The CARF Protein MM_0565 Affects Transcription of the Casposon-Encoded cas1-solo Gene in Methanosarcina mazei Gö1
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Plasmids
2.2. Generation of Plasmids and Construction of Mutant Strains
2.3. Growth of M. mazei
2.4. RNA Extraction
2.5. RACE (Rapid Amplification of cDNA Ends) Analysis
2.6. Quantitative RT-PCR
2.7. Construction of cDNA Libraries for RNA-Seq and Illumina Sequencing
2.8. Bioinformatical Analysis of the RNA-Seq Data
2.9. Purification of Heterologously Expressed His6-MM_0565
2.10. Size-Exclusion Chromatography
2.11. Protein Analysis by Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis (SDS-PAGE)
2.12. Protein Thermal Shift Assay
2.13. Western Blot Analysis
2.14. Microscale Thermophoresis (MST)
2.15. Electrophoretic Mobility Shift Assays (EMSA)
3. Results
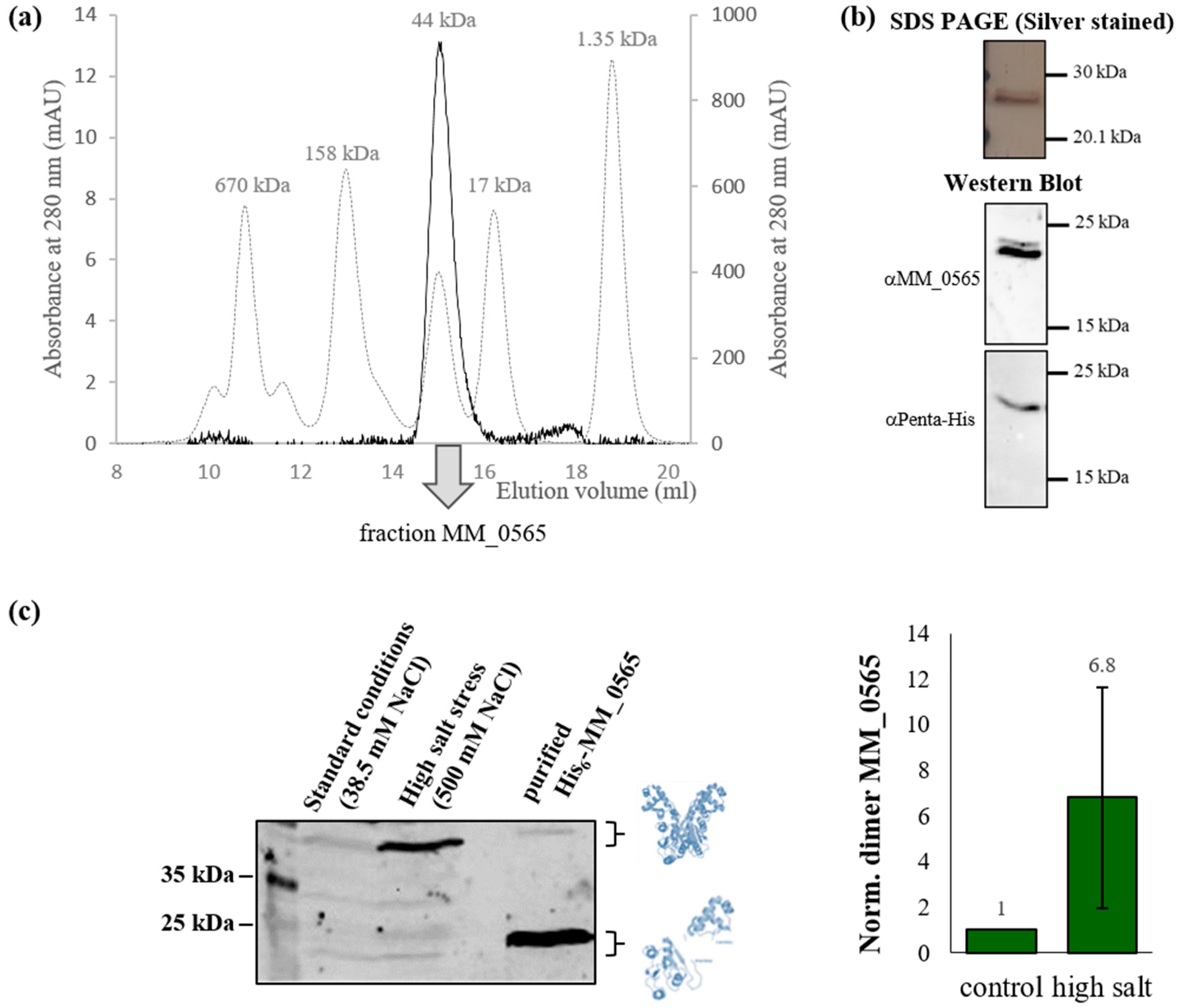
3.1. MM_0565 Is a CRISPR-Cas-Associated Dimeric DNA-Binding Protein
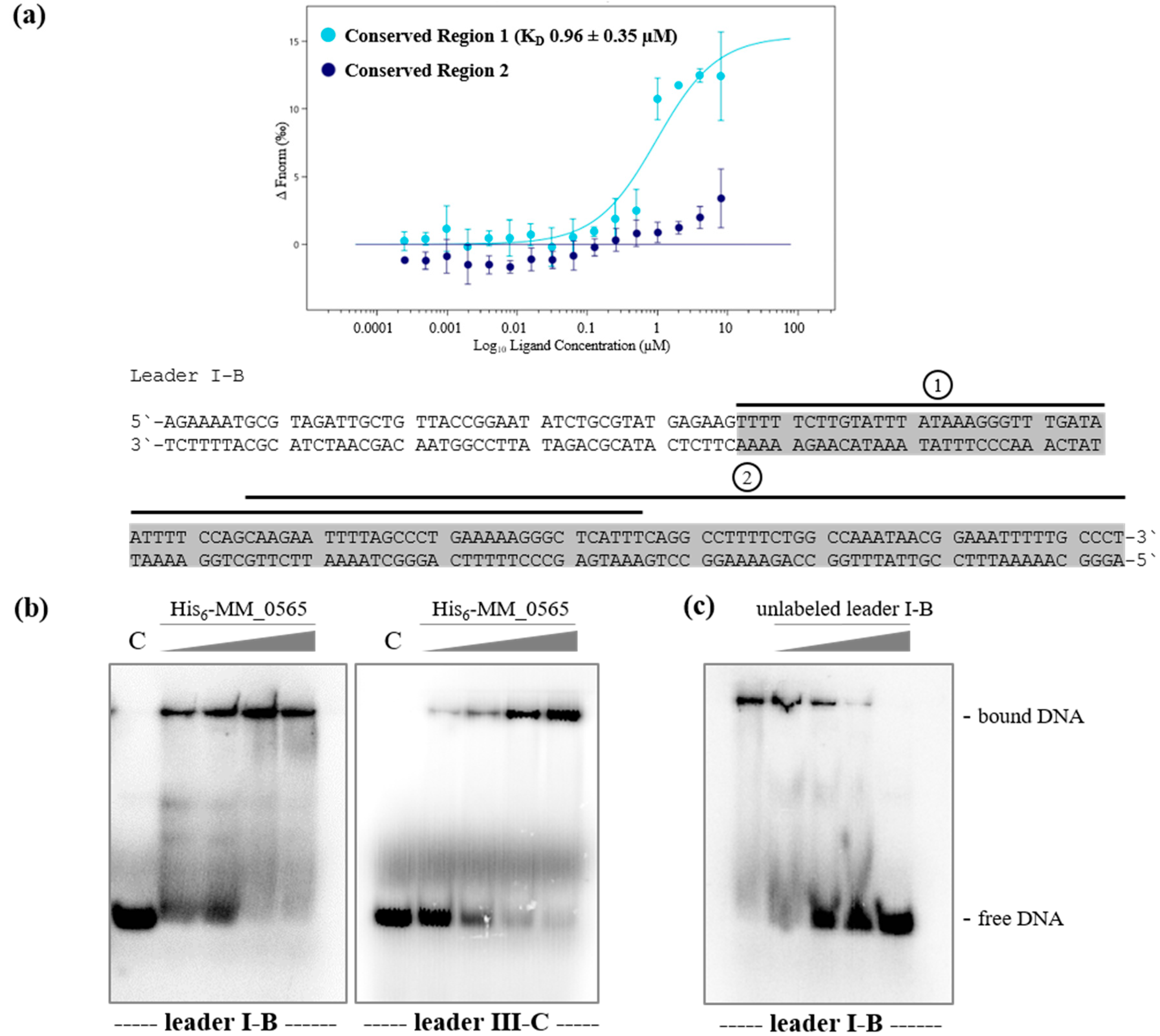
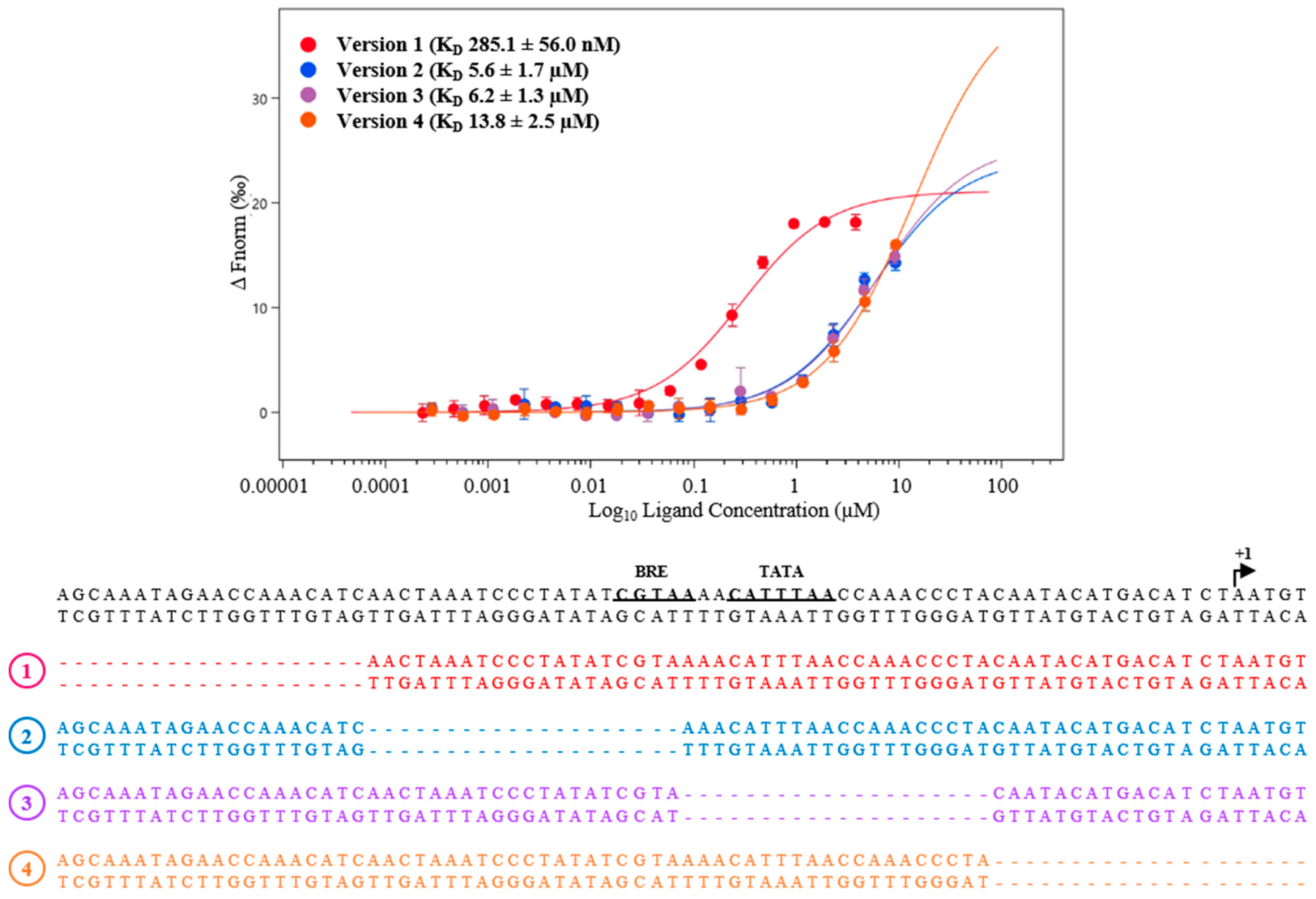
3.2. Regulation under Salt Stress and Binding of MM_0565 to the Leader Regions of Both CRISPR Arrays
