The Structure of Blood Coagulation Factor XIII Is Adapted to Oxidation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation of Human FXIII and Fibrinogen
2.2. Sample Preparation
2.3. Covalent Linking of Fibrin Chains
2.4. Exposure of FXIII at the Different Stages of Its Activation to Hypochlorite
2.5. Enzymatic Digestion and HPLC-MS/MS Analysis
2.6. Data Processing
3. Results
3.1. Effect of Oxidation on the Enzymatic Activity of FXIII
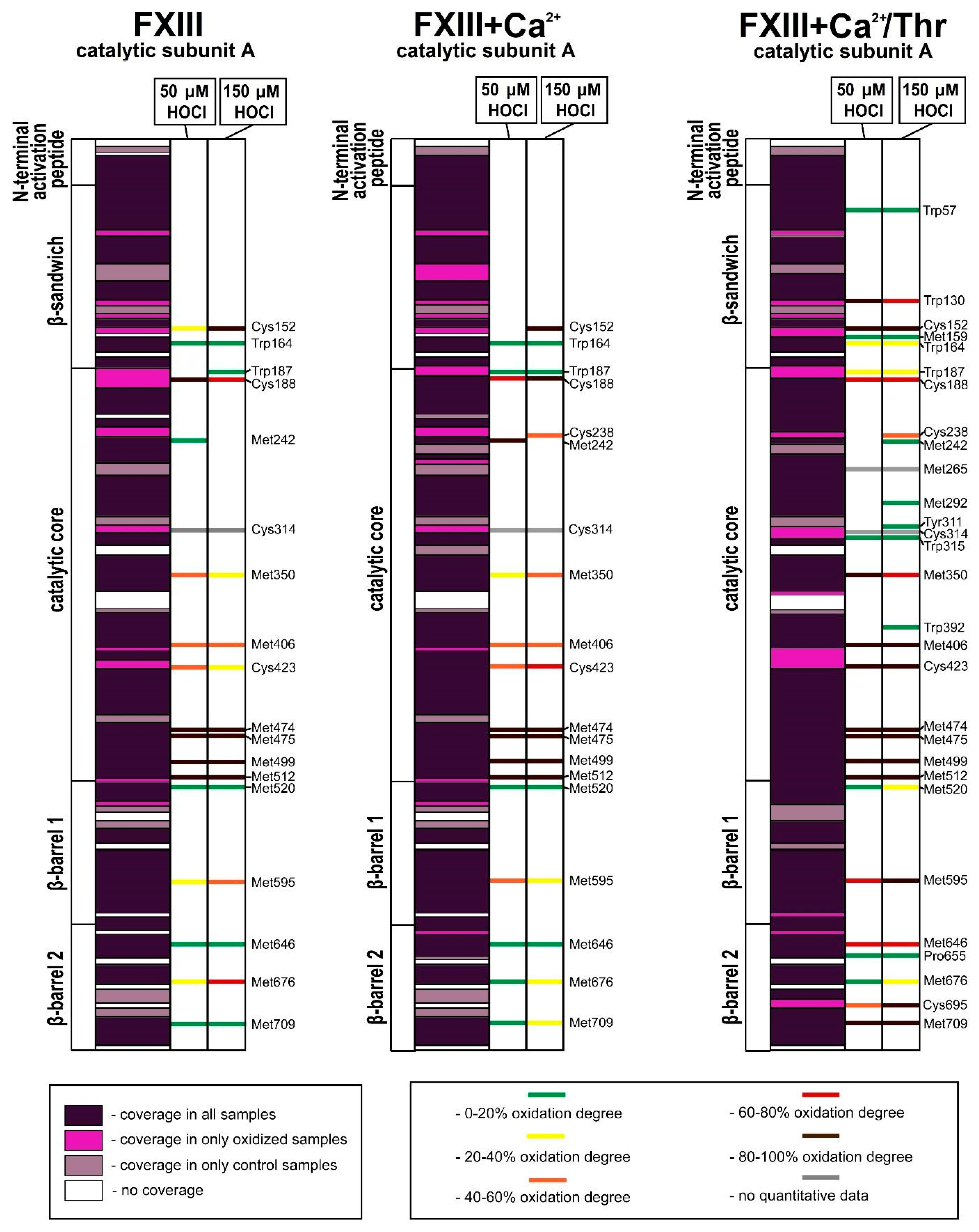
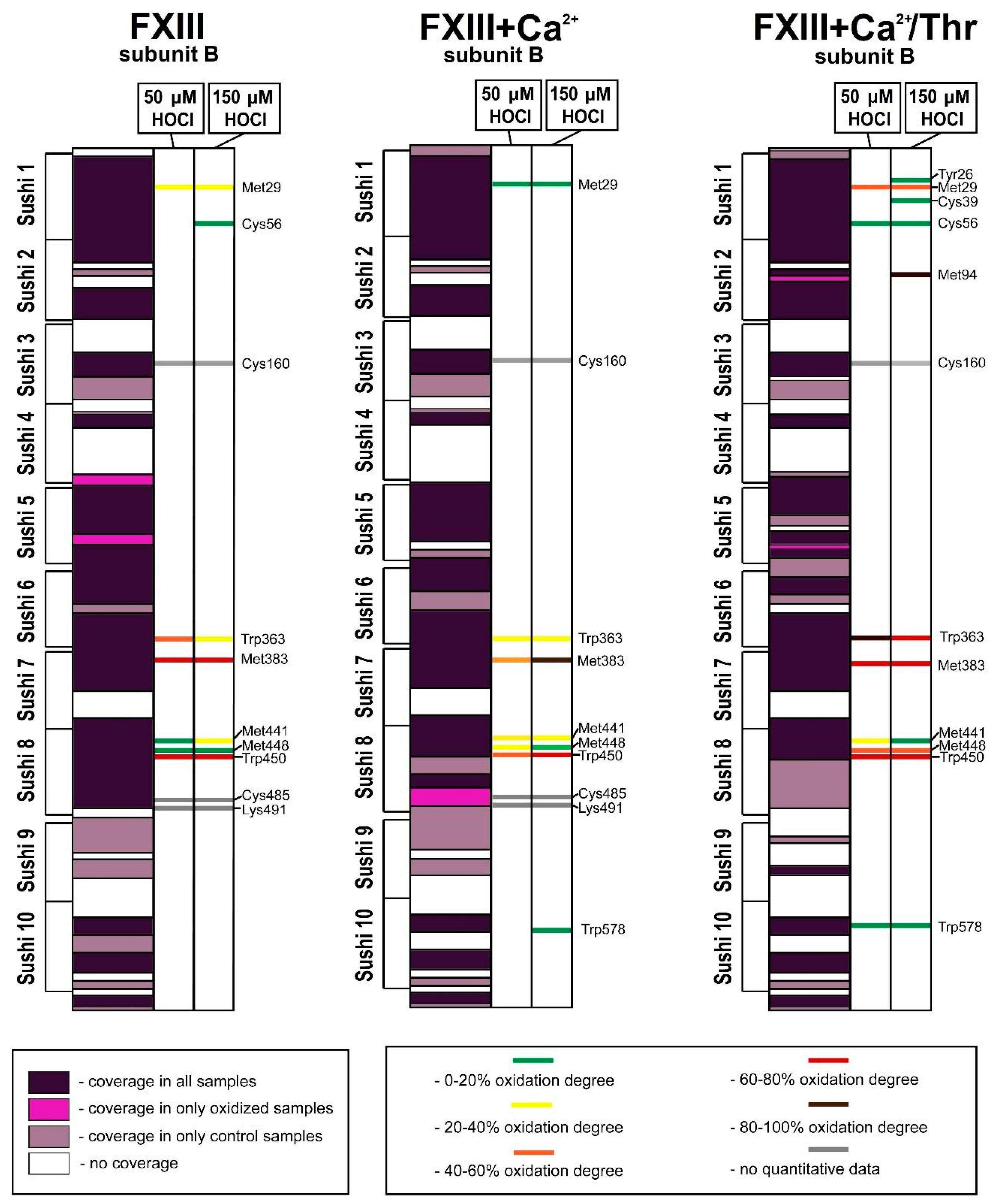
3.2. Analysis of Native and Oxidized FXIII Using High-Resolution Tandem Mass-Spectrometry (LC–MS/MS)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Muszbek, Z.; Bereczky, Z.; Bagoly, Z.; Komaromi, I.; Katona, E. Factor XIII: A coagulation factor with multiple plasmatic and cellular functions. Physiol. Rev. 2011, 91, 931–972. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Souri, M.; Kaetsu, H.; Ichinose, A. Sushi domains in the subunit-B of factor XIII responsible for oligomer assembly. Biomolecules 2008, 47, 8656–8664. [Google Scholar] [CrossRef] [PubMed]
- Ichinose, A.; McMullen, B.A.; Fujikawa, K.; Davie, E.W. Amino acid sequence of the B subunit of human coagulation factor XIII, a protein composed of ten repetitive segments. Biomolecules 1986, 25, 4633–4638. [Google Scholar] [CrossRef]
- Schroeder, V.; Kohler, H.P. Factor XIII: Structure and function. In Seminars in Thrombosis and Hemostasis; Thieme Medical Publishers: New York, NY, USA, 2016. [Google Scholar] [CrossRef]
- Bagoly, Z.; Muszbek, L. Factor XIII: What does it look like? J. Thromb. Haemost. 2019, 17, 714–716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anokhin, B.A.; Stribinskis, V.; Dean, W.L.; Maurer, M.C. Activation of factor XIII is accompained by a change in oligomerization state. FEBS J. 2017, 284, 3849–3861. [Google Scholar] [CrossRef] [PubMed]
- Protopopova, A.D.; Ramirez, A.; Klinov, D.V.; Litvinov, R.I.; Weisel, J.W. Factor XIII topology: Organization of B subunits and changes with activation studied with single-molecule atomic force microscopy. J. Thromb. Haemost. 2019, 17, 737–748. [Google Scholar] [CrossRef]
- Singh, S.; Nazabal, A.; Kaniyappan, S.; Pellequer, J.L.; Wolberg, A.S.; Imhof, D.; Oldenburg, J.; Biswas, A. The plasma factor XIII heterotetrameric complex structure: Unexpected unequal pairing within a symmetric complex. Biomolecules 2019, 9, 765. [Google Scholar] [CrossRef] [Green Version]
