Structural Basis of Specific Glucoimidazole and Mannoimidazole Binding by Os3BGlu7
Abstract
1. Introduction
2. Materials and Methods
2.1. Protein Expression, Purification, Crystallization and Structure Determination
2.2. System Preparation for Molecular Modeling
2.3. Molecular Dynamics Simulations
2.4. Replica Exchange Molecular Dynamics of the Unbound Glycoside Inhibitors
3. Results and Discussion
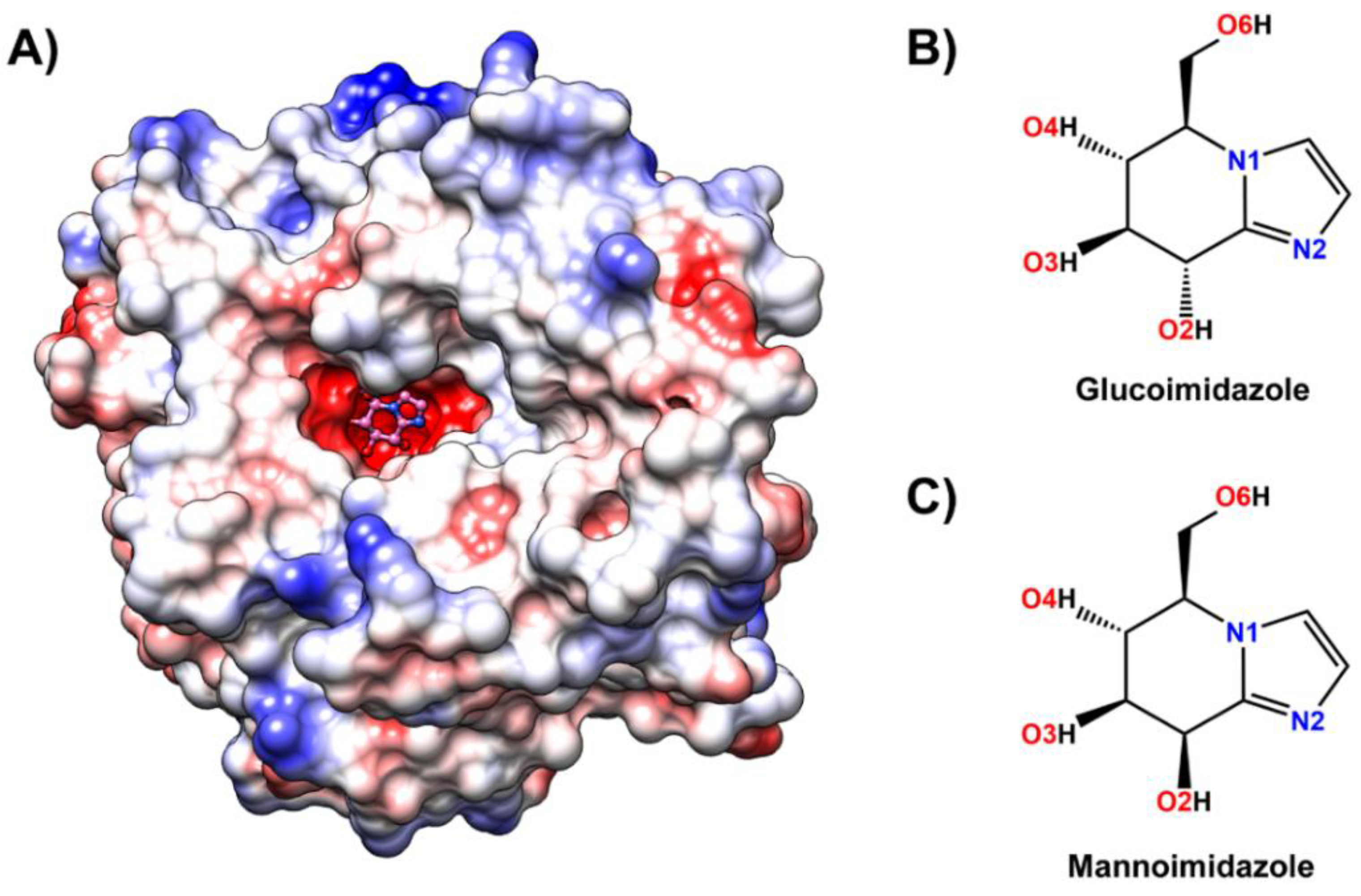
3.1. Structure of Rice Os3BGlu7 in Complex with Inhibitor
3.2. Conformational Dynamics of Protein—Inhibitor Complex
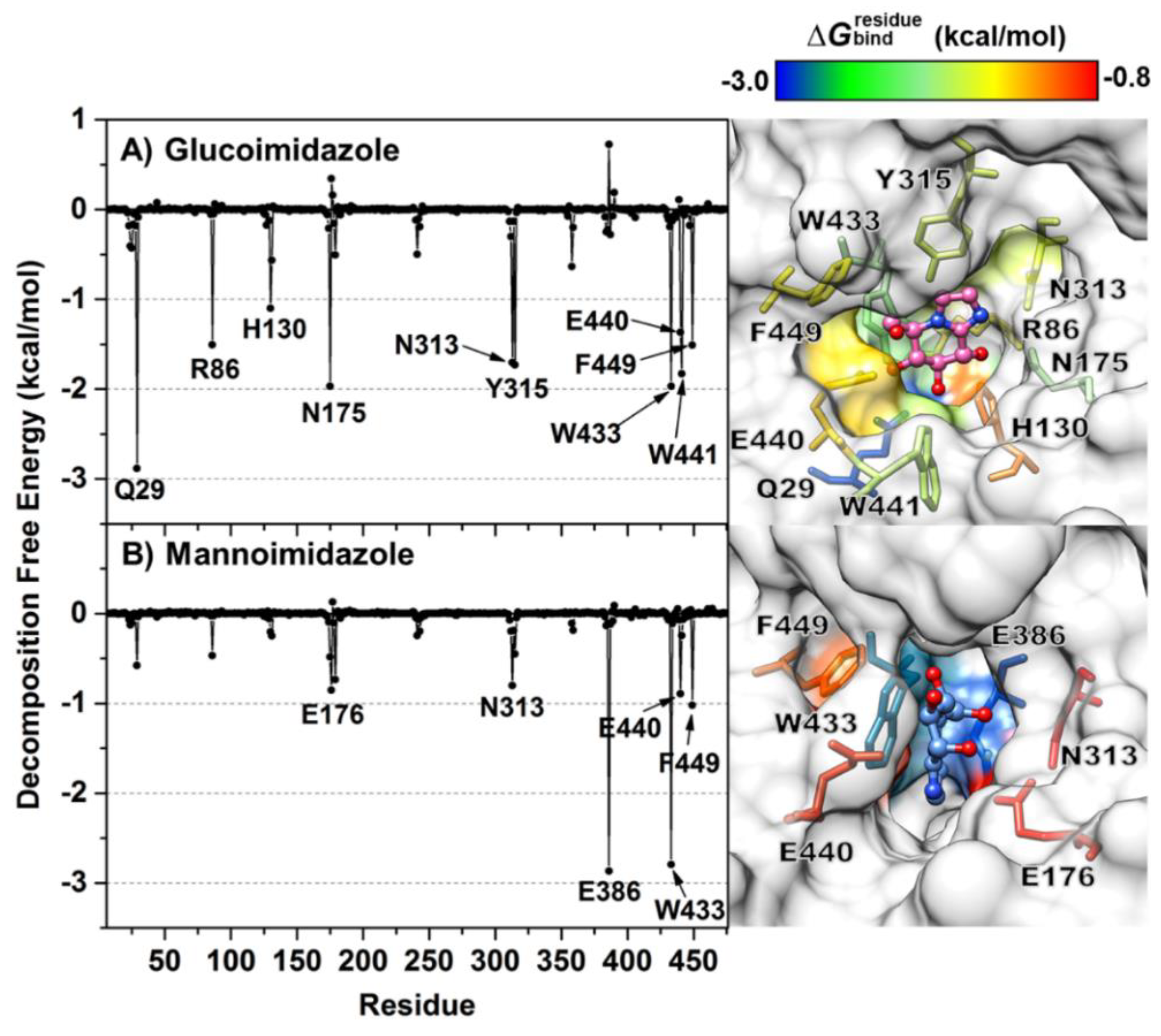
3.3. Binding Affinity of Protein—Inhibitor Complex
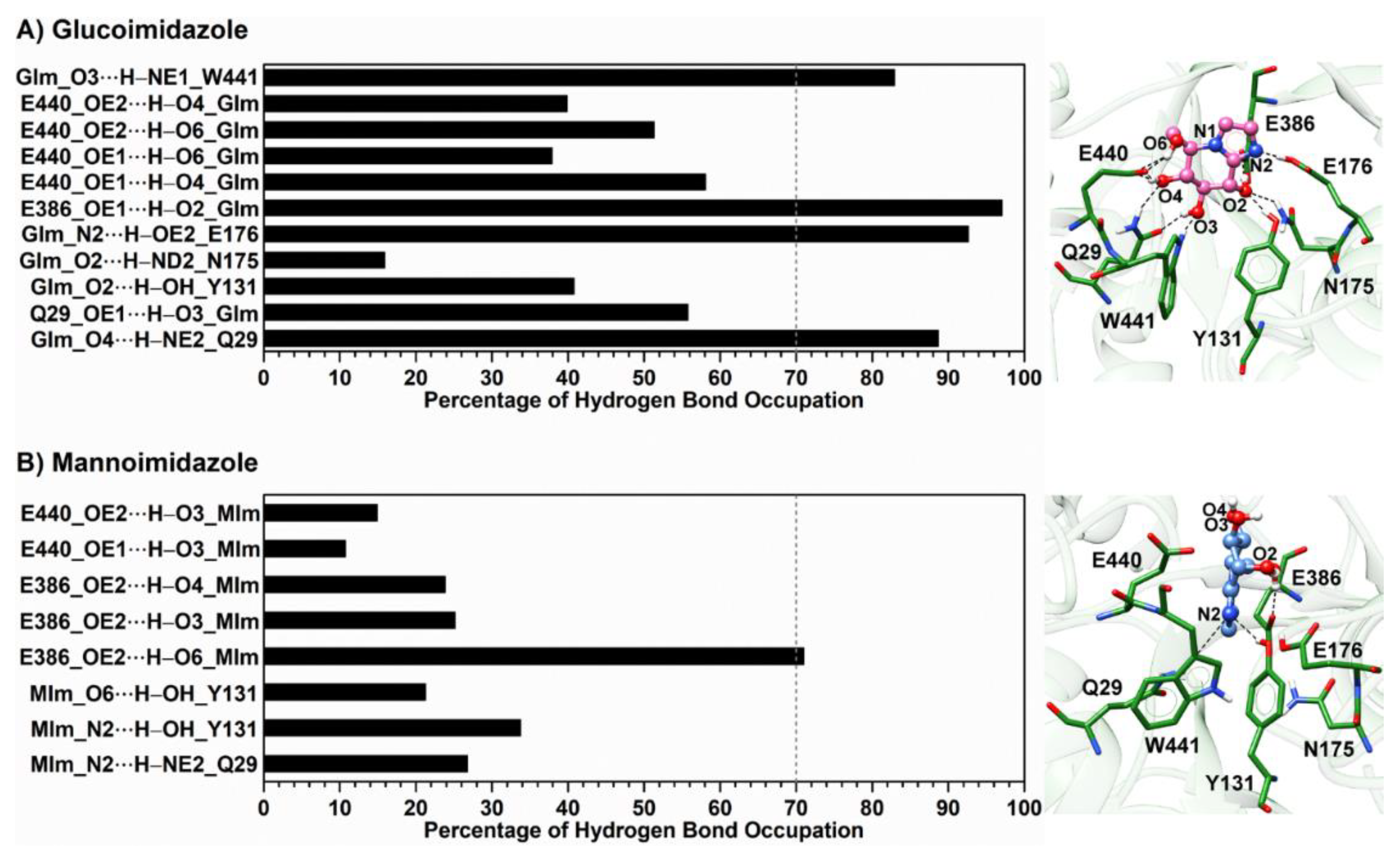
3.4. Inhibitor Binding Pattern
3.5. Ligand-Binding Pocket Volume and Water Accessibility in the Binding Pocket
3.6. Sugar Ring Conformation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gilbert, H.J. The biochemistry and structural biology of plant cell wall deconstruction. Plant Physiol. 2010, 153, 444–455. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, H.J.; Knox, J.P.; Boraston, A.B. Advances in understanding the molecular basis of plant cell wall polysaccharide recognition by carbohydrate-binding modules. Curr. Opin. Struct. Biol. 2013, 23, 669–677. [Google Scholar] [CrossRef] [PubMed]
- Cantarel, B.I.; Coutinho, P.M.; Rancurel, C.; Bernard, T.; Lombard, V.; Henrissat, B. The Carbohydrate-Active EnZymes database (CAZy): An expert resource for glycogenomics. Nucleic Acids Res. 2009, 37, D233–D238. [Google Scholar] [CrossRef] [PubMed]
- Ketudat Cairns, J.R.; Esen, A. β-Glucosidases. Cell. Mol. Life Sci. 2010, 67, 3389–3405. [Google Scholar] [CrossRef]
- Ketudat Cairns, J.R.; Mahong, B.; Baiya, S.; Jeon, J.S. β-Glucosidases: Multitasking, Moonlighting or Simply Misunderstood? Plant Sci. 2015, 241, 246–259. [Google Scholar] [CrossRef] [PubMed]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef]
- Henrissat, B.; Davies, G. Structural and sequence-based classification of glycoside hydrolases. Curr. Opin. Struct. Biol. 1997, 7, 637–644. [Google Scholar] [CrossRef]
- Czjzek, M.; Cicek, M.; Zamboni, V.; Bevan, D.R.; Henrissat, B.; Esen, A. The mechanism of substrate (aglycone) specificity in β-glucosidases is revealed by crystal structures of mutant maize β-glucosidase-DIMBOA, -DIMBOAGIc, and -dhurrin complexes. Proc. Natl. Acad. Sci. USA 2000, 97, 13555–13560. [Google Scholar] [CrossRef]
- Tribolo, S.; Berrin, J.G.; Kroon, P.A.; Czjzek, M.; Juge, N. The Crystal Structure of Human Cytosolic β-Glucosidase Unravels the Substrate Aglycone Specificity of a Family 1 Glycoside Hydrolase. J. Mol. Biol. 2007, 370, 964–975. [Google Scholar] [CrossRef]
