Transcriptome Analysis Reveals Potential Roles of Abscisic Acid and Polyphenols in Adaptation of Onobrychis viciifolia to Extreme Environmental Conditions in the Qinghai-Tibetan Plateau
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Total RNA Extraction, and Construction and Sequencing of mRNA Libraries
2.3. Transcriptome Sequencing Data Processing and Analysis
2.4. Identification and Functional Annotation of DEGs
2.5. Phylogenetic Analysis
2.6. RT-qPCR
2.7. Determination of the Contents of Total Flavonoids, Total Anthocyanins and Other Flavonoid Compounds
3. Results
3.1. Transcriptome Profiling of O. viciifolia Grown under Two Different Natural Environmental Conditions
3.2. Annotation of Assembled Unigenes
3.3. DEGs from O. viciifolia Plants Grown at Different Altitudes
3.4. Functional Enrichment Analysis of DEGs
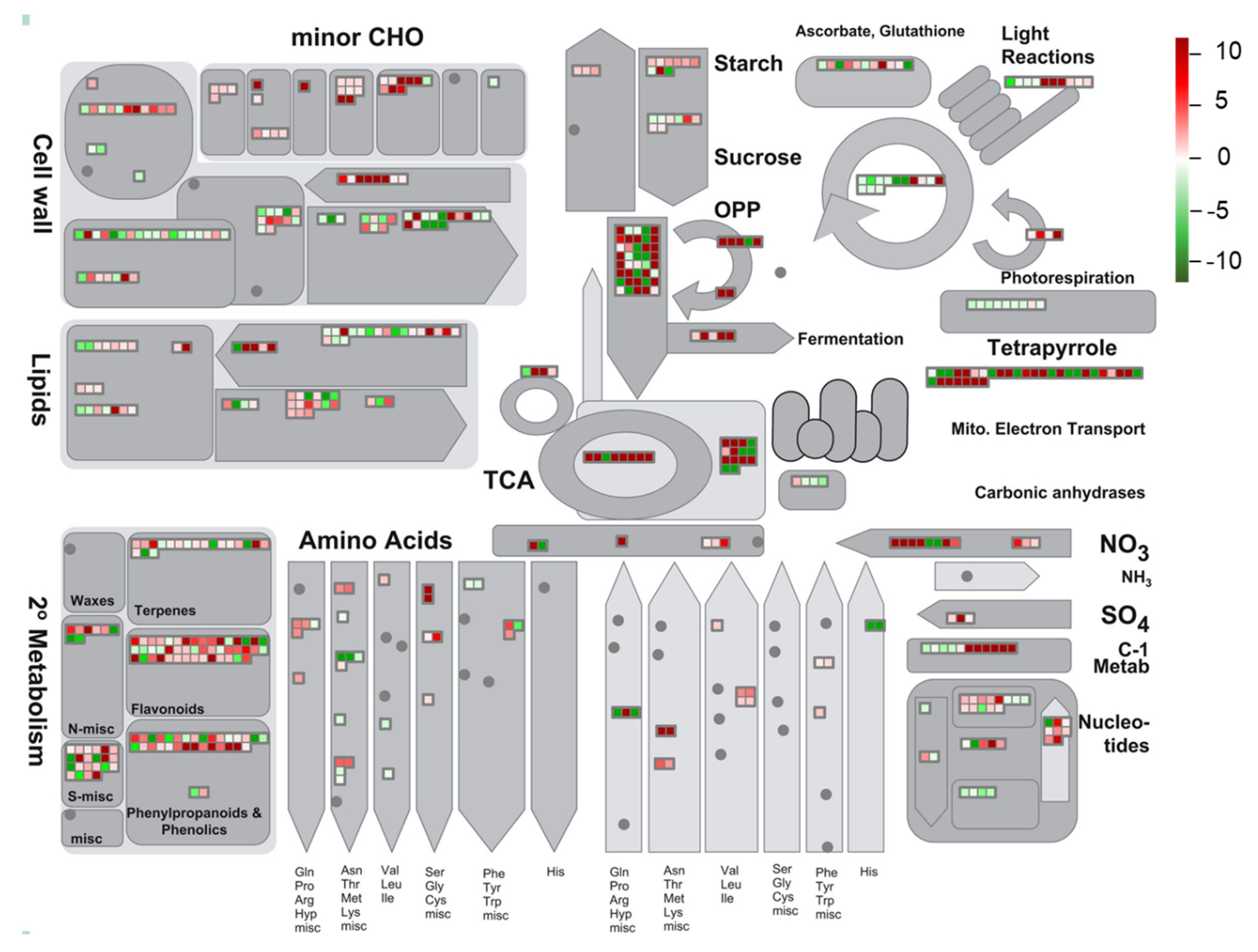
3.5. Global Environmental Stress Responses Revealed by MapMan Analysis
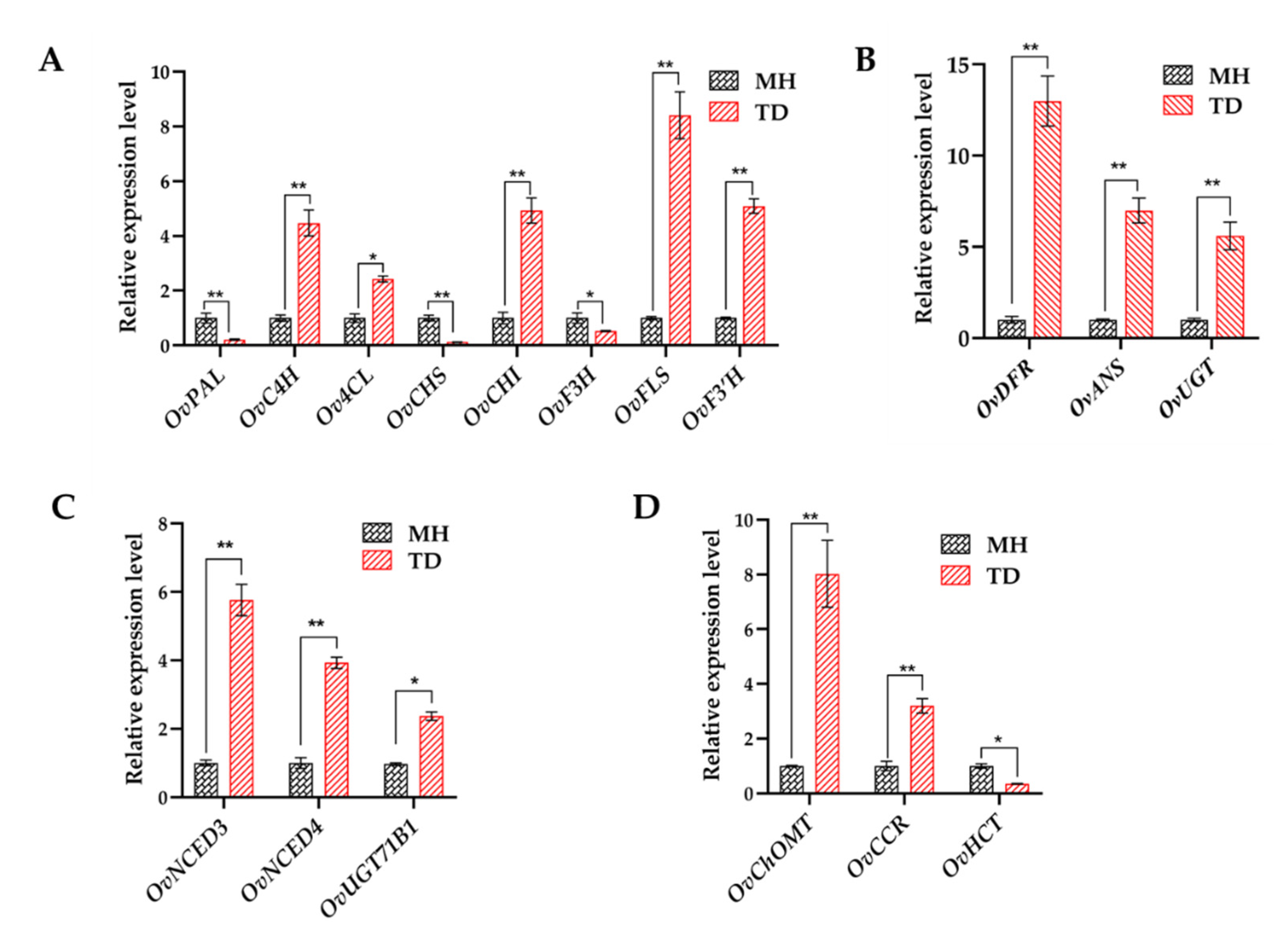
3.6. Responses of DEGs Involved in Phytohormone Pathways
3.7. Responses of DEGs Encoding TFs
3.8. Quantitative Analyses of Flavonoids and Anthocyanins
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Abdelrahman, M.; Burritt, D.J.; Gupta, A.; Tsujimoto, H.; Tran, L.P. Heat stress effects on source-sink relationships and metabolome dynamics in wheat. J. Exp. Bot. 2020, 71, 543–554. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, N.J.; Urwin, P.E. The interaction of plant biotic and abiotic stresses: From genes to the field. J. Exp. Bot. 2012, 63, 3523–3543. [Google Scholar] [CrossRef] [PubMed]
- Cramer, G.R.; Urano, K.; Delrot, S.; Pezzotti, M.; Shinozaki, K. Effects of abiotic stress on plants: A systems biology perspective. BMC Plant Biol. 2011, 11, 163. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Sinha, R.; Fernandes, J.L.; Abdelrahman, M.; Burritt, D.J.; Tran, L.P. Phytohormones regulate convergent and divergent responses between individual and combined drought and pathogen infection. Crit. Rev. Biotechnol. 2020, 40, 320–340. [Google Scholar] [CrossRef]
- Mohanta, T.K.; Bashir, T.; Hashem, A.; Abd Allah, E.F. Systems biology approach in plant abiotic stresses. Plant Physiol. Biochem. 2017, 121, 58–73. [Google Scholar] [CrossRef]
- Bray, E.A. Response to abiotic stress. Biochem. Mol. Biol. Plants 2000, 1158–1203. [Google Scholar]
- Atkinson, N.J.; Lilley, C.J.; Urwin, P.E. Identification of genes involved in the response of Arabidopsis to simultaneous biotic and abiotic stresses. Plant Physiol. 2013, 162, 2028–2041. [Google Scholar] [CrossRef]
- Iyer, N.J.; Tang, Y.; Mahalingam, R. Physiological, biochemical and molecular responses to a combination of drought and ozone in Medicago truncatula. Plant Cell. Environ. 2013, 36, 706–720. [Google Scholar] [CrossRef]
- Koussevitzky, S.; Suzuki, N.; Huntington, S.; Armijo, L.; Sha, W.; Cortes, D.; Shulaev, V.; Mittler, R. Ascorbate peroxidase 1 plays a key role in the response of Arabidopsis thaliana to stress combination. J. Biol. Chem. 2008, 283, 34197–34203. [Google Scholar] [CrossRef]
- Rasmussen, S.; Barah, P.; Suarez-Rodriguez, M.C.; Bressendorff, S.; Friis, P.; Costantino, P.; Bones, A.M.; Nielsen, H.B.; Mundy, J. Transcriptome responses to combinations of stresses in Arabidopsis. Plant Physiol. 2013, 161, 1783–1794. [Google Scholar] [CrossRef]