- Stadtman, E.R. Protein oxidation in aging and age-related diseases. Ann. N. Y. Acad. Sci. 2001, 928, 22–38. [Google Scholar] [CrossRef]
- Rosenfeld, M.A.; Bychkova, A.V.; Shchegolikhin, A.N.; Leonova, V.B.; Biryukova, M.I.; Kostanova, E.A. Ozone-induced oxidative modification of plasma fibrin-stabilizing factor. BBA Proteins Proteom. 2013, 1834, 2470–2479. [Google Scholar] [CrossRef]
- Vasilyeva, A.D.; Bychkova, A.V.; Bugrova, A.E.; Indeykina, M.I.; Chikunova, A.P.; Leonova, V.B.; Kostanova, E.A.; Biryukova, M.; Konstantinova, M.L.; Kononikhin, A.; et al. Modification of the catalytic subunit of plasma fibrin-stabilizing factor under induced oxidation. In Doklady Biochem. Biophys.; Springer: Berlin/Heidelberg, Germany, 2017. [Google Scholar] [CrossRef]
- Vasilyeva, A.; Yurina, L.; Indeykina, M.; Bychkova, A.; Bugrova, A.; Biryukova, M.; Kononikhin, A.; Nikolaev, E.; Rosenfeld, M. Oxidation-induced modifications of the catalytic subunits of plasma fibrin-stabilizing factor at the different stages of its activation identified by mass spectrometry. BBA Proteins Proteom. 2018, 1866, 875–884. [Google Scholar] [CrossRef]
- Loria, V.; Dato, I.; Graziani, F.; Biasucci, L.M. Myeloperoxidase: A new biomarker of inflammation in ischemic heart disease and acute coronary syndromes. Mediat. Inflamm. 2008, 2008, 135625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klebanoff, S.J. Myeloperoxidase: Friend and foe. J. Leukoc. Biol. 2005. 77, 598–625. [CrossRef]
- Clark, R.A.; Stone, P.J.; El Hag, A.; Calore, J.D.; Franzblau, C. Myeloperoxidase-catalyzed inactivation of alpha 1-protease inhibitor by human neutrophils. J. Leukoc. Biol. 1981, 256, 3348–3353. [Google Scholar]
- Summers, F.A.; Morgan, P.E.; Davies, M.J.; Hawkins, C.L. Identification of plasma proteins that are susceptible to thiol oxidation by hypochlorous acid and N-chloramines. Chem. Res. Toxicol. 2008, 21, 1832–1840. [Google Scholar] [CrossRef] [PubMed]
- Gugliucci, A. Hypochlorous acid is a potent inactivator of human plasminogen at concentrations secreted by activated granulocytes. Clin. Chem. Lab. Med. 2008, 46, 1403–1409. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, T.; Zia, M.K.; Ali, S.S.; Ahsan, H.; Khan, F.H. Insight into the interactions of proteinase inhibitor- alpha-2-macroglobulin with hypochlorite. Int. J. Biol. Macromol. 2018, 117, 401–406. [Google Scholar] [CrossRef] [PubMed]
- Vlasova, I.I.; Sokolov, A.V.; Kostevich, V.A.; Mikhalchik, E.V.; Vasilyev, V.B. Myeloperoxidase-induced oxidation of albumin and ceruloplasmin: Role of tyrosines. Biochemistry 2019, 84, 652–662. [Google Scholar] [CrossRef]