- Verdoucq, L.; Morinière, J.; Bevan, D.R.; Esen, A.; Vasella, A.; Henrissat, B.; Czjzek, M. Structural determinants of substrate specificity in family 1 β-glucosidases. Novel insights from the crystal structure of Sorghum dhurrinase-1, a plant β-glucosidase with strict specificity, in complex with its natural substrate. J. Biol. Chem. 2004, 279, 31796–31803. [Google Scholar] [CrossRef]
- Chuenchor, W.; Pengthaisong, S.; Robinson, R.C.; Yuvaniyama, J.; Svasti, J.; Ketudat Cairns, J.R. The structural basis of oligosaccharide binding by rice BGlu1 beta-glucosidase. J. Struct. Biol. 2011, 173, 169–179. [Google Scholar] [CrossRef]
- Vuong, T.V.; Wilson, D.B. Glycoside hydrolases: Catalytic base/nucleophile diversity. Biotechnol. Bioeng. 2010, 107, 195–205. [Google Scholar] [CrossRef]
- Vocadlo, D.J.; Davies, G.J. Mechanistic insights into glycosidase chemistry. Curr. Opin. Chem. Biol. 2008, 12, 539–555. [Google Scholar] [CrossRef]
- Woods, R.J.; Bowen, J.P.; Andrews, C.W.; Woods, R.J.; Bowen, J.P. Molecular Mechanical Investigations of the Properties of Oxocarbenium Ions. 2. Application to Glycoside Hydrolysis. J. Am. Chem. Soc. 1992, 114, 859–864. [Google Scholar] [CrossRef]
- Davies, G.J.; Planas, A.; Rovira, C. Conformational analyses of the reaction coordinate of glycosidases. Acc. Chem. Res. 2012, 45, 308–316. [Google Scholar] [CrossRef] [PubMed]
- Isorna, P.; Polaina, J.; Latorre-García, L.; Cañada, F.J.; González, B.; Sanz-Aparicio, J. Crystal Structures of Paenibacillus polymyxa β-Glucosidase B Complexes Reveal the Molecular Basis of Substrate Specificity and Give New Insights into the Catalytic Machinery of Family I Glycosidases. J. Mol. Biol. 2007, 371, 1204–1218. [Google Scholar] [CrossRef] [PubMed]
- Nijikken, Y.; Tsukada, T.; Igarashi, K.; Samejima, M.; Wakagi, T.; Shoun, H.; Fushinobu, S. Crystal structure of intracellular family 1 β-glucosidase BGL1A from the basidiomycete Phanerochaete chrysosporium. FEBS Lett. 2007, 581, 1514–1520. [Google Scholar] [CrossRef]
- Svasti, J.; Chuenchor, W.; Opassiri, R.; Pengthaisong, S.; Yuvaniyama, J.; Ketudat Cairns, J.R. Purification, crystallization and preliminary X-ray analysis of rice BGlu1 β-glucosidase with and without 2-deoxy-2-fluoro-β-d-glucoside. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2006, 62, 798–801. [Google Scholar] [CrossRef]