- Rizhsky, L.; Liang, H.; Shuman, J.; Shulaev, V.; Davletova, S.; Mittler, R. When defense pathways collide. The response of Arabidopsis to a combination of drought and heat stress. Plant Physiol. 2004, 134, 1683–1696. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, N.; Rivero, R.M.; Shulaev, V.; Blumwald, E.; Mittler, R. Abiotic and biotic stress combinations. New Phytologist 2014, 203, 32–43. [Google Scholar] [CrossRef] [PubMed]
- Hewezi, T.; Leger, M.; Gentzbittel, L. A comprehensive analysis of the combined effects of high light and high temperature stresses on gene expression in sunflower. Ann. Bot. 2008, 102, 127–140. [Google Scholar] [CrossRef] [PubMed]
- Imadi, S.R.; Kazi, A.G.; Ahanger, M.A.; Gucel, S.; Ahmad, P. Plant transcriptomics and responses to environmental stress: An overview. J. Genet. 2015, 94, 525–537. [Google Scholar] [CrossRef]
- Nejat, N.; Ramalingam, A.; Mantri, N. Advances in Transcriptomics of Plants. Adv. Biochem. Eng. Biotechnol. 2018, 164, 161–185. [Google Scholar]
- Banerjee, A.; Roychoudhury, A. Abscisic-acid-dependent basic leucine zipper (bZIP) transcription factors in plant abiotic stress. Protoplasma 2017, 254, 3–16. [Google Scholar] [CrossRef]
- Jiang, J.; Ma, S.; Ye, N.; Jiang, M.; Cao, J.; Zhang, J. WRKY transcription factors in plant responses to stresses. J. Integr. Plant Biol. 2017, 59, 86–101. [Google Scholar] [CrossRef]
- Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AP2/ERF family transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 2012, 1819, 86–96. [Google Scholar] [CrossRef]
- Muller, M.; Munne-Bosch, S. Ethylene Response Factors: A Key Regulatory Hub in Hormone and Stress Signaling. Plant Physiol. 2015, 169, 32–41. [Google Scholar] [CrossRef]
- Zhu, J.K. Abiotic Stress Signaling and Responses in Plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef]
- Verma, V.; Ravindran, P.; Kumar, P.P. Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 2016, 16, 86. [Google Scholar] [CrossRef] [PubMed]
- Kollist, H.; Zandalinas, S.I.; Sengupta, S.; Nuhkat, M.; Kangasjarvi, J.; Mittler, R. Rapid Responses to Abiotic Stress: Priming the Landscape for the Signal Transduction Network. Trends Plant Sci. 2019, 24, 25–37. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Huang, F.; Wu, N.; Li, X.; Hu, H.; Xiong, L. Integrative Regulation of Drought Escape through ABA-Dependent and -Independent Pathways in Rice. Mol. Plant 2018, 11, 584–597. [Google Scholar] [CrossRef] [PubMed]
- Shinozaki, K.; Yamaguchi-Shinozaki, K. Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 2007, 58, 221–227. [Google Scholar] [CrossRef] [PubMed]
- Tran, L.S.; Nakashima, K.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Plant gene networks in osmotic stress response: From genes to regulatory networks. Methods Enzymol. 2007, 428, 109–128. [Google Scholar] [PubMed]
- Malisch, C.S.; Lüscher, A.; Baert, N.; Engström, M.T.; Studer, B.; Fryganas, C.; Suter, D.; Mueller-Harvey, I.; Salminen, J.P. Large Variability of Proanthocyanidin Content and Composition in Sainfoin (Onobrychis viciifolia). J. Agric. Food Chem. 2015, 63, 10234–10242. [Google Scholar] [CrossRef]
- Barrau, E.; Fabre, N.; Fouraste, I.; Hoste, H. Effect of bioactive compounds from Sainfoin (Onobrychis viciifolia Scop.) on the in vitro larval migration of Haemonchus contortus: Role of tannins and flavonol glycosides. Parasitology 2005, 131, 531–538. [Google Scholar] [CrossRef]