- Weigandt, K.M.; White, N.; Chung, D.; Ellingson, E.; Wang, Y.; Fu, X.; Pozzo, D.C. Fibrin clot structure and mechanics associated with specific oxidation of methionine residues in fibrinogen. Biophys. J. 2012, 103, 2399–2407. [Google Scholar] [CrossRef] [Green Version]
- Lorand, L.; Credo, R.B.; Janus, T.J. Factor XIII (fibrin-stabilizing factor). In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 1981. [Google Scholar]
- Loewy, A.G.; Dunathan, K.; Kriel, R.; Wolfinger, H.L. Fibrinase: I. Purification of substrate and enzyme. J. Biol. Chem. 1961, 236, 2625–2633. [Google Scholar]
- Blombäck, B.B.; Blombäck, M. Purification of human and bovine fibrinogen. Arch. Chem. 1956, 10, 415–443. [Google Scholar] [CrossRef] [Green Version]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Rosenfeld, M.A.; Vasileva, M.V. Mechanism of aggregation of fibrinogen molecules: The influence of fibrin-stabilizing factor. Biomed. Sci. 1991, 2, 155–161. [Google Scholar]
- Svendsen, L.; Blombäck, B.; Blombäck, M.; Olsson, P.I. Synthetic chromogenic substrates to determine of trypsin, thrombin and thrombin-like enzymes. Thromb. Res. 1972, 1, 267–278. [Google Scholar] [CrossRef]
- Colombo, G.; Cleric, M.; Giustarini, D.; Portinaro, N.; Badalamenti, S.; Rossi, R.; Milzani, A. Dalle-Donne I A central role for intermolecular dityrosine cross-linking of fibrinogen in high molecular weight advanced oxidation protein product (AOPP) formation. BBA Gen. Subj. 2015, 1850, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Morris, J.C. The acid ionisation constant of HOCl from 5°C to 35°C. J. Phys. Chem. 1966, 70, 3198–3805. [Google Scholar] [CrossRef]
- Ishihama, Y.; Rappsilber, J.; Andersen, J.S.; Mann, M. Microcolumns with self-assembled particle frits for proteomics. J. Chromatogr. A 2002, 979, 233–239. [Google Scholar] [CrossRef]
- Yurina, L.; Vasilyeva, A.; Indeykina, M.; Bugrova, A.; Biryukova, M.; Kononikhin, A.; Nikolaev, E.; Rosenfeld, M. Ozone-induced damage of fibrinogen molecules: Identification of oxidation sites by high-resolution mass spectrometry. Free Radic. Res. 2019, 53, 430–455. [Google Scholar] [CrossRef]
- Han, X.; He, L.; Xin, L.; Shan, B.; Ma, B. PeaksPTM: Mass spectrometry-based identification of peptides with unspecified modifications. J. Proteome Res. 2011, 10, 2930–2936. [Google Scholar] [CrossRef]
