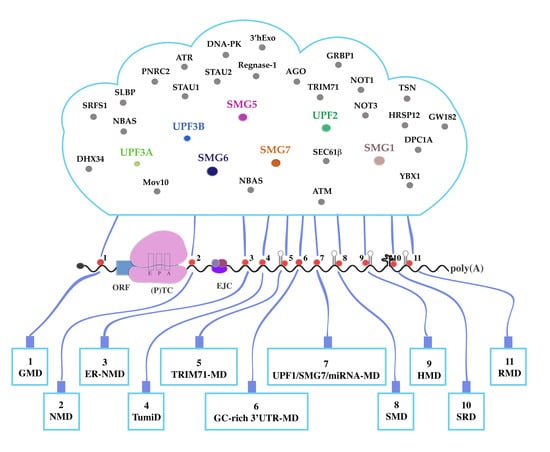
UPF1-Mediated RNA Decay—Danse Macabre in a Cloud
Abstract
:1. Nonsense-Mediated RNA Decay
2. The Termination Phase of NMD
3. The Decay Phase of NMD
4. NMD Occurs on Transcripts with Nuclear or Cytoplasmic Cap-Binding Factors and in Each Round of Translation
5. Human UPF1 Does Not Directly Interact with the Release Factors
6. If Not by the eRFs, How Is UPF1 Recruited to NMD-Target Transcripts?
7. UPF1–PABP Competition and Distance between the TC and PABP
8. Is Termination at a PTC Really Slower than at an NTC?
9. UPF3B and ABCE1 Functions in Early and Late Translation Termination
10. Are There Any Indispensable Trans-Acting Factors or Cis-Acting Features in NMD?
10.1. UPF2, UPF3B, SMG1, and the EJC
10.2. Decay-Inducing Factors
10.3. Translation Termination
10.4. Target RNA Features
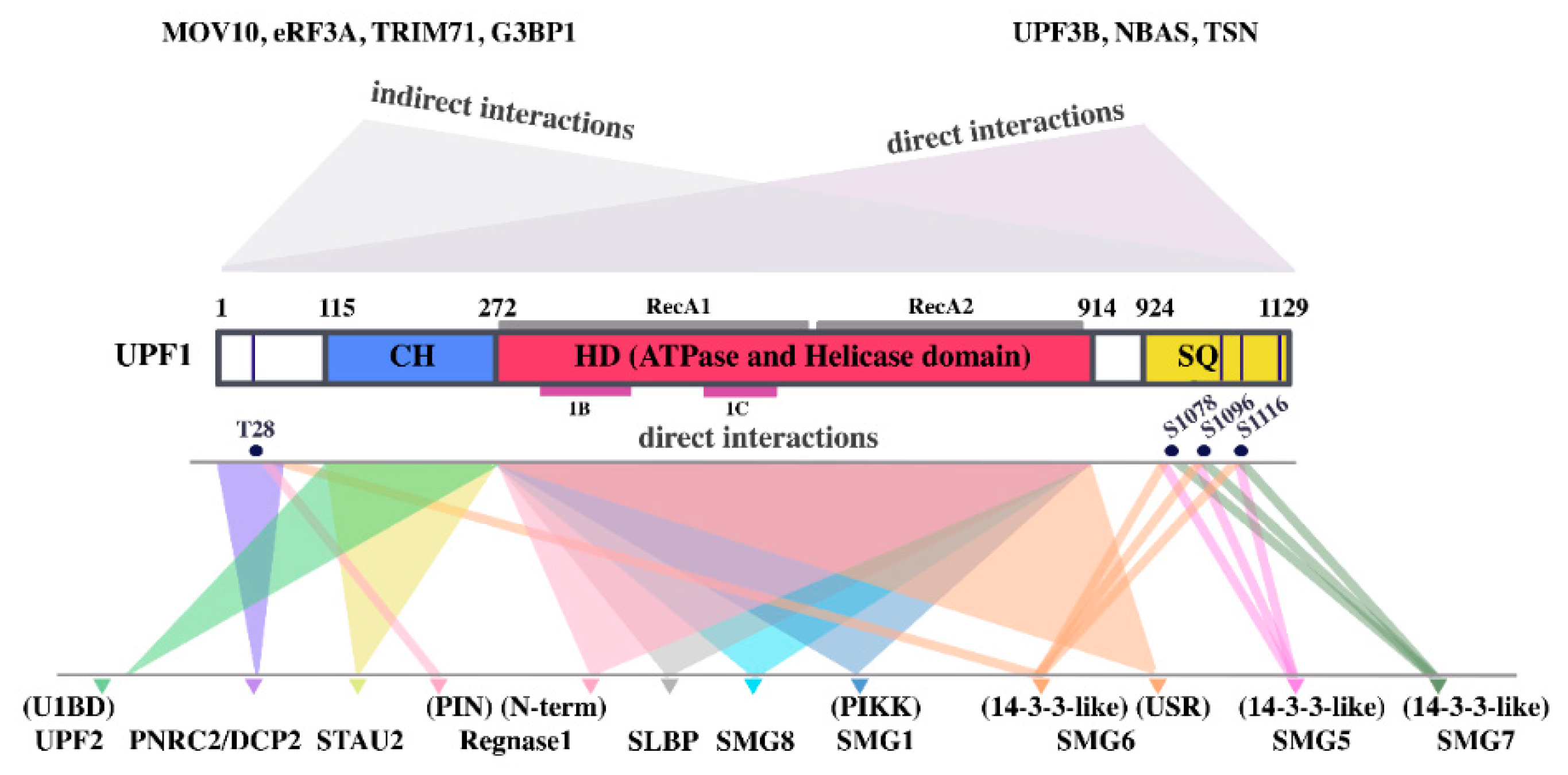
10.5. UPF1
11. Staufen-Mediated mRNA Decay (SMD)
12. Histone mRNA Decay
13. Regnase-1 Mediated mRNA Decay
14. TRIM71-Mediated mRNA Decay (TRIM71-MD)
15. GC-Rich-3′UTR Mediated mRNA Decay (GC-rich 3′UTR-MD)
16. UPF1/SMG7/miRNA-Mediated mRNA Decay (UPF1/SMG7/miRNA-MD)
17. Structure-Mediated RNA Decay (SRD)
18. Tudor-Mediated miRNA Decay (TumiD)
19. Glucocorticoid Receptor-Mediated mRNA Decay (GMD)
20. A Growing List
21. Crosstalk between Decay Pathways
22. Targets of More Than One UPF1-Dependent Degradation Pathway?
23. To NMD or Not to NMD?
24. A New Metaphor to Understand the UMD Family
Funding
Acknowledgments
Conflicts of Interest
References
- Bhuvanagiri, M.; Schlitter, A.M.; Hentze, M.W.; Kulozik, A.E. NMD: RNA biology meets human genetic medicine. The Biochemical journal 2010, 430, 365–377. [Google Scholar] [CrossRef] [Green Version]
- Miller, J.N.; Pearce, D.A. Nonsense-mediated decay in genetic disease: Friend or foe? Mutation research. Reviews in mutation research 2014, 762, 52–64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lindeboom, R.G.H.; Vermeulen, M.; Lehner, B.; Supek, F. The impact of nonsense-mediated mRNA decay on genetic disease, gene editing and cancer immunotherapy. Nature genetics 2019, 51, 1645–1651. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.S.; Wilkinson, M.F.; Gecz, J. Nonsense-mediated mRNA decay: Inter-individual variability and human disease. Neuroscience and biobehavioral reviews 2014, 46, 175–186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nasif, S.; Contu, L.; Mühlemann, O. Beyond quality control: The role of nonsense-mediated mRNA decay (NMD) in regulating gene expression. Semin Cell Dev Biol 2018, 75, 78–87. [Google Scholar] [CrossRef] [PubMed]
- Karousis, E.D.; Mühlemann, O. Nonsense-Mediated mRNA Decay Begins Where Translation Ends. Cold Spring Harb. Perspect. Biol. 2019, 11. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.K.; Maquat, L.E. UPFront and center in RNA decay: UPF1 in nonsense-mediated mRNA decay and beyond. RNA 2019, 25, 407–422. [Google Scholar] [CrossRef] [Green Version]
- Kishor, A.; Fritz, S.E.; Hogg, J.R. Nonsense-mediated mRNA decay: The challenge of telling right from wrong in a complex transcriptome. Wiley Interdiscip Rev. RNA 2019, 10, e1548. [Google Scholar] [CrossRef]
- Dyle, M.C.; Kolakada, D.; Cortazar, M.A.; Jagannathan, S. How to get away with nonsense: Mechanisms and consequences of escape from nonsense-mediated RNA decay. Wiley Interdiscip Rev. RNA 2020, 11, e1560. [Google Scholar] [CrossRef]
- Kurosaki, T.; Popp, M.W.; Maquat, L.E. Quality and quantity control of gene expression by nonsense-mediated mRNA decay. Nature reviews. Mol. Cell biology 2019, 20, 406–420. [Google Scholar] [CrossRef]
- Raimondeau, E.; Bufton, J.C.; Schaffitzel, C. New insights into the interplay between the translation machinery and nonsense-mediated mRNA decay factors. Biochem. Soc. Trans. 2018, 46, 503–512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lloyd, J.P.B. The evolution and diversity of the nonsense-mediated mRNA decay pathway. F1000Res 2018, 7, 1299. [Google Scholar] [CrossRef] [PubMed]
- Goetz, A.E.; Wilkinson, M. Stress and the nonsense-mediated RNA decay pathway. CMLS 2017, 74, 3509–3531. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, R.; Nogueira, G.; da Costa, P.J.; Pinto, F.; Romao, L. Nonsense-Mediated mRNA Decay in Development, Stress and Cancer. Adv. Exp. Med. Biol. 2019, 1157, 41–83. [Google Scholar] [CrossRef]
- Ottens, F.; Gehring, N.H. Physiological and pathophysiological role of nonsense-mediated mRNA decay. Pflugers Arch. 2016, 468, 1013–1028. [Google Scholar] [CrossRef]
- Nickless, A.; Bailis, J.M.; You, Z. Control of gene expression through the nonsense-mediated RNA decay pathway. Cell Biosci 2017, 7, 26. [Google Scholar] [CrossRef]
- Neu-Yilik, G.; Gehring, N.H.; Hentze, M.W.; Kulozik, A.E. Nonsense-mediated mRNA decay: From vacuum cleaner to Swiss army knife. Genome Boail. 2004, 5, 218. [Google Scholar] [CrossRef] [Green Version]
- Lykke-Andersen, S.; Jensen, T.H. Nonsense-mediated mRNA decay: An intricate machinery that shapes transcriptomes. J. Mol. Cell Biol. 2015. [Google Scholar] [CrossRef] [Green Version]
- Jaffrey, S.R.; Wilkinson, M.F. Nonsense-mediated RNA decay in the brain: Emerging modulator of neural development and disease. Nat. Rev. Neurosci. 2018, 19, 715–728. [Google Scholar] [CrossRef]
- Karam, R.; Lou, C.H.; Kroeger, H.; Huang, L.; Lin, J.H.; Wilkinson, M.F. The unfolded protein response is shaped by the NMD pathway. EMBO reports 2015, 16, 599–609. [Google Scholar] [CrossRef]
- Karam, R.; Wengrod, J.; Gardner, L.B.; Wilkinson, M.F. Regulation of nonsense-mediated mRNA decay: Implications for physiology and disease. BBA 2013, 1829, 624–633. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lou, C.H.; Shum, E.Y.; Wilkinson, M.F. RNA degradation drives stem cell differentiation. EMBO 2015, 34, 1606–1608. [Google Scholar] [CrossRef] [Green Version]
- Colombo, M.; Karousis, E.D.; Bourquin, J.; Bruggmann, R.; Mühlemann, O. Transcriptome-wide identification of NMD-targeted human mRNAs reveals extensive redundancy between SMG6- and SMG7-mediated degradation pathways. RNA 2017, 23, 189–201. [Google Scholar] [CrossRef] [Green Version]
- Lykke-Andersen, S.; Chen, Y.; Ardal, B.R.; Lilje, B.; Waage, J.; Sandelin, A.; Jensen, T.H. Human nonsense-mediated RNA decay initiates widely by endonucleolysis and targets snoRNA host genes. Genes Dev. 2014, 28, 2498–2517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilusz, J.E.; Wilusz, J. Nonsense-mediated RNA decay: At the ‘cutting edge’ of regulated snoRNA production. Genes Dev. 2014, 28, 2447–2449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hartford, C.C.R.; Lal, A. When Long Noncoding Becomes Protein Coding. Mol. Cell Biol. 2020, 40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kashima, I.; Yamashita, A.; Izumi, N.; Kataoka, N.; Morishita, R.; Hoshino, S.; Ohno, M.; Dreyfuss, G.; Ohno, S. Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev. 2006, 20, 355–367. [Google Scholar] [CrossRef] [Green Version]
- Amrani, N.; Ganesan, R.; Kervestin, S.; Mangus, D.A.; Ghosh, S.; Jacobson, A. A faux 3’-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature 2004, 432, 112–118. [Google Scholar] [CrossRef]
- Le Hir, H.; Izaurralde, E.; Maquat, L.E.; Moore, M.J. The spliceosome deposits multiple proteins 20-24 nucleotides upstream of mRNA exon-exon junctions. EMBO 2000, 19, 6860–6869. [Google Scholar] [CrossRef] [Green Version]
