Biomolecules. 2020; 10(6):851. https://doi.org/10.3390/biom10060851
Chicago/Turabian StyleWu, Yuanting, Qianjin Kang, Li-Li Zhang, and Linquan Bai. 2020. "Subtilisin-Involved Morphology Engineering for Improved Antibiotic Production in Actinomycetes" Biomolecules 10, no. 6: 851. https://doi.org/10.3390/biom10060851
APA StyleWu, Y., Kang, Q., Zhang, L.-L., & Bai, L. (2020). Subtilisin-Involved Morphology Engineering for Improved Antibiotic Production in Actinomycetes. Biomolecules, 10(6), 851. https://doi.org/10.3390/biom10060851