3.3. MM_0565 Globally Affects Transcription in M. mazei
4. Discussion
4.1. MM_0565, Structural Features and Its Link to CRISPR-Cas Systems
4.2. MM_0565 Has No Direct Effect on Type I-B cas Gene Transcription
4.3. MM_0565 Might Influence Activity of CRISPR-Based Immunity by Binding to the Leader
4.4. MM_565 Highlights the Position of Casposons as Ancesterors of CRISPR-Cas-Encoded Cas1 Proteins
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Brezgin, S.; Kostyusheva, A.; Kostyushev, D.; Chulanov, V. Dead Cas Systems: Types, Principles, and Applications. Int. J. Mol. Sci. 2019, 20, 6041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khosravi, S.; Ishii, T.; Dreissig, S.; Houben, A. Application and prospects of CRISPR/Cas9-based methods to trace defined genomic sequences in living and fixed plant cells. Chromosome Res. 2019, 28, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Khanzadi, M.N.; Khan, A.A. CRISPR/Cas9: Nature’s gift to prokaryotes and an auspicious tool in genome editing. J. Basic Microbiol. 2019, 60, 91–102. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Grishin, N.V.; Shabalina, S.A.; Wolf, Y.I.; Koonin, E.V. A putative RNA-interference-based immune system in prokaryotes: Computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Boil. Direct 2006, 1, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barrangou, R.; Horvath, P. CRISPR: New Horizons in Phage Resistance and Strain Identification. Annu. Rev. Food Sci. Technol. 2012, 3, 143–162. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Makarova, K.S.; Zhang, F. Diversity, classification and evolution of CRISPR-Cas systems. Curr. Opin. Microbiol. 2017, 37, 67–78. [Google Scholar] [CrossRef]
- Makarova, K.S.; Wolf, Y.I.; Alkhnbashi, O.S.; Costa, F.; Shah, S.A.; Saunders, S.J.; Barrangou, R.; Brouns, S.J.J.; Charpentier, E.; Haft, D.H.; et al. An updated evolutionary classification of CRISPR–Cas systems. Nat. Rev. Genet. 2015, 13, 722–736. [Google Scholar] [CrossRef] [Green Version]
- Mohanraju, P.; Makarova, K.S.; Zetsche, B.; Zhang, F.; Koonin, E.V.; Van Der Oost, J. Diverse evolutionary roots and mechanistic variations of the CRISPR-Cas systems. Science 2016, 353, aad5147. [Google Scholar] [CrossRef] [Green Version]