- Le Hir, H.; Gatfield, D.; Izaurralde, E.; Moore, M.J. The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO 2001, 20, 4987–4997. [Google Scholar] [CrossRef] [Green Version]
- Schlautmann, L.P.; Gehring, N.H. A Day in the Life of the Exon Junction Complex. Biomolecules 2020, 10, 866. [Google Scholar] [CrossRef] [PubMed]
- Chamieh, H.; Ballut, L.; Bonneau, F.; Le Hir, H. NMD factors UPF2 and UPF3 bridge UPF1 to the exon junction complex and stimulate its RNA helicase activity. Nat. Struct. Mol. Biol. 2008, 15, 85–93. [Google Scholar] [CrossRef]
- Longman, D.; Hug, N.; Keith, M.; Anastasaki, C.; Patton, E.E.; Grimes, G.; Caceres, J.F. DHX34 and NBAS form part of an autoregulatory NMD circuit that regulates endogenous RNA targets in human cells, zebrafish and Caenorhabditis elegans. Nucleic Acids Res. 2013, 41, 8319–8331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baird, T.D.; Cheng, K.C.; Chen, Y.C.; Buehler, E.; Martin, S.E.; Inglese, J.; Hogg, J.R. ICE1 promotes the link between splicing and nonsense-mediated mRNA decay. eLife 2018, 7. [Google Scholar] [CrossRef] [PubMed]
- Aznarez, I.; Nomakuchi, T.T.; Tetenbaum-Novatt, J.; Rahman, M.A.; Fregoso, O.; Rees, H.; Krainer, A.R. Mechanism of Nonsense-Mediated mRNA Decay Stimulation by Splicing Factor SRSF1. Cell Rep. 2018, 23, 2186–2198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hug, N.; Caceres, J.F. The RNA helicase DHX34 activates NMD by promoting a transition from the surveillance to the decay-inducing complex. Cell Rep. 2014, 8, 1845–1856. [Google Scholar] [CrossRef] [Green Version]
- Gregersen, L.H.; Schueler, M.; Munschauer, M.; Mastrobuoni, G.; Chen, W.; Kempa, S.; Dieterich, C.; Landthaler, M. MOV10 Is a 5’ to 3’ RNA helicase contributing to UPF1 mRNA target degradation by translocation along 3’ UTRs. Mol. Cell 2014, 54, 573–585. [Google Scholar] [CrossRef] [Green Version]
- Yamashita, A.; Izumi, N.; Kashima, I.; Ohnishi, T.; Saari, B.; Katsuhata, Y.; Muramatsu, R.; Morita, T.; Iwamatsu, A.; Hachiya, T.; et al. SMG-8 and SMG-9, two novel subunits of the SMG-1 complex, regulate remodeling of the mRNA surveillance complex during nonsense-mediated mRNA decay. Genes Dev. 2009, 23, 1091–1105. [Google Scholar] [CrossRef] [Green Version]
- Amrani, N.; Dong, S.; He, F.; Ganesan, R.; Ghosh, S.; Kervestin, S.; Li, C.; Mangus, D.A.; Spatrick, P.; Jacobson, A. Aberrant termination triggers nonsense-mediated mRNA decay. Biochem. Soc. Trans. 2006, 34, 39–42. [Google Scholar] [CrossRef]
- Bühler, M.; Steiner, S.; Mohn, F.; Paillusson, A.; Mühlemann, O. EJC-independent degradation of nonsense immunoglobulin-mu mRNA depends on 3’ UTR length. Nat. Struct. Mol. Biol. 2006, 13, 462–464. [Google Scholar] [CrossRef]
- Eberle, A.B.; Stalder, L.; Mathys, H.; Orozco, R.Z.; Mühlemann, O. Posttranscriptional gene regulation by spatial rearrangement of the 3’ untranslated region. PLoS Biol. 2008, 6, e92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Behm-Ansmant, I.; Gatfield, D.; Rehwinkel, J.; Hilgers, V.; Izaurralde, E. A conserved role for cytoplasmic poly(A)-binding protein 1 (PABPC1) in nonsense-mediated mRNA decay. EMBO 2007, 26, 1591–1601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, G.; Rebbapragada, I.; Lykke-Andersen, J. A competition between stimulators and antagonists of Upf complex recruitment governs human nonsense-mediated mRNA decay. PLoS Biol. 2008, 6, e111. [Google Scholar] [CrossRef]
- Ivanov, P.V.; Gehring, N.H.; Kunz, J.B.; Hentze, M.W.; Kulozik, A.E. Interactions between UPF1, eRFs, PABP and the exon junction complex suggest an integrated model for mammalian NMD pathways. EMBO 2008, 27, 736–747. [Google Scholar] [CrossRef] [Green Version]
- Silva, A.L.; Ribeiro, P.; Inacio, A.; Liebhaber, S.A.; Romao, L. Proximity of the poly(A)-binding protein to a premature termination codon inhibits mammalian nonsense-mediated mRNA decay. RNA 2008, 14, 563–576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gowravaram, M.; Bonneau, F.; Kanaan, J.; Maciej, V.D.; Fiorini, F.; Raj, S.; Croquette, V.; Le Hir, H.; Chakrabarti, S. A conserved structural element in the RNA helicase UPF1 regulates its catalytic activity in an isoform-specific manner. Nucleic Acids Res. 2018, 46, 2648–2659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chakrabarti, S.; Jayachandran, U.; Bonneau, F.; Fiorini, F.; Basquin, C.; Domcke, S.; Le Hir, H.; Conti, E. Molecular mechanisms for the RNA-dependent ATPase activity of Upf1 and its regulation by Upf2. Mol. Cell 2011, 41, 693–703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fiorini, F.; Bonneau, F.; Le Hir, H. Biochemical characterization of the RNA helicase UPF1 involved in nonsense-mediated mRNA decay. Methods Enzymol 2012, 511, 255–274. [Google Scholar] [CrossRef]
- Franks, T.M.; Singh, G.; Lykke-Andersen, J. Upf1 ATPase-dependent mRNP disassembly is required for completion of nonsense- mediated mRNA decay. Cell 2010, 143, 938–950. [Google Scholar] [CrossRef] [Green Version]
- Weng, Y.; Czaplinski, K.; Peltz, S.W. Genetic and biochemical characterization of mutations in the ATPase and helicase regions of the Upf1 protein. Mol. Cell Biol. 1996, 16, 5477–5490. [Google Scholar] [CrossRef] [Green Version]
- Cheng, Z.; Morisawa, G.; Song, H. Biochemical characterization of human Upf1 helicase. Methods Mol. Biol. 2010, 587, 327–338. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.; Muhlrad, D.; Lim, M.K.; Parker, R.; Song, H. Structural and functional insights into the human Upf1 helicase core. EMBO 2007, 26, 253–264. [Google Scholar] [CrossRef] [PubMed]
- Kanaan, J.; Raj, S.; Decourty, L.; Saveanu, C.; Croquette, V.; Le Hir, H. UPF1-like helicase grip on nucleic acids dictates processivity. Nat. Commun. 2018, 9, 3752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dehecq, M.; Decourty, L.; Namane, A.; Proux, C.; Kanaan, J.; Le Hir, H.; Jacquier, A.; Saveanu, C. Nonsense-mediated mRNA decay involves two distinct Upf1-bound complexes. EMBO 2018, 37. [Google Scholar] [CrossRef] [PubMed]
- Fiorini, F.; Boudvillain, M.; Le Hir, H. Tight intramolecular regulation of the human Upf1 helicase by its N- and C-terminal domains. Nucleic Acids Res. 2013, 41, 2404–2415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fiorini, F.; Bagchi, D.; Le Hir, H.; Croquette, V. Human Upf1 is a highly processive RNA helicase and translocase with RNP remodelling activities. Nat. Commun. 2015, 6, 7581. [Google Scholar] [CrossRef] [Green Version]
- Gleghorn, M.L.; Maquat, L.E. UPF1 learns to relax and unwind. Mol. Cell 2011, 41, 621–623. [Google Scholar] [CrossRef]
- He, F.; Ganesan, R.; Jacobson, A. Intra- and intermolecular regulatory interactions in Upf1, the RNA helicase central to nonsense-mediated mRNA decay in yeast. Mol. Cell Biol. 2013, 33, 4672–4684. [Google Scholar] [CrossRef] [Green Version]
- Clerici, M.; Mourao, A.; Gutsche, I.; Gehring, N.H.; Hentze, M.W.; Kulozik, A.; Kadlec, J.; Sattler, M.; Cusack, S. Unusual bipartite mode of interaction between the nonsense-mediated decay factors, UPF1 and UPF2. EMBO 2009, 28, 2293–2306. [Google Scholar] [CrossRef] [Green Version]