- Chuenchor, W.; Pengthaisong, S.; Robinson, R.C.; Yuvaniyama, J.; Oonanant, W.; Bevan, D.R.; Esen, A.; Chen, C.J.; Opassiri, R.; Svasti, J.; et al. Structural Insights into Rice BGlu1 β-Glucosidase Oligosaccharide Hydrolysis and Transglycosylation. J. Mol. Biol. 2008, 377, 1200–1215. [Google Scholar] [CrossRef] [PubMed]
- Barrett, T.; Suresh, C.G.; Tolley, S.P.; Dodson, E.J.; Hughes, M.A. The crystal structure of a cyanogenic β-glucosidase from white clover, a family 1 glycosyl hydrolase. Structure 1995, 3, 951–960. [Google Scholar] [CrossRef]
- Opassiri, R.; Ketudat Cairns, J.R.; Akiyama, T.; Wara-Aswapati, O.; Svasti, J.; Esen, A. Characterization of a rice β-glucosidase highly expressed in flower and germinating shoot. Plant Sci. 2003, 165, 627–638. [Google Scholar] [CrossRef]
- Opassiri, R.; Hua, Y.; Wara-Aswapati, O.; Akiyama, T.; Svasti, J.; Esen, A.; Ketudat Cairns, J.R. β-Glucosidase, exo-β-glucanase and pyridoxine transglucosylase activities of rice BGlu1. Biochem. J. 2004, 379, 125–131. [Google Scholar] [CrossRef] [PubMed]
- Hommalai, G.; Withers, S.G.; Chuenchor, W.; Ketudat Cairns, J.R.; Svasti, J. Enzymatic synthesis of cello-oligosaccharides by rice BGlu1 β-glucosidase glycosynthase mutants. Glycobiology 2007, 17, 744–753. [Google Scholar] [CrossRef] [PubMed]
- Hrmova, M.; Harvey, A.J.; Wang, J.; Shirley, N.J.; Jones, G.P.; Stone, B.A.; Høj, P.B.; Fincher, G.B. Barley β-d-glucan exohydrolases with β-d-glucosidase activity: Purification, characterization, and determination of primary structure from a cDNA clone. J. Biol. Chem. 1996, 271, 5277–5286. [Google Scholar] [CrossRef]
- Hrmova, M.; Varghese, J.N.; Høj, P.B.; Fincher, G.B. Crystallization and preliminary X-ray analysis of β-glucan exohydrolase isoenzyme ExoI from barley (Hordeum vulgare). Acta Crystallogr. Sect. D Biol. Crystallogr. 1998, 54, 687–689. [Google Scholar] [CrossRef]
- Kuntothom, T.; Luang, S.; Harvey, A.J.; Fincher, G.B.; Opassiri, R.; Hrmova, M.; Ketudat Cairns, J.R. Rice family GH1 glycoside hydrolases with β-d-glucosidase and β-d-mannosidase activities. Arch. Biochem. Biophys. 2009, 491, 85–95. [Google Scholar] [CrossRef]
- Williams, R.J.; Iglesias-Fernández, J.; Stepper, J.; Jackson, A.; Thompson, A.J.; Lowe, E.C.; White, J.M.; Gilbert, H.J.; Rovira, C.; Davies, G.J.; et al. Combined inhibitor free-energy landscape and structural analysis reports on the mannosidase conformational coordinate. Angew. Chem. Int. Ed. 2014, 53, 1087–1091. [Google Scholar] [CrossRef]
- Sinnott, M.L. The Principle of Least Nuclear Motion and the Theory of Stereoelectronic Control. Adv. Phys. Org. Chem. 1988, 24, 113–204. [Google Scholar] [CrossRef]