- Mueller-Harvey, I. Unravelling the conundrum of tannins in animal nutrition and health. J. Sci. Food Agric. 2006, 86, 2010–2037. [Google Scholar] [CrossRef]
- Lu, Y.; Sun, Y.; Foo, L.Y.; McNabb, W.C.; Molan, A.L. Phenolic glycosides of forage legume Onobrychis viciifolia. Phytochemistry 2000, 55, 67–75. [Google Scholar] [CrossRef]
- Regos, I.; Urbanella, A.; Treutter, D. Identification and Quantification of Phenolic Compounds from the Forage Legume Sainfoin (Onobrychis viciifolia). J. Agric. Food Chem. 2009, 57, 5843–5852. [Google Scholar] [CrossRef]
- Stringano, E.; Cramer, R.; Hayes, W.; Smith, C.; Gibson, T.; Mueller-Harvey, I. Deciphering the complexity of sainfoin (Onobrychis viciifolia) proanthocyanidins by MALDI-TOF mass spectrometry with a judicious choice of isotope patterns and matrixes. Anal. Chem. 2011, 83, 4147–4153. [Google Scholar] [CrossRef]
- Veitch, N.C.; Regos, I.; Kite, G.C.; Treutter, D. Acylated flavonol glycosides from the forage legume, Onobrychis viciifolia (sainfoin). Phytochemistry 2011, 72, 423–429. [Google Scholar] [CrossRef]
- Malisch, C.S.; Salminen, J.P.; Kölliker, R.; Engström, M.T.; Suter, D.; Studer, B.; Lüscher, A. Drought Effects on Proanthocyanidins in Sainfoin (Onobrychis viciifolia Scop.) Are Dependent on the Plant’s Ontogenetic Stage. J. Agric. Food Chem. 2016, 64, 9307–9316. [Google Scholar] [CrossRef]
- Scharenberg, A.; Arrigo, Y.; Gutzwiller, A.; Soliva, C.R.; Wyss, U.; Kreuzer, M.; Dohme, F. Palatability in sheep and in vitro nutritional value of dried and ensiled sainfoin (Onobrychis viciifolia) birdsfoot trefoil (Lotus corniculatus), and chicory (Cichorium intybus). Arch. Anim. Nutr. 2007, 61, 481–496. [Google Scholar] [CrossRef]
- Liu, X.M.; Zhang, R.; Dou, W.; Gao, Z.Q.; Bu-man, A.; Meng, X.J. Study on adaptability of Onobrychis viciaefolia cv. Gansu and Medicago sativa cv. Algonguin in alpine pastoral area. Grassland Turf 2010, 30, 51–54. [Google Scholar]
- Cao, M.; Jin, Y.; Liu, N.; Ji, W. Effects of the Qinghai-Tibetan Plateau uplift and environmental changes on phylogeographic structure of the Daurian Partridge (Perdix dauuricae) in China. Mol. Phylogenet. Evol. 2012, 65, 823–830. [Google Scholar] [CrossRef]
- Ge, R.L.; Cai, Q.; Shen, Y.Y.; San, A.; Ma, L.; Zhang, Y.; Yi, X.; Chen, Y.; Yang, L.; Huang, Y.; et al. Draft genome sequence of the Tibetan antelope. Nat. Commun. 2013, 4, 1858. [Google Scholar] [CrossRef]
- Wang, P.; Lassoie, J.P.; Morreale, S.J.; Dong, S. A critical review of socioeconomic and natural factors in ecological degradation on the Qinghai-Tibetan Plateau, China. Rangel. J. 2015, 37, 1–9. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef]
- Jin, J.; Tian, F.; Yang, D.C.; Meng, Y.Q.; Kong, L.; Luo, J.; Gao, G. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017, 45, D1040–D1045. [Google Scholar] [CrossRef] [PubMed]
- Lohse, M.; Nagel, A.; Herter, T.; May, P.; Schroda, M.; Zrenner, R.; Tohge, T.; Fernie, A.R.; Stitt, M.; Usadel, B. Mercator: A fast and simple web server for genome scale functional annotation of plant sequence data. Plant Cell Environ. 2014, 37, 1250–1258. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Abu Bakar, M.F.; Mohamed, M.; Rahmat, A.; Fry, J. Phytochemicals and antioxidant activity of different parts of bambangan (Mangifera pajang) and tarap (Artocarpus odoratissimus). Food Chem. 2009, 113, 479–483. [Google Scholar] [CrossRef]