- Serin, G.; Gersappe, A.; Black, J.D.; Aronoff, R.; Maquat, L.E. Identification and characterization of human orthologues to Saccharomyces cerevisiae Upf2 protein and Upf3 protein (Caenorhabditis elegans SMG-4). Mol. Cell Biol. 2001, 21, 209–223. [Google Scholar] [CrossRef] [Green Version]
- Kunz, J.B.; Neu-Yilik, G.; Hentze, M.W.; Kulozik, A.E.; Gehring, N.H. Functions of hUpf3a and hUpf3b in nonsense-mediated mRNA decay and translation. RNA 2006, 12, 1015–1022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, W.K.; Bhalla, A.D.; Le Hir, H.; Nguyen, L.S.; Huang, L.; Gecz, J.; Wilkinson, M.F. A UPF3-mediated regulatory switch that maintains RNA surveillance. Nat. Struct. Mol. Biol. 2009, 16, 747–753. [Google Scholar] [CrossRef]
- Shum, E.Y.; Jones, S.H.; Shao, A.; Dumdie, J.; Krause, M.D.; Chan, W.K.; Lou, C.H.; Espinoza, J.L.; Song, H.W.; Phan, M.H.; et al. The Antagonistic Gene Paralogs Upf3a and Upf3b Govern Nonsense-Mediated RNA Decay. Cell 2016, 165, 382–395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, A.; Shao, S.; Murray, J.; Hegde, R.S.; Ramakrishnan, V. Structural basis for stop codon recognition in eukaryotes. Nature 2015, 524, 493–496. [Google Scholar] [CrossRef] [PubMed]
- Alkalaeva, E.Z.; Pisarev, A.V.; Frolova, L.Y.; Kisselev, L.L.; Pestova, T.V. In vitro reconstitution of eukaryotic translation reveals cooperativity between release factors eRF1 and eRF3. Cell 2006, 125, 1125–1136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nurenberg-Goloub, E.; Tampe, R. Ribosome recycling in mRNA translation, quality control, and homeostasis. Biol Chem 2019, 401, 47–61. [Google Scholar] [CrossRef] [PubMed]
- Hellen, C.U.T. Translation Termination and Ribosome Recycling in Eukaryotes. Cold Spring Harb. Perspect. Biol. 2018, 10. [Google Scholar] [CrossRef]
- He, F.; Jacobson, A. Nonsense-Mediated mRNA Decay: Degradation of Defective Transcripts Is Only Part of the Story. Annu. Rev. Genet. 2015, 49, 339–366. [Google Scholar] [CrossRef] [Green Version]
- Serdar, L.D.; Whiteside, D.L.; Baker, K.E. ATP hydrolysis by UPF1 is required for efficient translation termination at premature stop codons. Nat. Commun. 2016, 7, 14021. [Google Scholar] [CrossRef]
- Ivanov, A.; Mikhailova, T.; Eliseev, B.; Yeramala, L.; Sokolova, E.; Susorov, D.; Shuvalov, A.; Schaffitzel, C.; Alkalaeva, E. PABP enhances release factor recruitment and stop codon recognition during translation termination. Nucleic Acids Res. 2016, 44, 7766–7776. [Google Scholar] [CrossRef]
- Mühlemann, O. Recognition of nonsense mRNA: Towards a unified model. Biochem. Soc. Trans. 2008, 36, 497–501. [Google Scholar] [CrossRef] [Green Version]
- Okada-Katsuhata, Y.; Yamashita, A.; Kutsuzawa, K.; Izumi, N.; Hirahara, F.; Ohno, S. N- and C-terminal Upf1 phosphorylations create binding platforms for SMG-6 and SMG-5:SMG-7 during NMD. Nucleic Acids Res. 2012, 40, 1251–1266. [Google Scholar] [CrossRef] [Green Version]
- Arias-Palomo, E.; Yamashita, A.; Fernandez, I.S.; Nunez-Ramirez, R.; Bamba, Y.; Izumi, N.; Ohno, S.; Llorca, O. The nonsense-mediated mRNA decay SMG-1 kinase is regulated by large-scale conformational changes controlled by SMG-8. Genes Dev. 2011, 25, 153–164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deniaud, A.; Karuppasamy, M.; Bock, T.; Masiulis, S.; Huard, K.; Garzoni, F.; Kerschgens, K.; Hentze, M.W.; Kulozik, A.E.; Beck, M.; et al. A network of SMG-8, SMG-9 and SMG-1 C-terminal insertion domain regulates UPF1 substrate recruitment and phosphorylation. Nucleic Acids Res. 2015, 43, 7600–7611. [Google Scholar] [CrossRef] [Green Version]
- Fernandez, I.S.; Yamashita, A.; Arias-Palomo, E.; Bamba, Y.; Bartolome, R.A.; Canales, M.A.; Teixido, J.; Ohno, S.; Llorca, O. Characterization of SMG-9, an essential component of the nonsense-mediated mRNA decay SMG1C complex. Nucleic Acids Res. 2011, 39, 347–358. [Google Scholar] [CrossRef] [PubMed]
- Melero, R.; Uchiyama, A.; Castano, R.; Kataoka, N.; Kurosawa, H.; Ohno, S.; Yamashita, A.; Llorca, O. Structures of SMG1-UPFs complexes: SMG1 contributes to regulate UPF2-dependent activation of UPF1 in NMD. Structure 2014, 22, 1105–1119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurosaki, T.; Li, W.; Hoque, M.; Popp, M.W.; Ermolenko, D.N.; Tian, B.; Maquat, L.E. A post-translational regulatory switch on UPF1 controls targeted mRNA degradation. Genes Dev. 2014, 28, 1900–1916. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Durand, S.; Franks, T.M.; Lykke-Andersen, J. Hyperphosphorylation amplifies UPF1 activity to resolve stalls in nonsense-mediated mRNA decay. Nat. Commun. 2016, 7, 12434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Isken, O.; Kim, Y.K.; Hosoda, N.; Mayeur, G.L.; Hershey, J.W.; Maquat, L.E. Upf1 phosphorylation triggers translational repression during nonsense-mediated mRNA decay. Cell 2008, 133, 314–327. [Google Scholar] [CrossRef] [Green Version]
- Flury, V.; Restuccia, U.; Bachi, A.; Mühlemann, O. Characterization of phosphorylation- and RNA-dependent UPF1 interactors by quantitative proteomics. J. Proteome Res. 2014, 13, 3038–3053. [Google Scholar] [CrossRef]
- Nicholson, P.; Josi, C.; Kurosawa, H.; Yamashita, A.; Mühlemann, O. A novel phosphorylation-independent interaction between SMG6 and UPF1 is essential for human NMD. Nucleic Acids Res. 2014, 42, 9217–9235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chakrabarti, S.; Bonneau, F.; Schussler, S.; Eppinger, E.; Conti, E. Phospho-dependent and phospho-independent interactions of the helicase UPF1 with the NMD factors SMG5-SMG7 and SMG6. Nucleic Acids Res. 2014, 42, 9447–9460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melero, R.; Hug, N.; Lopez-Perrote, A.; Yamashita, A.; Caceres, J.F.; Llorca, O. The RNA helicase DHX34 functions as a scaffold for SMG1-mediated UPF1 phosphorylation. Nat. Commun. 2016, 7, 10585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jonas, S.; Weichenrieder, O.; Izaurralde, E. An unusual arrangement of two 14-3-3-like domains in the SMG5-SMG7 heterodimer is required for efficient nonsense-mediated mRNA decay. Genes Dev. 2013, 27, 211–225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anders, K.R.; Grimson, A.; Anderson, P. SMG-5, required for C.elegans nonsense-mediated mRNA decay, associates with SMG-2 and protein phosphatase 2A. EMBO 2003, 22, 641–650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiu, S.Y.; Serin, G.; Ohara, O.; Maquat, L.E. Characterization of human Smg5/7a: A protein with similarities to Caenorhabditis elegans SMG5 and SMG7 that functions in the dephosphorylation of Upf1. RNA 2003, 9, 77–87. [Google Scholar] [CrossRef] [Green Version]
- Eberle, A.B.; Lykke-Andersen, S.; Mühlemann, O.; Jensen, T.H. SMG6 promotes endonucleolytic cleavage of nonsense mRNA in human cells. Nat. Struct Mol Biol 2009, 16, 49–55. [Google Scholar] [CrossRef]
- Ottens, F.; Boehm, V.; Sibley, C.R.; Ule, J.; Gehring, N.H. Transcript-specific characteristics determine the contribution of endo- and exonucleolytic decay pathways during the degradation of nonsense-mediated decay substrates. RNA 2017, 23, 1224–1236. [Google Scholar] [CrossRef]
- Boehm, V.; Haberman, N.; Ottens, F.; Ule, J.; Gehring, N.H. 3’ UTR length and messenger ribonucleoprotein composition determine endocleavage efficiencies at termination codons. Cell Rep. 2014, 9, 555–568. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, S.A.; Foley, P.L.; Jeong, D.H.; Rymarquis, L.A.; Doyle, F.; Tenenbaum, S.A.; Belasco, J.G.; Green, P.J. Identification of SMG6 cleavage sites and a preferred RNA cleavage motif by global analysis of endogenous NMD targets in human cells. Nucleic Acids Res. 2015, 43, 309–323. [Google Scholar] [CrossRef]