- Verrastro, I.; Pasha, S.; Jensen, K.T.; Pitt, A.R.; Spickett, C.M. Mass spectrometry-based methods for identifying oxidized proteins in disease: Advances and challenges. Biomolecules 2015, 5, 378–411. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Xin, L.; Shan, B.; Chen, W.; Xie, M.; Yuen, D.; Zhang, W.; Zhang, Z.; Lajoie, G.A.; Ma, B. PEAKS DB: De Novo Sequencing Assisted Database Search for Sensitive and Accurate Peptide Identification. Mol. Cell. Proteom. 2012, 11, M111.010587. [Google Scholar] [CrossRef] [Green Version]
- Chen, R.; Doolittle, R.F. Cross-linking sites in human and bovine fibrin. Biochemistry 1971, 10, 4487–4491. [Google Scholar] [CrossRef]
- Sobel, J.H.; Gawinowicz, M.A. Identification of the α chain lysine donor sites involved in factor XIIIa fibrin cross-linking. J. Biol. Chem. 1996, 271, 19288–19297. [Google Scholar] [CrossRef] [Green Version]
- Drozdz, R.; Naskalski, J.W.; Sznajd, J. Oxidation of amino acids and peptides in with myeloperoxidase, chloride and hydrogen peroxide. BBA Protein Struct. Mol. Enzymol. 1988, 957, 47–52. [Google Scholar] [CrossRef]
- Hawkins, C.L.; Pattison, D.I.; Davies, M.J. Hypochlorite-induced oxidation of amino acids, peptides and proteins. Amino Acids 2003, 25, 259–274. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.; Kim, G.; Levine, R. Methionine in Proteins:It’s not just for protein initiation anymore. Neurochem. Res. 2019, 44, 247–257. [Google Scholar] [CrossRef] [PubMed]
- Luo, S.; Levine, R.L. Methionine in proteins defends against oxidative stress. FASEB J. 2008, 23, 464–472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reddy, V.Y.; Desrochers, P.E.; Pizzo, S.V.; Gonias, S.L.; Sahakian, J.A.; Levine, R.L.; Weiss, S.J. Oxidative dissociation of human alpha 2-macroglobulin tetramers into dysfunctional dimers. J. Biol. Chem. 1994, 269, 4683–4691. [Google Scholar]
- Van Patten, S.M.; Hanson, E.; Bernasconi, R.; Zhang, K.; Manavalan, P.; Cole, E.S.; McPherson, J.M.; Edmunds, T. Oxidation of methionine residues in antithrombin. Effects on biological activity and heparin binding. J. Biol. Chem. 1999, 274, 10268–10276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kornfelt, T.; Persson, E.; Palm, L. Oxidation of methionine residues in coagulation factor VIIa. Arch. Biochem. Biophys. 1999, 363, 43–54. [Google Scholar] [CrossRef] [PubMed]
- Shah, D.D.; Zhang, J.; Maity, H.; Mallela, K.M.G. Effect of photo-degradation on the structure, stability, aggregation, and function of an IgG1 monoclonal antibody. Int. J. Pharm. 2018, 547, 438–449. [Google Scholar] [CrossRef]