- Tankrathok, A.; Iglesias-Fernández, J.; Williams, R.J.; Pengthaisong, S.; Baiya, S.; Hakki, Z.; Robinson, R.C.; Hrmova, M.; Rovira, C.; Williams, S.J.; et al. A Single Glycosidase Harnesses Different Pyranoside Ring Transition State Conformations for Hydrolysis of Mannosides and Glucosides. ACS Catal. 2015, 5, 6041–6051. [Google Scholar] [CrossRef]
- Granier, T.; Panday, N.; Vasella, A. Structure-activity relations for imidazo-pyridine-type inhibitors of β-d-glucosidases. Helv. Chim. Acta 1997, 80, 979–987. [Google Scholar] [CrossRef]
- Otwinowski, Z.; Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997, 276, 307–326. [Google Scholar] [CrossRef] [PubMed]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004, 60, 2126–2132. [Google Scholar] [CrossRef] [PubMed]
- Cremer, D.; Pople, J.A. A General Definition of Ring Puckering Coordinates. J. Am. Chem. Soc. 1975, 97, 1354–1358. [Google Scholar] [CrossRef]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Gordon, J.C.; Myers, J.B.; Folta, T.; Shoja, V.; Heath, L.S.; Onufriev, A. H++: A server for estimating p Ka s and adding missing hydrogens to macromolecules. Nucleic Acids Res. 2005, 33, W368–W371. [Google Scholar] [CrossRef]
- McIntosh, L.P.; Hand, G.; Johnson, P.E.; Joshi, M.D.; Korner, M.; Plesniak, L.A.; Ziser, L.; Wakarchuk, W.W.; Withers, S.G. The pK(a) of the general acid/base carboxyl group of a glycosidase cycles during catalysis: A 13C-NMR study of Bacillus circulans xylanase. Biochemistry 1996, 35, 9958–9966. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Gaussian 09; Gaussian, Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and testing of a general Amber force field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Åqvist, J. Ion-water interaction potentials derived from free energy perturbation simulations. J. Phys. Chem. 1990, 94, 8021–8024. [Google Scholar] [CrossRef]
- Case, D.A.; Darden, T.A.; Cheatham, T.E.; Simmerling, C.L.; Wang, J.; Duke, R.E.; Luo, R.; Walker, R.C.; Zhang, W.; Merz, K.M.; et al. AMBER 16; University of California: San Francisco, CA, USA, 2016. [Google Scholar]
- Ryckaert, J.-P.; Ciccotti, G.; Berendsen, H.J.C. Numerical integration of the cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J. Comput. Phys. 1977, 23, 327–341. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Uberuaga, B.P.; Anghel, M.; Voter, A.F. Synchronization of trajectories in canonical molecular-dynamics simulations: Observation, explanation, and exploitation. J. Chem. Phys. 2004, 120, 6363–6374. [Google Scholar] [CrossRef] [PubMed]