- Huntzinger, E.; Kashima, I.; Fauser, M.; Sauliere, J.; Izaurralde, E. SMG6 is the catalytic endonuclease that cleaves mRNAs containing nonsense codons in metazoan. RNA 2008, 14, 2609–2617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karamyshev, A.L.; Karamysheva, Z.N. Lost in Translation: Ribosome-Associated mRNA and Protein Quality Controls. Front. Genet. 2018, 9, 431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Graille, M.; Seraphin, B. Surveillance pathways rescuing eukaryotic ribosomes lost in translation. Nat. Rev. Mol Cell Biol 2012, 13, 727–735. [Google Scholar] [CrossRef] [PubMed]
- Lykke-Andersen, J.; Bennett, E.J. Protecting the proteome: Eukaryotic cotranslational quality control pathways. J. Cell Biol. 2014, 204, 467–476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gerbracht, J.V.; Boehm, V.; Britto-Borges, T.; Kallabis, S.; Wiederstein, J.L.; Ciriello, S.; Aschemeier, D.U.; Krüger, M.; Frese, C.K.; Altmüller, J.; et al. CASC3 promotes transcriptome-wide activation of nonsense-mediated decay by the exon junction complex. Biorxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Loh, B.; Jonas, S.; Izaurralde, E. The SMG5-SMG7 heterodimer directly recruits the CCR4-NOT deadenylase complex to mRNAs containing nonsense codons via interaction with POP2. Genes Dev. 2013, 27, 2125–2138. [Google Scholar] [CrossRef] [Green Version]
- Cho, H.; Kim, K.M.; Kim, Y.K. Human proline-rich nuclear receptor coregulatory protein 2 mediates an interaction between mRNA surveillance machinery and decapping complex. Mol. Cell 2009, 33, 75–86. [Google Scholar] [CrossRef]
- Nicholson, P.; Gkratsou, A.; Josi, C.; Colombo, M.; Mühlemann, O. Dissecting the functions of SMG5, SMG7, and PNRC2 in nonsense-mediated mRNA decay of human cells. RNA 2018, 24, 557–573. [Google Scholar] [CrossRef] [Green Version]
- Lykke-Andersen, J. Identification of a human decapping complex associated with hUpf proteins in nonsense-mediated decay. Mol. Cell Biol. 2002, 22, 8114–8121. [Google Scholar] [CrossRef] [Green Version]
- Cho, H.; Han, S.; Choe, J.; Park, S.G.; Choi, S.S.; Kim, Y.K. SMG5-PNRC2 is functionally dominant compared with SMG5-SMG7 in mammalian nonsense-mediated mRNA decay. Nucleic Acids Res. 2013, 41, 1319–1328. [Google Scholar] [CrossRef] [Green Version]
- Kurosaki, T.; Maquat, L.E. Molecular autopsy provides evidence for widespread ribosome-phased mRNA fragmentation. Nat. Struct. Mol. Biol. 2018, 25, 299–301. [Google Scholar] [CrossRef] [PubMed]
- Da Costa, P.J.; Menezes, J.; Saramago, M.; Garcia-Moreno, J.F.; Santos, H.A.; Gama-Carvalho, M.; Arraiano, C.M.; Viegas, S.C.; Romao, L. A role for DIS3L2 over natural nonsense-mediated mRNA decay targets in human cells. Biochem. Biophys. Res. Commun. 2019, 518, 664–671. [Google Scholar] [CrossRef] [PubMed]
- Morozov, I.Y.; Jones, M.G.; Gould, P.D.; Crome, V.; Wilson, J.B.; Hall, A.J.; Rigden, D.J.; Caddick, M.X. mRNA 3’ tagging is induced by nonsense-mediated decay and promotes ribosome dissociation. Mol. Cell Biol. 2012, 32, 2585–2595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brogna, S.; McLeod, T.; Petric, M. The Meaning of NMD: Translate or Perish. Trends Genet. 2016, 32, 395–407. [Google Scholar] [CrossRef] [Green Version]
- Celik, A.; He, F.; Jacobson, A. NMD monitors translational fidelity 24/7. Curr. Genet. 2017, 63, 1007–1010. [Google Scholar] [CrossRef] [Green Version]
- Maquat, L.E.; Tarn, W.Y.; Isken, O. The pioneer round of translation: Features and functions. Cell 2010, 142, 368–374. [Google Scholar] [CrossRef] [Green Version]
- Rufener, S.C.; Mühlemann, O. eIF4E-bound mRNPs are substrates for nonsense-mediated mRNA decay in mammalian cells. Nat. Struct. Mol. Biol. 2013, 20, 710–717. [Google Scholar] [CrossRef]
- Durand, S.; Lykke-Andersen, J. Nonsense-mediated mRNA decay occurs during eIF4F-dependent translation in human cells. Nat. Struct. Mol. Biol. 2013, 20, 702–709. [Google Scholar] [CrossRef]
- Hentze, M.W.; Izaurralde, E. Making sense of nonsense. Nat. Struct. Mol. Biol. 2013, 20, 651–653. [Google Scholar] [CrossRef]
- Gao, Q.; Das, B.; Sherman, F.; Maquat, L.E. Cap-binding protein 1-mediated and eukaryotic translation initiation factor 4E-mediated pioneer rounds of translation in yeast. Proceedings of the PANS 2005, 102, 4258–4263. [Google Scholar] [CrossRef] [Green Version]
- Maderazo, A.B.; Belk, J.P.; He, F.; Jacobson, A. Nonsense-containing mRNAs that accumulate in the absence of a functional nonsense-mediated mRNA decay pathway are destabilized rapidly upon its restitution. Mol. Cell Biol. 2003, 23, 842–851. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoek, T.A.; Khuperkar, D.; Lindeboom, R.G.H.; Sonneveld, S.; Verhagen, B.M.P.; Boersma, S.; Vermeulen, M.; Tanenbaum, M.E. Single-Molecule Imaging Uncovers Rules Governing Nonsense-Mediated mRNA Decay. Mol. Cell 2019, 75, 324–339. [Google Scholar] [CrossRef] [PubMed]
- Celik, A.; Kervestin, S.; Jacobson, A. NMD: At the crossroads between translation termination and ribosome recycling. Biochimie 2015, 114, 2–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.; Czaplinski, K.; Rao, Y.; Peltz, S.W. The role of Upf proteins in modulating the translation read-through of nonsense-containing transcripts. EMBO 2001, 20, 880–890. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Czaplinski, K.; Ruiz-Echevarria, M.J.; Paushkin, S.V.; Han, X.; Weng, Y.; Perlick, H.A.; Dietz, H.C.; Ter-Avanesyan, M.D.; Peltz, S.W. The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes Dev. 1998, 12, 1665–1677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Min, E.E.; Roy, B.; Amrani, N.; He, F.; Jacobson, A. Yeast Upf1 CH domain interacts with Rps26 of the 40S ribosomal subunit. RNA 2013, 19, 1105–1115. [Google Scholar] [CrossRef] [Green Version]
- Neu-Yilik, G.; Raimondeau, E.; Eliseev, B.; Yeramala, L.; Amthor, B.; Deniaud, A.; Huard, K.; Kerschgens, K.; Hentze, M.W.; Schaffitzel, C.; et al. Dual function of UPF3B in early and late translation termination. EMBO 2017, 36, 2968–2986. [Google Scholar] [CrossRef]
- Schuller, A.P.; Zinshteyn, B.; Enam, S.U.; Green, R. Directed hydroxyl radical probing reveals Upf1 binding to the 80S ribosomal E site rRNA at the L1 stalk. Nucleic Acids Res. 2018, 46, 2060–2073. [Google Scholar] [CrossRef] [Green Version]
- Gao, Z.; Wilkinson, M. An RNA decay factor wears a new coat: UPF3B modulates translation termination. F1000 Res. 2017, 6, 2159. [Google Scholar] [CrossRef] [Green Version]
- Mühlemann, O.; Karousis, E.D. New functions in translation termination uncovered for NMD factor UPF3B. EMBO 2017, 36, 2928–2930. [Google Scholar] [CrossRef]
- Hogg, J.R.; Goff, S.P. Upf1 senses 3’UTR length to potentiate mRNA decay. Cell 2010, 143, 379–389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zünd, D.; Gruber, A.R.; Zavolan, M.; Mühlemann, O. Translation-dependent displacement of UPF1 from coding sequences causes its enrichment in 3’ UTRs. Nat. Struct. Mol. Biol. 2013, 20, 936–943. [Google Scholar] [CrossRef] [PubMed]