- Zhishen, J.; Mengcheng, T.; Jianming, W. The determination of flavonoid contents in mulberry and their scavenging effects on superoxide radicals. Food Chem. 1999, 64, 555–559. [Google Scholar] [CrossRef]
- Montiel, J.; Szűcs, A.; Boboescu, I.Z.; Gherman, V.D.; Kondorosi, É.; Kereszt, A. Terminal Bacteroid Differentiation Is Associated With Variable Morphological Changes in Legume Species Belonging to the Inverted Repeat-Lacking Clade. Mol. Plant Microbe Interact. 2016, 29, 210–219. [Google Scholar] [CrossRef]
- Wang, J.; Sun, P.; Li, Y.; Liu, Y.; Yu, J.; Ma, X.; Sun, S.; Yang, N.; Xia, R.; Lei, T.; et al. Hierarchically Aligning 10 Legume Genomes Establishes a Family-Level Genomics Platform. Plant Physiol. 2017, 174, 284–300. [Google Scholar] [CrossRef]
- Wang, F.; Xu, Z.; Fan, X.; Zhou, Q.; Cao, J.; Ji, G.; Jing, S.; Feng, B.; Wang, T. Transcriptome Analysis Reveals Complex Molecular Mechanisms Underlying UV Tolerance of Wheat (Triticum aestivum, L.). J. Agric. Food Chem. 2019, 67, 563–577. [Google Scholar] [CrossRef]
- Hut, R.A.; Beersma, D.G. Evolution of time-keeping mechanisms: Early emergence and adaptation to photoperiod. Philos. Trans. R Soc. Lond. B Biol. Sci. 2011, 366, 2141–2154. [Google Scholar] [CrossRef]
- D’Amico-Damiao, V.; Carvalho, R.F. Cryptochrome-Related Abiotic Stress Responses in Plants. Front. Plant Sci. 2018, 9, 1897. [Google Scholar] [CrossRef]
- Jiang, B.; Shi, Y.; Zhang, X.; Xin, X.; Qi, L.; Guo, H.; Li, J.; Yang, S. PIF3 is a negative regulator of the CBF pathway and freezing tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 2017, 114, E6695–E6702. [Google Scholar] [CrossRef] [PubMed]
- Nakamichi, N.; Takao, S.; Kudo, T.; Kiba, T.; Wang, Y.; Kinoshita, T.; Sakakibara, H. Improvement of Arabidopsis Biomass and Cold, Drought and Salinity Stress Tolerance by Modified Circadian Clock-Associated PSEUDO-RESPONSE REGULATORs. Plant Cell. Physiol. 2016, 57, 1085–1097. [Google Scholar] [CrossRef] [PubMed]
- Nakabayashi, R.; Saito, K. Integrated metabolomics for abiotic stress responses in plants. Curr. Opin. Plant Biol. 2015, 24, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Shahzad, B.; Rehman, A.; Bhardwaj, R.; Landi, M.; Zheng, B. Response of Phenylpropanoid Pathway and the Role of Polyphenols in Plants under Abiotic Stress. Molecules 2019, 24, 2452. [Google Scholar] [CrossRef]
- Li, Q.; Yu, H.M.; Meng, X.F.; Lin, J.S.; Li, Y.J.; Hou, B.K. Ectopic expression of glycosyltransferase UGT76E11 increases flavonoid accumulation and enhances abiotic stress tolerance in Arabidopsis. Plant Biol. (Stuttg.) 2018, 20, 10–19. [Google Scholar] [CrossRef]
- Li, W.; Herrera-Estrella, L.; Tran, L.P. Do Cytokinins and Strigolactones Crosstalk during Drought Adaptation? Trends Plant Sci. 2019, 24, 669–672. [Google Scholar] [CrossRef]
- Mostofa, M.G.; Li, W.; Nguyen, K.H.; Fujita, M.; Tran, L.P. Strigolactones in plant adaptation to abiotic stresses: An emerging avenue of plant research. Plant Cell. Environ. 2018, 41, 2227–2243. [Google Scholar] [CrossRef]
- Sarkar, T.; Thankappan, R.; Mishra, G.P.; Nawade, B.D. Advances in the development and use of DREB for improved abiotic stress tolerance in transgenic crop plants. Physiol. Mol. Biol. Plants 2019, 25, 1323–1334. [Google Scholar] [CrossRef]