- Berendsen, H.J.C.; Postma, J.P.M.; Van Gunsteren, W.F.; Dinola, A.; Haak, J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984, 81, 3684–3690. [Google Scholar] [CrossRef]
- Roe, D.R.; Cheatham, T.E. PTRAJ and CPPTRAJ: Software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 2013, 9, 3084–3095. [Google Scholar] [CrossRef]
- Miller Iii, B.R.; McGee, T.D.; Swails, J.M.; Homeyer, N.; Gohlke, H.; Roitberg, A.E. MMPBSA.py: An efficient program for end-state free energy calculations. J. Chem. Theory Comput. 2012, 8, 3314–3321. [Google Scholar] [CrossRef] [PubMed]
- Naïm, M.; Bhat, S.; Rankin, K.N.; Dennis, S.; Chowdhury, S.F.; Siddiqi, I.; Drabik, P.; Sulea, T.; Bayly, C.I.; Jakalian, A.; et al. Solvated Interaction Energy (SIE) for scoring protein-ligand binding affinities. 1. Exploring the parameter space. J. Chem. Inf. Model. 2007, 47, 122–133. [Google Scholar] [CrossRef] [PubMed]
- Sulea, T.; Cui, Q.; Purisima, E.O. Solvated interaction energy (SIE) for scoring protein-ligand binding affinities. 2. benchmark in the CSAR-2010 scoring exercise. J. Chem. Inf. Model. 2011, 51, 2066–2081. [Google Scholar] [CrossRef] [PubMed]
- Woods, C.J.; Malaisree, M.; Hannongbua, S.; Mulholland, A.J. A water-swap reaction coordinate for the calculation of absolute protein-ligand binding free energies. J. Chem. Phys. 2011, 134. [Google Scholar] [CrossRef] [PubMed]
- Woods, C.J.; Malaisree, M.; Michel, J.; Long, B.; McIntosh-Smith, S.; Mulholland, A.J. Rapid decomposition and visualisation of protein-ligand binding free energies by residue and by water. Faraday Discuss. 2014, 169, 477–499. [Google Scholar] [CrossRef] [PubMed]
- Woods, C.J.; Malaisree, M.; Long, B.; McIntosh-Smith, S.; Mulholland, A.J. Computational assay of h7n9 influenza neuraminidase reveals r292k mutation reduces drug binding affinity. Sci. Rep. 2013, 3. [Google Scholar] [CrossRef] [PubMed]
- Woods, C.J.; Essex, J.W.; King, M.A. The development of replica-exchange-based free-energy methods. J. Phys. Chem. B 2003, 107, 13703–13710. [Google Scholar] [CrossRef]
- Woods, C.J.; Essex, J.W.; King, M.A. Enhanced configurational sampling in binding free-energy calculations. J. Phys. Chem. B 2003, 107, 13711–13718. [Google Scholar] [CrossRef]
- Khuntawee, W.; Kunaseth, M.; Rungnim, C.; Intagorn, S.; Wolschann, P.; Kungwan, N.; Rungrotmongkol, T.; Hannongbua, S. Comparison of Implicit and Explicit Solvation Models for Iota-Cyclodextrin Conformation Analysis from Replica Exchange Molecular Dynamics. J. Chem. Inf. Model. 2017, 57, 778–786. [Google Scholar] [CrossRef]