- Kurosaki, T.; Maquat, L.E. Rules that govern UPF1 binding to mRNA 3’ UTRs. Proceedings of the PANS 2013, 110, 3357–3362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hurt, J.A.; Robertson, A.D.; Burge, C.B. Global analyses of UPF1 binding and function reveal expanded scope of nonsense-mediated mRNA decay. Genome Res. 2013, 23, 1636–1650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.R.; Pratt, G.A.; Martinez, F.J.; Yeo, G.W.; Lykke-Andersen, J. Target Discrimination in Nonsense-Mediated mRNA Decay Requires Upf1 ATPase Activity. Mol. Cell 2015, 59, 413–425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johns, L.; Grimson, A.; Kuchma, S.L.; Newman, C.L.; Anderson, P. Caenorhabditis elegans SMG-2 selectively marks mRNAs containing premature translation termination codons. Mol. Cell Biol. 2007, 27, 5630–5638. [Google Scholar] [CrossRef] [Green Version]
- Johansson, M.J.; He, F.; Spatrick, P.; Li, C.; Jacobson, A. Association of yeast Upf1p with direct substrates of the NMD pathway. Proceedings of the PANS 2007, 104, 20872–20877. [Google Scholar] [CrossRef] [Green Version]
- Bhattacharya, A.; Czaplinski, K.; Trifillis, P.; He, F.; Jacobson, A.; Peltz, S.W. Characterization of the biochemical properties of the human Upf1 gene product that is involved in nonsense-mediated mRNA decay. RNA 2000, 6, 1226–1235. [Google Scholar] [CrossRef] [Green Version]
- Fritz, S.E.; Ranganathan, S.; Wang, C.D.; Hogg, J.R. The RNA-binding protein PTBP1 promotes ATPase-dependent dissociation of the RNA helicase UPF1 to protect transcripts from nonsense-mediated mRNA decay. J. Biol. Chem. 2020. [Google Scholar] [CrossRef]
- Kishor, A.; Ge, Z.; Hogg, J.R. hnRNP L-dependent protection of normal mRNAs from NMD subverts quality control in B cell lymphoma. EMBO 2019, 38. [Google Scholar] [CrossRef]
- Ge, Z.; Quek, B.L.; Beemon, K.L.; Hogg, J.R. Polypyrimidine tract binding protein 1 protects mRNAs from recognition by the nonsense-mediated mRNA decay pathway. eLife 2016, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurosaki, T.; Maquat, L.E. Nonsense-mediated mRNA decay in humans at a glance. J. Cell Sci. 2016, 129, 461–467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, A.K.; Choudhury, S.R.; De, S.; Zhang, J.; Kissane, S.; Dwivedi, V.; Ramanathan, P.; Petric, M.; Orsini, L.; Hebenstreit, D.; et al. The RNA helicase UPF1 associates with mRNAs co-transcriptionally and is required for the release of mRNAs from gene loci. eLife 2019, 8. [Google Scholar] [CrossRef] [PubMed]
- Kervestin, S.; Li, C.; Buckingham, R.; Jacobson, A. Testing the faux-UTR model for NMD: Analysis of Upf1p and Pab1p competition for binding to eRF3/Sup35p. Biochimie 2012, 94, 1560–1571. [Google Scholar] [CrossRef] [Green Version]
- Roque, S.; Cerciat, M.; Gaugue, I.; Mora, L.; Floch, A.G.; de Zamaroczy, M.; Heurgue-Hamard, V.; Kervestin, S. Interaction between the poly(A)-binding protein Pab1 and the eukaryotic release factor eRF3 regulates translation termination but not mRNA decay in Saccharomyces cerevisiae. RNA 2015, 21, 124–134. [Google Scholar] [CrossRef] [Green Version]
- Meaux, S.; van Hoof, A.; Baker, K.E. Nonsense-mediated mRNA decay in yeast does not require PAB1 or a poly(A) tail. Mol. Cell 2008, 29, 134–140. [Google Scholar] [CrossRef] [Green Version]
- Fatscher, T.; Boehm, V.; Weiche, B.; Gehring, N.H. The interaction of cytoplasmic poly(A)-binding protein with eukaryotic initiation factor 4G suppresses nonsense-mediated mRNA decay. RNA 2014, 20, 1579–1592. [Google Scholar] [CrossRef] [Green Version]
- Joncourt, R.; Eberle, A.B.; Rufener, S.C.; Mühlemann, O. Eukaryotic initiation factor 4G suppresses nonsense-mediated mRNA decay by two genetically separable mechanisms. PLoS ONE 2014, 9, e104391. [Google Scholar] [CrossRef] [Green Version]
- Wells, S.E.; Hillner, P.E.; Vale, R.D.; Sachs, A.B. Circularization of mRNA by eukaryotic translation initiation factors. Mol. Cell 1998, 2, 135–140. [Google Scholar] [CrossRef] [Green Version]
- Sugimoto, Y.; Vigilante, A.; Darbo, E.; Zirra, A.; Militti, C.; D’Ambrogio, A.; Luscombe, N.M.; Ule, J. hiCLIP reveals the in vivo atlas of mRNA secondary structures recognized by Staufen 1. Nature 2015, 519, 491–494. [Google Scholar] [CrossRef] [Green Version]
- Hilleren, P.; Parker, R. mRNA surveillance in eukaryotes: Kinetic proofreading of proper translation termination as assessed by mRNP domain organization? RNA 1999, 5, 711–719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peixeiro, I.; Inacio, A.; Barbosa, C.; Silva, A.L.; Liebhaber, S.A.; Romao, L. Interaction of PABPC1 with the translation initiation complex is critical to the NMD resistance of AUG-proximal nonsense mutations. Nucleic Acids Res. 2012, 40, 1160–1173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karousis, E.D.; Gurzeler, L.-A.; Annibaldis, G.; Dreos, R.; Mühlemann, O. Human NMD ensues independently of stable ribosome stalling. Biorxiv 2019. [Google Scholar] [CrossRef] [Green Version]
- Tate, W.P.; Cridge, A.G.; Brown, C.M. ‘Stop’ in protein synthesis is modulated with exquisite subtlety by an extended RNA translation signal. Biochem. Soc. Trans. 2018, 46, 1615–1625. [Google Scholar] [CrossRef] [PubMed]
- Dominissini, D.; Moshitch-Moshkovitz, S.; Schwartz, S.; Salmon-Divon, M.; Ungar, L.; Osenberg, S.; Cesarkas, K.; Jacob-Hirsch, J.; Amariglio, N.; Kupiec, M.; et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012, 485, 201–206. [Google Scholar] [CrossRef]
- Meyer, K.D.; Saletore, Y.; Zumbo, P.; Elemento, O.; Mason, C.E.; Jaffrey, S.R. Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell 2012, 149, 1635–1646. [Google Scholar] [CrossRef] [Green Version]
- Choe, J.; Lin, S.; Zhang, W.; Liu, Q.; Wang, L.; Ramirez-Moya, J.; Du, P.; Kim, W.; Tang, S.; Sliz, P.; et al. mRNA circularization by METTL3-eIF3h enhances translation and promotes oncogenesis. Nature 2018, 561, 556–560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pisarev, A.V.; Hellen, C.U.; Pestova, T.V. Recycling of eukaryotic posttermination ribosomal complexes. Cell 2007, 131, 286–299. [Google Scholar] [CrossRef] [Green Version]
- Lopez-Perrote, A.; Castano, R.; Melero, R.; Zamarro, T.; Kurosawa, H.; Ohnishi, T.; Uchiyama, A.; Aoyagi, K.; Buchwald, G.; Kataoka, N.; et al. Human nonsense-mediated mRNA decay factor UPF2 interacts directly with eRF3 and the SURF complex. Nucleic Acids Res. 2016, 44, 1909–1923. [Google Scholar] [CrossRef] [Green Version]
- Annibaldis, G.; Dreos, R.; Domanski, M.; Carl, S.; Mühlemann, O. Ribosome recycling factor ABCE1 depletion inhibits nonsense-mediated mRNA decay by promoting stop codon readthrough. Biorxiv 2019. [Google Scholar] [CrossRef] [Green Version]
- Gehring, N.H.; Kunz, J.B.; Neu-Yilik, G.; Breit, S.; Viegas, M.H.; Hentze, M.W.; Kulozik, A.E. Exon-junction complex components specify distinct routes of nonsense-mediated mRNA decay with differential cofactor requirements. Mol. Cell 2005, 20, 65–75. [Google Scholar] [CrossRef] [PubMed]
- Fatscher, T.; Boehm, V.; Gehring, N.H. Mechanism, factors, and physiological role of nonsense-mediated mRNA decay. Cell. Mol. Life Sci. 2015. [Google Scholar] [CrossRef] [PubMed]
- Chan, W.K.; Huang, L.; Gudikote, J.P.; Chang, Y.F.; Imam, J.S.; MacLean, J.A., 2nd; Wilkinson, M.F. An alternative branch of the nonsense-mediated decay pathway. EMBO 2007, 26, 1820–1830. [Google Scholar] [CrossRef] [Green Version]