- El-Esawi, M.A.; Alayafi, A.A. Overexpression of StDREB2 Transcription Factor Enhances Drought Stress Tolerance in Cotton (Gossypium barbadense L.). Genes (Basel) 2019, 10, 142. [Google Scholar] [CrossRef]
- Li, X.; Liang, Y.; Gao, B.; Mijiti, M.; Bozorov, T.A.; Yang, H.; Zhang, D.; Wood, A.J. ScDREB10, an A-5c type of DREB Gene of the Desert Moss Syntrichia caninervis, Confers Osmotic and Salt Tolerances to Arabidopsis. Genes (Basel) 2019, 10, 146. [Google Scholar] [CrossRef]
- Yang, Y.; Al-Baidhani, H.H.J.; Harris, J.; Riboni, M.; Li, Y.; Mazonka, I.; Bazanova, N.; Chirkova, L.; Sarfraz Hussain, S.; Hrmova, M.; et al. DREB/CBF expression in wheat and barley using the stress-inducible promoters of HD-Zip I genes: Impact on plant development, stress tolerance and yield. Plant Biotechnol. J. 2020, 18, 829–844. [Google Scholar] [CrossRef] [PubMed]
- Coego, A.; Brizuela, E.; Castillejo, P.; Ruíz, S.; Koncz, C.; del Pozo, J.C.; Piñeiro, M.; Jarillo, J.A.; Paz-Ares, J.; León, J. The TRANSPLANTA collection of Arabidopsis lines: A resource for functional analysis of transcription factors based on their conditional overexpression. Plant J. 2014, 77, 944–953. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Li, W.; Gao, X.; Xu, M.; Guo, Y. DEAR4, a Member of DREB/CBF Family, Positively Regulates Leaf Senescence and Response to Multiple Stressors in Arabidopsis thaliana. Front. Plant Sci. 2020, 11, 367. [Google Scholar] [CrossRef] [PubMed]
- Mora-Ortiz, M.; Swain, M.T.; Vickers, M.J.; Hegarty, M.J.; Kelly, R.; Smith, L.M.; Skøt, L. De-novo transcriptome assembly for gene identification, analysis, annotation, and molecular marker discovery in Onobrychis viciifolia. BMC Genomics 2016, 17, 756. [Google Scholar] [CrossRef] [PubMed]
- Etterson, J.R. Evolutionary potential of Chamaecrista fasciculata in relation to climate change. I. Clinal patterns of selection along an environmental gradient in the great plains. Evolution 2004, 58, 1446–1458. [Google Scholar] [CrossRef] [PubMed]
- Fustier, M.A.; Martínez-Ainsworth, N.E.; Aguirre-Liguori, J.A.; Venon, A.; Corti, H.; Rousselet, A.; Dumas, F.; Dittberner, H.; Camarena, M.G.; Grimanelli, D.; et al. Common gardens in teosintes reveal the establishment of a syndrome of adaptation to altitude. PLoS Genet. 2019, 15, e1008512. [Google Scholar] [CrossRef]
- Leger, E.A.; Rice, K.J. Assessing the speed and predictability of local adaptation in invasive California poppies (Eschscholzia californica). J. Evol. Biol. 2007, 20, 1090–1103. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, J.; Lai, L.; Jiang, L.; Zhuang, P.; Zhang, L.; Zheng, Y.; Baskin, J.M.; Baskin, C.C. Geographic variation in seed traits within and among forty-two species of Rhododendron (Ericaceae) on the Tibetan plateau: Relationships with altitude, habitat, plant height, and phylogeny. Ecol. Evol. 2014, 4, 1913–1923. [Google Scholar] [CrossRef]
- Kollas, C.; Körner, C.; Randin, C.F. Spring frost and growing season length co-control the cold range limits of broad-leaved trees. J. Biogeogr. 2014, 41, 773–783. [Google Scholar] [CrossRef]
- Suter, L.; Rüegg, M.; Zemp, N.; Hennig, L.; Widmer, A. Gene regulatory variation mediates flowering responses to vernalization along an altitudinal gradient in Arabidopsis. Plant Physiol. 2014, 166, 1928–1942. [Google Scholar] [CrossRef]