- Patriksson, A.; Van Der Spoel, D. A temperature predictor for parallel tempering simulations. Phys. Chem. Chem. Phys. 2008, 10, 2073–2077. [Google Scholar] [CrossRef]
- Heightman, T.D.; Vasella, A.T. Recent insights into inhibition, structure, and mechanism of configuration-retaining glycosidases. Angew. Chem. Int. Ed. 1999, 38, 750–770. [Google Scholar] [CrossRef]
- Gloster, T.M.; Roberts, S.; Perugino, G.; Rossi, M.; Moracci, M.; Panday, N.; Terinek, M.; Vasella, A.; Davies, G.J. Structural, kinetic, and thermodynamic analysis of glucoimidazole-derived glycosidase inhibitors. Biochemistry 2006, 45, 11879–11884. [Google Scholar] [CrossRef]
- Pengthaisong, S.; Ketudat Cairns, J.R. Effects of active site cleft residues on oligosaccharide binding, hydrolysis, and glycosynthase activities of rice BGlu1 and its mutants. Protein Sci. 2014, 23, 1738–1752. [Google Scholar] [CrossRef]
- Kabsch, W.; Sander, C. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 1983, 22, 2577–2637. [Google Scholar] [CrossRef]
- Tidor, B.; Karplus, M. The contribution of vibrational entropy to molecular association: The dimerization of insulin. J. Mol. Biol. 1994, 238, 405–414. [Google Scholar] [CrossRef]
- Panman, W.; Nutho, B.; Chamni, S.; Dokmaisrijan, S.; Kungwan, N.; Rungrotmongkol, T. Computational screening of fatty acid synthase inhibitors against thioesterase domain. J. Biomol. Struct. Dyn. 2018, 36, 4114–4125. [Google Scholar] [CrossRef]
- Kammarabutr, J.; Mahalapbutr, P.; Nutho, B.; Kungwan, N.; Rungrotmongkol, T. Low susceptibility of asunaprevir towards R155K and D168A point mutations in HCV NS3/4A protease: A molecular dynamics simulation. J. Mol. Graph. Model. 2019, 89, 122–130. [Google Scholar] [CrossRef] [PubMed]
- Su, P.C.; Tsai, C.C.; Mehboob, S.; Hevener, K.E.; Johnson, M.E. Comparison of radii sets, entropy, QM methods, and sampling on MM-PBSA, MM-GBSA, and QM/MM-GBSA ligand binding energies of F. tularensis enoyl-ACP reductase (FabI). J. Comput. Chem. 2015, 36, 1859–1873. [Google Scholar] [CrossRef] [PubMed]
- Phanich, J.; Threeracheep, S.; Kungwan, N.; Rungrotmongkol, T.; Hannongbua, S. Glycan binding and specificity of viral influenza neuraminidases by classical molecular dynamics and replica exchange molecular dynamics simulations. J. Biomol. Struct. Dyn. 2019, 37, 3354–3365. [Google Scholar] [CrossRef] [PubMed]
- Phanich, J.; Rungrotmongkol, T.; Kungwan, N.; Hannongbua, S. Role of R292K mutation in influenza H7N9 neuraminidase toward oseltamivir susceptibility: MD and MM/PB(GB)SA study. J. Comput. Aided Mol. Des. 2016, 30, 917–926. [Google Scholar] [CrossRef]