- Metze, S.; Herzog, V.A.; Ruepp, M.D.; Mühlemann, O. Comparison of EJC-enhanced and EJC-independent NMD in human cells reveals two partially redundant degradation pathways. RNA 2013, 19, 1432–1448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bao, J.; Tang, C.; Yuan, S.; Porse, B.T.; Yan, W. UPF2, a nonsense-mediated mRNA decay factor, is required for prepubertal Sertoli cell development and male fertility by ensuring fidelity of the transcriptome. Development 2015, 142, 352–362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, J.L.; Stoica, L.; Liu, Y.; Zhu, P.J.; Bhattacharya, A.; Buffington, S.A.; Huq, R.; Eissa, N.T.; Larsson, O.; Porse, B.T.; et al. Inhibition of Upf2-Dependent Nonsense-Mediated Decay Leads to Behavioral and Neurophysiological Abnormalities by Activating the Immune Response. Neuron 2019, 104, 665–679. [Google Scholar] [CrossRef]
- Nguyen, L.S.; Jolly, L.; Shoubridge, C.; Chan, W.K.; Huang, L.; Laumonnier, F.; Raynaud, M.; Hackett, A.; Field, M.; Rodriguez, J.; et al. Transcriptome profiling of UPF3B/NMD-deficient lymphoblastoid cells from patients with various forms of intellectual disability. Mol. Psychiatry 2012, 17, 1103–1115. [Google Scholar] [CrossRef] [Green Version]
- MacDonald, C.C.; Grozdanov, P.N. Nonsense in the testis: Multiple roles for nonsense-mediated decay revealed in male reproduction. Biol. Reprod. 2017, 96, 939–947. [Google Scholar] [CrossRef]
- Carter, M.S.; Li, S.; Wilkinson, M.F. A splicing-dependent regulatory mechanism that detects translation signals. EMBO 1996, 15, 5965–5975. [Google Scholar] [CrossRef]
- Thermann, R.; Neu-Yilik, G.; Deters, A.; Frede, U.; Wehr, K.; Hagemeier, C.; Hentze, M.W.; Kulozik, A.E. Binary specification of nonsense codons by splicing and cytoplasmic translation. EMBO 1998, 17, 3484–3494. [Google Scholar] [CrossRef]
- Nagy, E.; Maquat, L.E. A rule for termination-codon position within intron-containing genes: When nonsense affects RNA abundance. Trends Biochem. Sci. 1998, 23, 198–199. [Google Scholar] [CrossRef]
- Mabin, J.W.; Woodward, L.A.; Patton, R.D.; Yi, Z.; Jia, M.; Wysocki, V.H.; Bundschuh, R.; Singh, G. The Exon Junction Complex Undergoes a Compositional Switch that Alters mRNP Structure and Nonsense-Mediated mRNA Decay Activity. Cell Rep. 2018, 25, 2431–2446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wen, J.; Brogna, S. Splicing-dependent NMD does not require the EJC in Schizosaccharomyces pombe. EMBO 2010, 29, 1537–1551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wen, J.; He, M.; Petric, M.; Marzi, L.; Wang, J.; Piechocki, K.; McLeod, T.; Singh, A.K.; Dwivedi, V.; Brogna, S. An intron proximal to a PTC enhances NMD in Saccharomyces cerevisiae. Biorxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Gatfield, D.; Unterholzner, L.; Ciccarelli, F.D.; Bork, P.; Izaurralde, E. Nonsense-mediated mRNA decay in Drosophila: At the intersection of the yeast and mammalian pathways. EMBO 2003, 22, 3960–3970. [Google Scholar] [CrossRef] [PubMed]
- Frizzell, K.A.; Rynearson, S.G.; Metzstein, M.M. Drosophila mutants show NMD pathway activity is reduced, but not eliminated, in the absence of Smg6. RNA 2012, 18, 1475–1486. [Google Scholar] [CrossRef] [Green Version]
- Harger, J.W.; Dinman, J.D. Evidence against a direct role for the Upf proteins in frameshifting or nonsense codon readthrough. RNA 2004, 10, 1721–1729. [Google Scholar] [CrossRef] [Green Version]
- Keeling, K.M.; Lanier, J.; Du, M.; Salas-Marco, J.; Gao, L.; Kaenjak-Angeletti, A.; Bedwell, D.M. Leaky termination at premature stop codons antagonizes nonsense-mediated mRNA decay in S. cerevisiae. RNA 2004, 10, 691–703. [Google Scholar] [CrossRef] [Green Version]
- Weng, Y.; Czaplinski, K.; Peltz, S.W. Identification and characterization of mutations in the UPF1 gene that affect nonsense suppression and the formation of the Upf protein complex but not mRNA turnover. Mol. Cell Biol. 1996, 16, 5491–5506. [Google Scholar] [CrossRef] [Green Version]
- Simms, C.L.; Thomas, E.N.; Zaher, H.S. Ribosome-based quality control of mRNA and nascent peptides. Wiley Interdiscip Rev. RNA 2017, 8. [Google Scholar] [CrossRef] [Green Version]
- Imamachi, N.; Salam, K.A.; Suzuki, Y.; Akimitsu, N. A GC-rich sequence feature in the 3’ UTR directs UPF1-dependent mRNA decay in mammalian cells. Genome Res. 2017, 27, 407–418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neu-Yilik, G.; Amthor, B.; Gehring, N.H.; Bahri, S.; Paidassi, H.; Hentze, M.W.; Kulozik, A.E. Mechanism of escape from nonsense-mediated mRNA decay of human beta-globin transcripts with nonsense mutations in the first exon. RNA 2011, 17, 843–854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Maquat, L.E. Evidence that translation reinitiation abrogates nonsense-mediated mRNA decay in mammalian cells. EMBO 1997, 16, 826–833. [Google Scholar] [CrossRef] [PubMed]
- Cohen, S.; Kramarski, L.; Levi, S.; Deshe, N.; Ben David, O.; Arbely, E. Nonsense mutation-dependent reinitiation of translation in mammalian cells. Nucleic Acids Res. 2019, 47, 6330–6338. [Google Scholar] [CrossRef]
- Pereira, F.J.; Teixeira, A.; Kong, J.; Barbosa, C.; Silva, A.L.; Marques-Ramos, A.; Liebhaber, S.A.; Romao, L. Resistance of mRNAs with AUG-proximal nonsense mutations to nonsense-mediated decay reflects variables of mRNA structure and translational activity. Nucleic Acids Res. 2015, 43, 6528–6544. [Google Scholar] [CrossRef]
- Toma, K.G.; Rebbapragada, I.; Durand, S.; Lykke-Andersen, J. Identification of elements in human long 3’ UTRs that inhibit nonsense-mediated decay. RNA 2015, 21, 887–897. [Google Scholar] [CrossRef] [Green Version]
- Weil, J.E.; Beemon, K.L. A 3’ UTR sequence stabilizes termination codons in the unspliced RNA of Rous sarcoma virus. RNA 2006, 12, 102–110. [Google Scholar] [CrossRef] [Green Version]
- Delhi, P.; Queiroz, R.; Inchaustegui, D.; Carrington, M.; Clayton, C. Is there a classical nonsense-mediated decay pathway in trypanosomes? PLoS ONE 2011, 6, e25112. [Google Scholar] [CrossRef]
- Gorgoni, B.; Zhao, Y.B.; Krishnan, J.; Stansfield, I. Destabilization of Eukaryote mRNAs by 5’ Proximal Stop Codons Can Occur Independently of the Nonsense-Mediated mRNA Decay Pathway. Cells 2019, 8, 800. [Google Scholar] [CrossRef] [Green Version]
- Gong, C.; Maquat, L.E. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3’ UTRs via Alu elements. Nature 2011, 470, 284–288. [Google Scholar] [CrossRef] [Green Version]
- Gong, C.; Tang, Y.; Maquat, L.E. mRNA-mRNA duplexes that autoelicit Staufen1-mediated mRNA decay. Nat. Struct. Mol. Biol. 2013, 20, 1214–1220. [Google Scholar] [CrossRef] [Green Version]
- Heber, S.; Gaspar, I.; Tants, J.N.; Gunther, J.; Moya, S.M.F.; Janowski, R.; Ephrussi, A.; Sattler, M.; Niessing, D. Staufen2-mediated RNA recognition and localization requires combinatorial action of multiple domains. Nat. Commun. 2019, 10, 1659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, H.; Han, S.; Park, O.H.; Kim, Y.K. SMG1 regulates adipogenesis via targeting of staufen1-mediated mRNA decay. BBA 2013, 1829, 1276–1287. [Google Scholar] [CrossRef] [PubMed]
- Gong, C.; Kim, Y.K.; Woeller, C.F.; Tang, Y.; Maquat, L.E. SMD and NMD are competitive pathways that contribute to myogenesis: Effects on PAX3 and myogenin mRNAs. Genes Dev. 2009, 23, 54–66. [Google Scholar] [CrossRef] [Green Version]