- Ito, S.; Niwa, Y.; Nakamichi, N.; Kawamura, H.; Yamashino, T.; Mizuno, T. Insight into Missing Genetic Links Between Two Evening-Expressed Pseudo-Response Regulator Genes TOC1 and PRR5 in the Circadian Clock-Controlled Circuitry in Arabidopsis thaliana. Plant Cell. Physiol. 2008, 49, 201–213. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.D.; Kim, M.R.; Kang, M.Y.; Cha, J.Y.; Han, S.H.; Nawkar, G.M.; Sakuraba, Y.; Lee, S.Y.; Imaizumi, T.; McClung, C.R.; et al. The F-box protein FKF1 inhibits dimerization of COP1 in the control of photoperiodic flowering. Nat. Commun. 2017, 8, 2259. [Google Scholar] [CrossRef] [PubMed]
- Nakamichi, N.; Kita, M.; Niinuma, K.; Ito, S.; Yamashino, T.; Mizoguchi, T.; Mizuno, T. Arabidopsis Clock-Associated Pseudo-Response Regulators PRR9, PRR7 and PRR5 Coordinately and Positively Regulate Flowering Time Through the Canonical CONSTANS-Dependent Photoperiodic Pathway. Plant Cell. Physiol. 2007, 48, 822–832. [Google Scholar] [CrossRef]
- Nakamichi, N.; Kudo, T.; Makita, N.; Kiba, T.; Kinoshita, T.; Sakakibara, H. Flowering time control in rice by introducing Arabidopsis clock-associated PSEUDO-RESPONSE REGULATOR 5. Biosci. Biotechnol. Biochem. 2020, 84, 970–979. [Google Scholar] [CrossRef] [PubMed]
- Oda, A.; Fujiwara, S.; Kamada, H.; Coupland, G.; Mizoguchi, T. Antisense suppression of the Arabidopsis PIF3 gene does not affect circadian rhythms but causes early flowering and increases FT expression. FEBS Lett. 2004, 557, 259–264. [Google Scholar] [CrossRef]
- Isah, T. Stress and defense responses in plant secondary metabolites production. Biol. Res. 2019, 52, 39. [Google Scholar] [CrossRef]
- Yang, L.; Wen, K.S.; Ruan, X.; Zhao, Y.X.; Wei, F.; Wang, Q. Response of Plant Secondary Metabolites to Environmental Factors. Molecules 2018, 23, 762. [Google Scholar] [CrossRef]
- Hossain, M.A.; Bhattacharjee, S.; Armin, S.M.; Qian, P.; Xin, W.; Li, H.Y.; Burritt, D.J.; Fujita, M.; Tran, L.S. Hydrogen peroxide priming modulates abiotic oxidative stress tolerance: Insights from ROS detoxification and scavenging. Front. Plant Sci. 2015, 6, 420. [Google Scholar] [CrossRef]
- Mostofa, M.G.; Rahman, M.M.; Siddiqui, M.N.; Fujita, M.; Tran, L.P. Salicylic acid antagonizes selenium phytotoxicity in rice: Selenium homeostasis, oxidative stress metabolism and methylglyoxal detoxification. J. Hazard. Mater. 2020, 394, 122572. [Google Scholar] [CrossRef]
- Li, W.; Nguyen, K.H.; Chu, H.D.; Watanabe, Y.; Osakabe, Y.; Sato, M.; Toyooka, K.; Seo, M.; Tian, L.; Tian, C.; et al. Comparative functional analyses of DWARF14 and KARRIKIN INSENSITIVE 2 in drought adaptation of Arabidopsis thaliana. Plant J. 2020. [Google Scholar] [CrossRef]
- Li, W.; Nguyen, K.H.; Tran, C.D.; Watanabe, Y.; Tian, C.; Yin, X.; Li, K.; Yang, Y.; Guo, J.; Miao, Y.; et al. Negative Roles of Strigolactone-Related SMXL6, 7 and 8 Proteins in Drought Resistance in Arabidopsis. Biomolecules 2020, 10, 607. [Google Scholar] [CrossRef]
- Zhang, L.; Li, X.; Ma, B.; Gao, Q.; Du, H.; Han, Y.; Li, Y.; Cao, Y.; Qi, M.; Zhu, Y.; et al. The Tartary Buckwheat Genome Provides Insights into Rutin Biosynthesis and Abiotic Stress Tolerance. Mol. Plant 2017, 10, 1224–1237. [Google Scholar] [CrossRef]
- Besseau, S.; Hoffmann, L.; Geoffroy, P.; Lapierre, C.; Pollet, B.; Legrand, M. Flavonoid accumulation in Arabidopsis repressed in lignin synthesis affects auxin transport and plant growth. Plant Cell 2007, 19, 148–162. [Google Scholar] [CrossRef]