- Marino, S.M.; Li, Y.; Fomenko, D.E.; Agisheva, N.; Cerny, R.L.; Gladyshev, V.N. Characterization of surface-exposed reactive cysteine residues in Saccharomyces cerevisiae. Biochemistry 2010, 49, 7709–7721. [Google Scholar] [CrossRef] [Green Version]
- Winterbourn, C.C. Comparative reactivities of various biological compounds with myeloperoxidase-hydrogen peroxide-chloride, and similarity of the oxidant to hypochlorite. BBA-Gen. Subj. 1985, 840, 204–210. [Google Scholar] [CrossRef]
- Saito, M.; Asakura, H.; Yoshida, T.; Ito, K.; Okafuji, K.; Matsuda, T. A familial factor XIII subunit B deficiency. Br. J. Haematol. 1990, 74, 290–294. [Google Scholar] [CrossRef] [PubMed]
- Rosenfeld, M.A.; Shchegolikhin, A.N.; Leonova, V.B.; Kostanova, E.A.; Biryukova, M.I.; Bychkova, A.V.; Konstantinova, M.L.; Vasilyeva, A.D. The oxidative modification of cellular fibrin-stabilizing factor. In Doklady Biochemistry and Biophysics; Pleiades Publishing, Inc.: New York, USA, 2016. [Google Scholar] [CrossRef]
- Misztal, T.; Golaszewska, A.; Tomasiak-Lozowska, M.M.; Iwanicka, M.; Marcinczyk, N.; Leszczynska, A.; Chabielska, E.; Rusak, T. The myeloperoxidase product, hypochlorous acid, reduces thrombus formation under flow and attenuates clot retraction and fibrinolysis in human blood. Free Radic. Biol. Med. 2019, 141, 426–437. [Google Scholar] [CrossRef] [PubMed]
- Yorifuji, H.; Anderson, K.; Lynch, G.W.; van de Water, L.; McDonagh, J. B protein of factor XIII: Differentiation between free B and complexed B. Blood 1988, 72, 1645–1650. [Google Scholar] [CrossRef] [Green Version]
- Stief, T.W.; Kurz, J.; Doss, M.O.; Fareed, J. Singlet oxygen inactivates fibrinogen, factor V, factor VIII, factor X, and platelet aggregation of human blood. Thromb. Res. 2000, 97, 473–480. [Google Scholar] [CrossRef]
- Souri, M.; Osaki, T.; Ichinose, A. The non-catalytic B subunit of coagulation actor XIII accelerates fibrin cross-linking. J. Biol. Chem. 2015, 290, 12027–12039. [Google Scholar] [CrossRef] [Green Version]
- Byrnes, J.R.; Wilson, C.; Boutelle, A.M.; Brandner, C.B.; Flick, M.J.; Philippou, H.; Wolberg, A.S. The interaction between fibrinogen and zymogen FXIII-A2B2 is mediated by fibrinogen residues γ390-396 and the FXIII-B subunits. Blood 2016, 128, 1969–1978. [Google Scholar] [CrossRef] [Green Version]
- Lishko, V.K.; Podolnikova, N.P.; Yakubenko, V.P.; Yakovlev, S.; Medved, L.; Yadav, S.P.; Ugarova, T.P. Multiple binding sites in fibrinogen for integrin alphaMbeta2 (Mac-1). J. Biol. Chem. 2004, 279, 44897–44906. [Google Scholar] [CrossRef] [Green Version]