- Gowravaram, M.; Schwarz, J.; Khilji, S.K.; Urlaub, H.; Chakrabarti, S. Insights into the assembly and architecture of a Staufen-mediated mRNA decay (SMD)-competent mRNP. Nat. Commun. 2019, 10, 5054. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marzluff, W.F.; Koreski, K.P. Birth and Death of Histone mRNAs. Trends Genet. 2017, 33, 745–759. [Google Scholar] [CrossRef] [PubMed]
- Zünd, D.; Mühlemann, O. Recent transcriptome-wide mapping of UPF1 binding sites reveals evidence for its recruitment to mRNA before translation. Translation 2013, 1, e26977. [Google Scholar] [CrossRef] [Green Version]
- Brooks, L., 3rd; Lyons, S.M.; Mahoney, J.M.; Welch, J.D.; Liu, Z.; Marzluff, W.F.; Whitfield, M.L. A multiprotein occupancy map of the mRNP on the 3’ end of histone mRNAs. RNA 2015, 21, 1943–1965. [Google Scholar] [CrossRef] [Green Version]
- Meaux, S.A.; Holmquist, C.E.; Marzluff, W.F. Role of oligouridylation in normal metabolism and regulated degradation of mammalian histone mRNAs. Philos Trans. R Soc Lond B Biol Sci 2018, 373. [Google Scholar] [CrossRef] [Green Version]
- Kaygun, H.; Marzluff, W.F. Regulated degradation of replication-dependent histone mRNAs requires both ATR and Upf1. Nat. Struct. Mol. Biol. 2005, 12, 794–800. [Google Scholar] [CrossRef]
- Choe, J.; Ahn, S.H.; Kim, Y.K. The mRNP remodeling mediated by UPF1 promotes rapid degradation of replication-dependent histone mRNA. Nucleic Acids Res. 2014, 42, 9334–9349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mullen, T.E.; Marzluff, W.F. Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5’ to 3’ and 3’ to 5’. Genes Dev. 2008, 22, 50–65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akira, S. Regnase-1, a ribonuclease involved in the regulation of immune responses. Cold Spring Harb. Symp. Quant. Biol. 2013, 78, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Mino, T.; Murakawa, Y.; Fukao, A.; Vandenbon, A.; Wessels, H.H.; Ori, D.; Uehata, T.; Tartey, S.; Akira, S.; Suzuki, Y.; et al. Regnase-1 and Roquin Regulate a Common Element in Inflammatory mRNAs by Spatiotemporally Distinct Mechanisms. Cell 2015, 161, 1058–1073. [Google Scholar] [CrossRef] [Green Version]
- Mino, T.; Iwai, N.; Endo, M.; Inoue, K.; Akaki, K.; Hia, F.; Uehata, T.; Emura, T.; Hidaka, K.; Suzuki, Y.; et al. Translation-dependent unwinding of stem-loops by UPF1 licenses Regnase-1 to degrade inflammatory mRNAs. Nucleic Acids Res. 2019, 47, 8838–8859. [Google Scholar] [CrossRef] [Green Version]
- Loedige, I.; Gaidatzis, D.; Sack, R.; Meister, G.; Filipowicz, W. The mammalian TRIM-NHL protein TRIM71/LIN-41 is a repressor of mRNA function. Nucleic Acids Res. 2013, 41, 518–532. [Google Scholar] [CrossRef] [Green Version]
- Ecsedi, M.; Grosshans, H. LIN-41/TRIM71: Emancipation of a miRNA target. Genes Dev. 2013, 27, 581–589. [Google Scholar] [CrossRef] [Green Version]
- Torres-Fernandez, L.A.; Jux, B.; Bille, M.; Port, Y.; Schneider, K.; Geyer, M.; Mayer, G.; Kolanus, W. The mRNA repressor TRIM71 cooperates with Nonsense-Mediated Decay factors to destabilize the mRNA of CDKN1A/p21. Nucleic Acids Res. 2019, 47, 11861–11879. [Google Scholar] [CrossRef]
- Van Nostrand, E.L.; Pratt, G.A.; Shishkin, A.A.; Gelboin-Burkhart, C.; Fang, M.Y.; Sundararaman, B.; Blue, S.M.; Nguyen, T.B.; Surka, C.; Elkins, K.; et al. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nature methods 2016, 13, 508–514. [Google Scholar] [CrossRef]
- Park, J.; Seo, J.W.; Ahn, N.; Park, S.; Hwang, J.; Nam, J.W. UPF1/SMG7-dependent microRNA-mediated gene regulation. Nat. Commun. 2019, 10, 4181. [Google Scholar] [CrossRef] [Green Version]
- Fischer, J.W.; Busa, V.F.; Shao, Y.; Leung, A.K.L. Structure-Mediated RNA Decay by UPF1 and G3BP1. Mol. Cell 2020, 78, 70–84. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Wei, X.; Peng, Y. Structure-Mediated Degradation of CircRNAs. Trends in Cell Bio. 2020. [Google Scholar] [CrossRef] [PubMed]
- Beaudoin, J.D.; Novoa, E.M.; Vejnar, C.E.; Yartseva, V.; Takacs, C.M.; Kellis, M.; Giraldez, A.J. Analyses of mRNA structure dynamics identify embryonic gene regulatory programs. Nat. Struct. Mol. Biol. 2018, 25, 677–686. [Google Scholar] [CrossRef] [PubMed]
- Su, Z.; Tang, Y.; Ritchey, L.E.; Tack, D.C.; Zhu, M.; Bevilacqua, P.C.; Assmann, S.M. Genome-wide RNA structurome reprogramming by acute heat shock globally regulates mRNA abundance. Proceedings of the PANS 2018, 115, 12170–12175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caudy, A.A.; Ketting, R.F.; Hammond, S.M.; Denli, A.M.; Bathoorn, A.M.; Tops, B.B.; Silva, J.M.; Myers, M.M.; Hannon, G.J.; Plasterk, R.H. A micrococcal nuclease homologue in RNAi effector complexes. Nature 2003, 425, 411–414. [Google Scholar] [CrossRef] [PubMed]
- Elbarbary, R.A.; Miyoshi, K.; Myers, J.R.; Du, P.; Ashton, J.M.; Tian, B.; Maquat, L.E. Tudor-SN-mediated endonucleolytic decay of human cell microRNAs promotes G1/S phase transition. Science 2017, 356, 859–862. [Google Scholar] [CrossRef] [Green Version]
- Elbarbary, R.A.; Miyoshi, K.; Hedaya, O.; Myers, J.R.; Maquat, L.E. UPF1 helicase promotes TSN-mediated miRNA decay. Genes Dev. 2017, 31, 1483–1493. [Google Scholar] [CrossRef]
- Cho, H.; Park, O.H.; Park, J.; Ryu, I.; Kim, J.; Ko, J.; Kim, Y.K. Glucocorticoid receptor interacts with PNRC2 in a ligand-dependent manner to recruit UPF1 for rapid mRNA degradation. Proceedings of the PANS 2015, 112, E1540–E1549. [Google Scholar] [CrossRef] [Green Version]
- Ishmael, F.T.; Fang, X.; Houser, K.R.; Pearce, K.; Abdelmohsen, K.; Zhan, M.; Gorospe, M.; Stellato, C. The human glucocorticoid receptor as an RNA-binding protein: Global analysis of glucocorticoid receptor-associated transcripts and identification of a target RNA motif. J. Immunol 2011, 186, 1189–1198. [Google Scholar] [CrossRef] [Green Version]
- Park, O.H.; Park, J.; Yu, M.; An, H.T.; Ko, J.; Kim, Y.K. Identification and molecular characterization of cellular factors required for glucocorticoid receptor-mediated mRNA decay. Genes Dev. 2016, 30, 2093–2105. [Google Scholar] [CrossRef] [Green Version]
- Park, O.H.; Ha, H.; Lee, Y.; Boo, S.H.; Kwon, D.H.; Song, H.K.; Kim, Y.K. Endoribonucleolytic Cleavage of m(6)A-Containing RNAs by RNase P/MRP Complex. Mol. Cell 2019, 74, 494–507. [Google Scholar] [CrossRef] [PubMed]
- Longman, D.; Jackson-Jones, K.A.; Maslon, M.M.; Murphy, L.C.; Young, R.S.; Stoddart, J.J.; Taylor, M.S.; Papadopoulos, D.K.; Cáceres, J.F. Identification of a Nonsense-Mediated Decay pathway at the Endoplasmic Reticulum. Biorxiv 2020. [Google Scholar] [CrossRef]
- Anastasaki, C.; Longman, D.; Capper, A.; Patton, E.E.; Caceres, J.F. Dhx34 and Nbas function in the NMD pathway and are required for embryonic development in zebrafish. Nucleic Acids Res. 2011, 39, 3686–3694. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sieber, J.; Hauer, C.; Bhuvanagiri, M.; Leicht, S.; Krijgsveld, J.; Neu-Yilik, G.; Hentze, M.W.; Kulozik, A.E. Proteomic Analysis Reveals Branch-specific Regulation of the Unfolded Protein Response by Nonsense-mediated mRNA Decay. Mol. Cell. Proteom. 2016, 15, 1584–1597. [Google Scholar] [CrossRef] [Green Version]
- Sakaki, K.; Kaufman, R.J. Interaction between quality control systems for ER protein folding and RNA biogenesis. Worm 2013, 2, e23005. [Google Scholar] [CrossRef]