- Milborrow, B.V. The pathway of biosynthesis of abscisic acid in vascular plants: A review of the present state of knowledge of ABA biosynthesis. J. Exp. Bot. 2001, 52, 1145–1164. [Google Scholar] [CrossRef]
- Osakabe, Y.; Osakabe, K.; Shinozaki, K.; Tran, L.S. Response of plants to water stress. Front. Plant Sci. 2014, 5, 86. [Google Scholar] [CrossRef]
- Osakabe, Y.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Tran, L.S. ABA control of plant macroelement membrane transport systems in response to water deficit and high salinity. New Phytol. 2014, 202, 35–49. [Google Scholar] [CrossRef]
- Tan, B.C.; Schwartz, S.H.; Zeevaart, J.A.; McCarty, D.R. Genetic control of abscisic acid biosynthesis in maize. Proc. Natl. Acad. Sci. USA 1997, 94, 12235–12240. [Google Scholar] [CrossRef]
- Kalladan, R.; Lasky, J.R.; Sharma, S.; Kumar, M.N.; Juenger, T.E.; Des Marais, D.L.; Verslues, P.E. Natural Variation in 9-Cis-Epoxycartenoid Dioxygenase 3 and ABA Accumulation. Plant Physiol. 2019, 179, 1620–1631. [Google Scholar] [CrossRef]
- Sato, H.; Takasaki, H.; Takahashi, F.; Suzuki, T.; Iuchi, S.; Mitsuda, N.; Ohme-Takagi, M.; Ikeda, M.; Seo, M.; Yamaguchi-Shinozaki, K.; et al. Arabidopsis thaliana NGATHA1 transcription factor induces ABA biosynthesis by activating NCED3 gene during dehydration stress. Proc. Natl. Acad. Sci. USA 2018, 115, E11178–E11187. [Google Scholar] [CrossRef]
- Huang, Y.; Guo, Y.; Liu, Y.; Zhang, F.; Wang, Z.; Wang, H.; Wang, F.; Li, D.; Mao, D.; Luan, S.; et al. 9-cis-Epoxycarotenoid Dioxygenase 3 Regulates Plant Growth and Enhances Multi-Abiotic Stress Tolerance in Rice. Front. Plant Sci. 2018, 9, 162. [Google Scholar] [CrossRef]
- Ori, N. Dissecting the Biological Functions of ARF and Aux/IAA Genes. Plant Cell 2019, 31, 1210–1211. [Google Scholar] [CrossRef]
- Stortenbeker, N.; Bemer, M. The SAUR gene family: The plant’s toolbox for adaptation of growth and development. J. Exp. Bot. 2018, 70, 17–27. [Google Scholar] [CrossRef]
- Chen, T.T.; Liu, F.F.; Xiao, D.W.; Jiang, X.Y.; Li, P.; Zhao, S.M.; Hou, B.K.; Li, Y.J. The Arabidopsis UDP-glycosyltransferase75B1, conjugates abscisic acid and affects plant response to abiotic stresses. Plant Mol. Biol. 2020, 102, 389–401. [Google Scholar] [CrossRef]
- Dong, T.; Hwang, I. Contribution of ABA UDP-glucosyltransferases in coordination of ABA biosynthesis and catabolism for ABA homeostasis. Plant Signal. Behav. 2014, 9, e28888. [Google Scholar] [CrossRef]
- Liu, Z.; Yan, J.P.; Li, D.K.; Luo, Q.; Yan, Q.; Liu, Z.B.; Ye, L.M.; Wang, J.M.; Li, X.F.; Yang, Y. UDP-glucosyltransferase71c5, a major glucosyltransferase, mediates abscisic acid homeostasis in Arabidopsis. Plant Physiol. 2015, 167, 1659–1670. [Google Scholar] [CrossRef]
- Priest, D.M.; Ambrose, S.J.; Vaistij, F.E.; Elias, L.; Higgins, G.S.; Ross, A.R.; Abrams, S.R.; Bowles, D.J. Use of the glucosyltransferase UGT71B6 to disturb abscisic acid homeostasis in Arabidopsis thaliana. Plant J. 2006, 46, 492–502. [Google Scholar] [CrossRef]
- Sun, Y.; Ji, K.; Liang, B.; Du, Y.; Jiang, L.; Wang, J.; Kai, W.; Zhang, Y.; Zhai, X.; Chen, P.; et al. Suppressing ABA uridine diphosphate glucosyltransferase (SlUGT75C1) alters fruit ripening and the stress response in tomato. Plant J. 2017, 91, 574–589. [Google Scholar] [CrossRef]