- Phanich, J.; Rungrotmongkol, T.; Sindhikara, D.; Phongphanphanee, S.; Yoshida, N.; Hirata, F.; Kungwan, N.; Hannongbua, S. A 3D-RISM/RISM study of the oseltamivir binding efficiency with the wild-type and resistance-associated mutant forms of the viral influenza B neuraminidase. Protein Sci. 2016, 25, 147–158. [Google Scholar] [CrossRef] [PubMed]
- Hanpaibool, C.; Leelawiwat, M.; Takahashi, K.; Rungrotmongkol, T. Source of oseltamivir resistance due to single E119D and double E119D/H274Y mutations in pdm09H1N1 influenza neuraminidase. J. Comput. Aided Mol. Des. 2019. [Google Scholar] [CrossRef] [PubMed]
- Kaper, T.; Van Heusden, H.H.; Van Loo, B.; Vasella, A.; Van der Oost, J.; De Vos, W.M. Substrate specificity engineering of β-mannosidase and β-glucosidase from Pyrococcus by exchange of unique active site residues. Biochemistry 2002, 41, 4147–4155. [Google Scholar] [CrossRef]
- Roberge, M.; Dupont, C.; Morosoli, R.; Shareck, F.; Kluepfel, D. Asparagine-127 of xylanase A from Streptomyces lividans, a key residue in glycosyl hydrolases of superfamily 4/7: Kinetic evidence for its involvement in stabilization of the catalytic intermediate. Protein Eng. 1997, 10, 399–403. [Google Scholar] [CrossRef] [PubMed]
- Wagner, J.R.; Sørensen, J.; Hensley, N.; Wong, C.; Zhu, C.; Perison, T.; Amaro, R.E. POVME 3.0: Software for Mapping Binding Pocket Flexibility. J. Chem. Theory Comput. 2017, 13, 4584–4592. [Google Scholar] [CrossRef]
- Vasella, A.; Davies, G.J.; Böhm, M. Glycosidase mechanisms. Curr. Opin. Chem. Biol. 2002, 6, 619–629. [Google Scholar] [CrossRef]
- Gloster, T.M.; Vocadlo, D.J. Developing inhibitors of glycan processing enzymes as tools for enabling glycobiology. Nat. Chem. Biol. 2012, 8, 683–694. [Google Scholar] [CrossRef]
- Burmeister, W.P.; Cottaz, S.; Rollin, P.; Vasella, A.; Henrissat, B. High resolution x-ray crystallography shows that ascorbate is a cofactor for myrosinase and substitutes for the function of the catalytic base. J. Biol. Chem. 2000, 275, 39385–39393. [Google Scholar] [CrossRef] [PubMed]
- Tailford, L.E.; Offen, W.A.; Smith, N.L.; Dumon, C.; Morland, C.; Gratien, J.; Heck, M.P.; Stick, R.V.; Blériot, Y.; Vasella, A.; et al. Structural and biochemical evidence for a boat-like transition state in β-mannosidases. Nat. Chem. Biol. 2008, 4, 306–312. [Google Scholar] [CrossRef] [PubMed]