- Carreras-Sureda, A.; Hetz, C. RNA metabolism: Putting the brake on the UPR. EMBO reports 2015, 16, 545–546. [Google Scholar] [CrossRef] [Green Version]
- Oren, Y.S.; McClure, M.L.; Rowe, S.M.; Sorscher, E.J.; Bester, A.C.; Manor, M.; Kerem, E.; Rivlin, J.; Zahdeh, F.; Mann, M.; et al. The unfolded protein response affects readthrough of premature termination codons. EMBO Mol. Med. 2014, 6, 685–701. [Google Scholar] [CrossRef] [Green Version]
- Yashiro, Y.; Tomita, K. Function and Regulation of Human Terminal Uridylyltransferases. Front. Genet. 2018, 9, 538. [Google Scholar] [CrossRef]
- Zigackova, D.; Vanacova, S. The role of 3’ end uridylation in RNA metabolism and cellular physiology. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2018, 373. [Google Scholar] [CrossRef] [Green Version]
- Menezes, M.R.; Balzeau, J.; Hagan, J.P. 3’ RNA Uridylation in Epitranscriptomics, Gene Regulation, and Disease. Front. Mol. Biosci. 2018, 5, 61. [Google Scholar] [CrossRef] [Green Version]
- Warkocki, Z.; Liudkovska, V.; Gewartowska, O.; Mroczek, S.; Dziembowski, A. Terminal nucleotidyl transferases (TENTs) in mammalian RNA metabolism. Philos. Trans. R Soc. Lond. B Biol. Sci. 2018, 373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boehm, V.; Gerbracht, J.V.; Marx, M.C.; Gehring, N.H. Interrogating the degradation pathways of unstable mRNAs with XRN1-resistant sequences. Nat. Commun. 2016, 7, 13691. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hashimoto, Y.; Takahashi, M.; Sakota, E.; Nakamura, Y. Nonstop-mRNA decay machinery is involved in the clearance of mRNA 5’-fragments produced by RNAi and NMD in Drosophila melanogaster cells. Biochem. Biophys. Res. Commun. 2017, 484, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Arribere, J.A.; Fire, A.Z. Nonsense mRNA suppression via nonstop decay. eLife 2018, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mao, R.; Yang, R.; Chen, X.; Harhaj, E.W.; Wang, X.; Fan, Y. Regnase-1, a rapid response ribonuclease regulating inflammation and stress responses. Cell Mol. Immunol. 2017, 14, 412–422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karousis, E.D.; Nasif, S.; Mühlemann, O. Nonsense-mediated mRNA decay: Novel mechanistic insights and biological impact. Wiley Interdiscip. Rev. RNA 2016, 7, 661–682. [Google Scholar] [CrossRef] [Green Version]
- Hug, N.; Longman, D.; Caceres, J.F. Mechanism and regulation of the nonsense-mediated decay pathway. Nucleic Acids Res. 2016, 44, 1483–1495. [Google Scholar] [CrossRef] [Green Version]
- Huang, L.; Lou, C.H.; Chan, W.; Shum, E.Y.; Shao, A.; Stone, E.; Karam, R.; Song, H.W.; Wilkinson, M.F. RNA homeostasis governed by cell type-specific and branched feedback loops acting on NMD. Mol. Cell 2011, 43, 950–961. [Google Scholar] [CrossRef] [Green Version]
- Wilkinson, M.F.; Cook-Andersen, H. Nonsense shielding: Protecting RNA from decay leads to cancer. EMBO 2019, 38. [Google Scholar] [CrossRef]
- Han, X.; Wei, Y.; Wang, H.; Wang, F.; Ju, Z.; Li, T. Nonsense-mediated mRNA decay: A ‘nonsense’ pathway makes sense in stem cell biology. Nucleic Acids Res. 2018, 46, 1038–1051. [Google Scholar] [CrossRef] [Green Version]
- Hong, D.; Park, T.; Jeong, S. Nuclear UPF1 Is Associated with Chromatin for Transcription-Coupled RNA Surveillance. Mol. Cells 2019, 42, 523–529. [Google Scholar] [CrossRef]
- Azzalin, C.M.; Lingner, J. The double life of UPF1 in RNA and DNA stability pathways. Cell Cycle 2006, 5, 1496–1498. [Google Scholar] [CrossRef]
- Celik, A.; Baker, R.; He, F.; Jacobson, A. High-resolution profiling of NMD targets in yeast reveals translational fidelity as a basis for substrate selection. RNA 2017, 23, 735–748. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heyer, E.E.; Moore, M.J. Redefining the Translational Status of 80S Monosomes. Cell 2016, 164, 757–769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brogna, S.; Wen, J. Nonsense-mediated mRNA decay (NMD) mechanisms. Nat. Struct. Mol. Biol. 2009, 16, 107–113. [Google Scholar] [CrossRef] [PubMed]
- Kay, L.E. Who Wrote the Book of Life? A History of the Genetic Code; Stanford University Press: Stanford, CA, USA, 2000; p. 472. [Google Scholar]
- Kuhn, T.S. The Structure of Scientific Revolutions, 2nd ed.; University of Chicago Press: Chicago, IL, USA, 1970. [Google Scholar]


| UMD | RNA Target | NMD Effectors | Translation Termination | Selected Other Effectors 1 | UPF1 Functions | UPF1 Phosphorylation | (p)-UPF1 Binding Factor | |
|---|---|---|---|---|---|---|---|---|
| ATPase | Helicase | |||||||
| NMD | mRNAs, (lncRNAs; snoRNA hosts) | UPF1+ (in variable combinations): UPF2, UPF3A, UPF3B, SMG1c, SMG6, SMG5/7 | yes | EJC, Mov10, DHX34, SRSF1, NBAS, long 3′UTR decapping and deadenylation factors, 3–5′ and 5′–3′ exonucleases, PNRC2 | yes | yes | yes | RNA? |
| SMD | mRNA | UPF1, UPF2, SMG1 | yes | STAU1, STAU2, dsRNA structures, PNRC2 | yes | yes | yes | UPF2 (STAU) |
| HMD | Histone mRNA | UPF1, SMG1 | yes | RNA stem loop SLBP, ATR, DNA–PK, 3‘hExo, PNRC2 | yes | yes | yes | 3′UTR of histone mRNAs |
| RMD | Proinflammatory cytokine transcripts | UPF1, SMG1 | yes | Stem loop, with Py–Pu-Py-loop, regnase-1 | yes | yes | yes | regnase-1 |
| SRD | mRNAs, circular RNAs | UPF1 | yes | GRBP1 | yes | yes | n.d. | Structured RNA |
| TRIM71-MD | CDKN1A/p21 mRNA | UPF1, SMG1, SMG7 | n.d. | TRIM71, TRIM71-binding stem loop in 3′UTR | n.d. | n.d. | implied 2 | TRIM71? |
| GC-rich 3′UTR-MD | mRNAs with GC-rich 3′UTRs | UPF1, UPF2, SMG1 | n.d. | n.d. | implied | implied | implied | GC-rich mRNA |
| UPF1/SMG7/miRNA-MD | mRNAs with CUG sequences embedded in miRNA seed sequences in 3′UTR | UPF1, SMG7, | n.d. | miRNA loaded AGO, NOT1, NOT3 | n.d. | no | n.d. | CUG embedded in 7mer miRNA seed sequence |
| TumiD | miRNA | UPF1 | no | Tudor staphylococcal/ micrococcal-like nuclease (TSN), AGO2, GW182 | n.d. | yes | No (by implication 2) | TSN |
| GMD | mRNA with GR-binding site often in 5′UTR | UPF1 | no | glucocorticoid, glucocorticoid receptor (GR), ATM, PNRC2, DCP1A, YBX1, HRSP12 | yes | yes | yes | GR-PNRC2 complex |
| ER NMD | ER-localized mRNAs | UPF1 (others?) | n.d. | NBAS, SEC61β | n.d. | n.d. | No (by implication) | NBAS |
| mRNA | NMD-Inducing Feature | 2nd UMD | UMD-Inducing Feature |
|---|---|---|---|
| PTGS2 | uORF | RMD | long structured 3′UTR |
| ATF3 | uORF, PTC introduced by alternative splicing | RMD | long structured 3′UTR |
| GADD45B | 3′UTR intron | long structured 3′UTR | |
| COL22A1 | PTC introduced by alternative splicing | RMD | long structured 3′UTR |
| PEA15 | uORF | UPF1/SMG7/ miRNA-MD | miRNA seed sequence complementarity in 3′UTR |
| NFKBIB | PTC introduced by alternative splicing | RMD | long structured 3′UTR |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lavysh, D.; Neu-Yilik, G. UPF1-Mediated RNA Decay—Danse Macabre in a Cloud. Biomolecules 2020, 10, 999. https://doi.org/10.3390/biom10070999
Lavysh D, Neu-Yilik G. UPF1-Mediated RNA Decay—Danse Macabre in a Cloud. Biomolecules. 2020; 10(7):999. https://doi.org/10.3390/biom10070999
Chicago/Turabian StyleLavysh, Daria, and Gabriele Neu-Yilik. 2020. "UPF1-Mediated RNA Decay—Danse Macabre in a Cloud" Biomolecules 10, no. 7: 999. https://doi.org/10.3390/biom10070999
APA StyleLavysh, D., & Neu-Yilik, G. (2020). UPF1-Mediated RNA Decay—Danse Macabre in a Cloud. Biomolecules, 10(7), 999. https://doi.org/10.3390/biom10070999