- Rijken, D.C.; Uitte de Willige, S. Inhibition of fibrinolysis by coagulation Factor XIII. BioMed Res. Int. 2017, 2017, 1209676. [Google Scholar] [CrossRef]
- Martinez, M.; Weisel, J.W.; Ischiropoulos, H. Functional impact of oxidative posttranslational modifications on fibrinogen and fibrin clots. Free Radic. Biol. Med. 2013, 65, 411–418. [Google Scholar] [CrossRef] [PubMed] [Green Version]





| Δm (Monoisotopic) | Modification | Composition | oxFXIII | oxFXIII + Ca2+ | oxFXIII + Ca2+/Thr | |||
|---|---|---|---|---|---|---|---|---|
| 50 μM HOCl/−OCl | 150 μM HOCl/–OCI | 50 μM HOCl/–OCl | 150 μM HOCl/–OCl | 50 μM HOCl/–OCl | 150 μM HOCl/–OCl | |||
| +15.994915 | Oxidation | O | 19 | 20 | 19 | 19 | 26 | 28 |
| +31.989828 | Dioxidation | O (2) | 1 | 1 | 1 | |||
| +47.984744 | Trioxidation | O (3) | 5 | 5 | 3 | 5 | 5 | 7 |
| +27.994915 | Formylation | C O | - | - | - | - | 1 | 1 |
| −48.003371 | Prompt loss of side chain from oxidized Methionine | H (−4) C (−1) S (−1) | 1 | 1 | 2 | 2 | 2 | 3 |
| +3.994915 | Tryptophan oxidation to kynurenin | C(−1) O | - | - | – | 2 | 1 | 2 |
| +13.979265 | Tryptophan oxidation to oxolactone | H(−2) O | 2 | 2 | 2 | 3 | 4 | 4 |
| +19.989829 | Tryptophan oxidation to hydroxykynurenin | C(−1) O(2) | 1 | - | 1 | - | - | - |
| +33.961028 | Chlorination of tyrosine residues | H(−1) Cl | - | - | - | - | - | 2 |
| +14.9632 | Lysine oxidation to a-aminoadipic acid | H(−3) N(−1) O(2) | 1 | 1 | 1 | 1 | - | - |
| −33.987721 | Dehydroalanine (from Cysteine) | H(−2) S(−1) | 2 | 2 | 2 | 3 | 1 | 3 |
| −27.994915 | Pyrrolidone from Proline | C(−1) O(−1) | - | - | - | - | 1 | 1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vasilyeva, A.; Yurina, L.; Shchegolikhin, A.; Indeykina, M.; Bugrova, A.; Kononikhin, A.; Nikolaev, E.; Rosenfeld, M. The Structure of Blood Coagulation Factor XIII Is Adapted to Oxidation. Biomolecules 2020, 10, 914. https://doi.org/10.3390/biom10060914
Vasilyeva A, Yurina L, Shchegolikhin A, Indeykina M, Bugrova A, Kononikhin A, Nikolaev E, Rosenfeld M. The Structure of Blood Coagulation Factor XIII Is Adapted to Oxidation. Biomolecules. 2020; 10(6):914. https://doi.org/10.3390/biom10060914
Chicago/Turabian StyleVasilyeva, Alexandra, Lyubov Yurina, Alexander Shchegolikhin, Maria Indeykina, Anna Bugrova, Alexey Kononikhin, Eugene Nikolaev, and Mark Rosenfeld. 2020. "The Structure of Blood Coagulation Factor XIII Is Adapted to Oxidation" Biomolecules 10, no. 6: 914. https://doi.org/10.3390/biom10060914