- Tanaka, K.; Hayashi, K.; Natsume, M.; Kamiya, Y.; Sakakibara, H.; Kawaide, H.; Kasahara, H. UGT74D1 catalyzes the glucosylation of 2-oxindole-3-acetic acid in the auxin metabolic pathway in Arabidopsis. Plant Cell Physiol. 2014, 55, 218–228. [Google Scholar] [CrossRef]
- Dean, J.V.; Delaney, S.P. Metabolism of salicylic acid in wild-type, ugt74f1 and ugt74f2 glucosyltransferase mutants of Arabidopsis thaliana. Physiol. Plant 2008, 132, 417–425. [Google Scholar] [CrossRef]









| Content | Total | TD-1 | TD-2 | TD-3 | MH-1 | MH-2 | MH-3 |
|---|---|---|---|---|---|---|---|
| Total raw reads | 57,014,250 | 44,392,764 | 81,922,056 | 69,023,986 | 53,001,304 | 44,778,542 | 48,966,846 |
| Total clean reads | 55,645,070 | 43,177,362 | 79,839,438 | 67,258,662 | 51,867,538 | 43,809,044 | 47,918,378 |
| GC percentage (%) | 43.25% | 43.43% | 44.07% | 44.08% | 43.67% | 43.94% | |
| Clean reads Q20 (%) | 97.51% | 97.66% | 97.66% | 97.76% | 97.76% | 97.80% | |
| Clean reads Q30 (%) | 93.59% | 93.95% | 93.95% | 94.09% | 94.14% | 94.22% | |
| Clean reads (%) | 97.26% | 97.46% | 97.44% | 97.86% | 97.83% | 97.86% | |
| Total mapped reads (%) | 88.04% | 88.71% | 89.16% | 89.54% | 88.91% | 89.33% | |
| Total expressed genes | 74,791 | 64,331 | 62,887 | 59,219 | 56,903 | 60,218 | 60,218 |
| Total transcripts | 218,482 | ||||||
| Total unigenes | 74,791 | ||||||
| N50 (bp) | 1458 | ||||||
| Mean length of unigenes (bp) | 820 |
| Annotated Database | Number of Unigenes | Percentage (%) |
|---|---|---|
| Annotated in Nr | 43,717 | 58.45 |
| Annotated in SwissPort | 32,973 | 44.09 |
| Annotated in KOG | 24,841 | 33.21 |
| Annotated in KEGG | 16,305 | 21.80 |
| Annotated in all databases | 46,508 | 62.18 |
| All assembled unigenes | 74,791 | 100 |
| Species | Number of Matched Unigenes | Percentage |
|---|---|---|
| Medicago truncatula | 11,592 | 24.92% |
| Cicer arietinum | 9758 | 20.98% |
| Glycine max | 3553 | 7.64% |
| Cajanus cajan | 3358 | 7.22% |
| Glycine soja | 1733 | 3.73% |
| Vigna angularis | 1113 | 3.39% |
| Vigna radiata | 1109 | 2.38% |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yin, H.; Zhou, H.; Wang, W.; Tran, L.-S.P.; Zhang, B. Transcriptome Analysis Reveals Potential Roles of Abscisic Acid and Polyphenols in Adaptation of Onobrychis viciifolia to Extreme Environmental Conditions in the Qinghai-Tibetan Plateau. Biomolecules 2020, 10, 967. https://doi.org/10.3390/biom10060967
Yin H, Zhou H, Wang W, Tran L-SP, Zhang B. Transcriptome Analysis Reveals Potential Roles of Abscisic Acid and Polyphenols in Adaptation of Onobrychis viciifolia to Extreme Environmental Conditions in the Qinghai-Tibetan Plateau. Biomolecules. 2020; 10(6):967. https://doi.org/10.3390/biom10060967
Chicago/Turabian StyleYin, Hengxia, Huakun Zhou, Wenying Wang, Lam-Son Phan Tran, and Benyin Zhang. 2020. "Transcriptome Analysis Reveals Potential Roles of Abscisic Acid and Polyphenols in Adaptation of Onobrychis viciifolia to Extreme Environmental Conditions in the Qinghai-Tibetan Plateau" Biomolecules 10, no. 6: 967. https://doi.org/10.3390/biom10060967
APA StyleYin, H., Zhou, H., Wang, W., Tran, L.-S. P., & Zhang, B. (2020). Transcriptome Analysis Reveals Potential Roles of Abscisic Acid and Polyphenols in Adaptation of Onobrychis viciifolia to Extreme Environmental Conditions in the Qinghai-Tibetan Plateau. Biomolecules, 10(6), 967. https://doi.org/10.3390/biom10060967