- Males, A.; Speciale, G.; Williams, S.J.; Davies, G.J. Distortion of mannoimidazole supports a: B 2,5 boat transition state for the family GH125 α-1,6-mannosidase from Clostridium perfringens. Org. Biomol. Chem. 2019, 17, 7863–7869. [Google Scholar] [CrossRef] [PubMed]
- Kuntothom, T.; Raab, M.; Tvaroška, I.; Fort, S.; Pengthaisong, S.; Cañada, J.; Calle, L.; Jiménez-Barbero, J.; Ketudat Cairns, J.R.; Hrmova, M. Binding of β-d-glucosides and β-d-mannosides by rice and barley β-d-glycosidases with distinct substrate specificities. Biochemistry 2010, 49, 8779–8793. [Google Scholar] [CrossRef] [PubMed]




| Energy Component | GIm Complex | MIm Complex |
|---|---|---|
| Gas term | ||
| ΔEvdW | −15.63 ± 0.06 | −19.65 ± 0.05 |
| ΔEele | −89.43 ± 0.09 | −34.05 ± 0.15 |
| ΔEMM | −105.06 ± 0.07 | −53.70 ± 0.15 |
| ΔEQM | −79.02 ± 0.08 | −30.78 ± 0.13 |
| −TΔS | 11.30 ± 0.40 | 13.79 ± 0.32 |
| Solvation term | ||
| PBSA | ||
| 86.43 ± 0.06 | 53.35 ± 0.14 | |
| −2.29 ± 0.00 | −2.60 ± 0.00 | |
| ΔGsol(PBSA) | 84.14 ± 0.06 | 50.75 ± 0.14 |
| GBSA | ||
| 77.38 ± 0.06 | 42.05 ± 0.12 | |
| −3.80 ± 0.00 | −3.56 ± 0.00 | |
| ΔGsol(GBSA) | 73.58 ± 0.06 | 38.48 ± 0.12 |
| QM-GBSA | ||
| 65.40 ± 0.05 | 34.35 ± 0.10 | |
| −3.80 ± 0.00 | −3.56 ± 0.00 | |
| ΔGsol(QM-GBSA) | 61.60 ± 0.05 | 30.79 ± 0.10 |
| Binding free energy | ||
| ΔGbind(MM/PBSA) | −9.62 | 10.84 |
| ΔGbind(MM/GBSA) | −20.18 | −1.43 |
| ΔGbind(QM/MM-GBSA) | −21.75 | −5.85 |
| ΔGbind(inhibition) 1 [29] | −13.00 | −7.90 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nutho, B.; Pengthaisong, S.; Tankrathok, A.; Lee, V.S.; Ketudat Cairns, J.R.; Rungrotmongkol, T.; Hannongbua, S. Structural Basis of Specific Glucoimidazole and Mannoimidazole Binding by Os3BGlu7. Biomolecules 2020, 10, 907. https://doi.org/10.3390/biom10060907
Nutho B, Pengthaisong S, Tankrathok A, Lee VS, Ketudat Cairns JR, Rungrotmongkol T, Hannongbua S. Structural Basis of Specific Glucoimidazole and Mannoimidazole Binding by Os3BGlu7. Biomolecules. 2020; 10(6):907. https://doi.org/10.3390/biom10060907
Chicago/Turabian StyleNutho, Bodee, Salila Pengthaisong, Anupong Tankrathok, Vannajan Sanghiran Lee, James R. Ketudat Cairns, Thanyada Rungrotmongkol, and Supot Hannongbua. 2020. "Structural Basis of Specific Glucoimidazole and Mannoimidazole Binding by Os3BGlu7" Biomolecules 10, no. 6: 907. https://doi.org/10.3390/biom10060907
APA StyleNutho, B., Pengthaisong, S., Tankrathok, A., Lee, V. S., Ketudat Cairns, J. R., Rungrotmongkol, T., & Hannongbua, S. (2020). Structural Basis of Specific Glucoimidazole and Mannoimidazole Binding by Os3BGlu7. Biomolecules, 10(6), 907. https://doi.org/10.3390/biom10060907