- Wiedenheft, B.; Sternberg, S.H.; Doudna, J.A. RNA-guided genetic silencing systems in bacteria and archaea. Nature 2012, 482, 331–338. [Google Scholar] [CrossRef]
- Hille, F.; Charpentier, E. CRISPR-Cas: Biology, mechanisms and relevance. Philos. Trans. R. Soc. B Boil. Sci. 2016, 371, 20150496. [Google Scholar] [CrossRef]
- Makarova, K.S.; Wolf, Y.I.; Iranzo, J.; Shmakov, S.A.; Alkhnbashi, O.S.; Brouns, S.J.J.; Charpentier, E.; Cheng, D.; Haft, D.H.; Horvath, P.; et al. Evolutionary classification of CRISPR–Cas systems: A burst of class 2 and derived variants. Nat. Rev. Genet. 2019, 18, 67–83. [Google Scholar] [CrossRef] [PubMed]
- Lillestøl, R.; Redder, P.; Garrett, R.A.; Brügger, K.I. A putative viral defence mechanism in archaeal cells. Archaea 2006, 2, 59–72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, T.-H.; Bachellerie, J.-P.; Rozhdestvensky, T.S.; Bortolin, M.-L.; Huber, H.; Drungowski, M.; Elge, T.; Brosius, J.; Hüttenhofer, A. Identification of 86 candidates for small non-messenger RNAs from the archaeon Archaeoglobus fulgidus. Proc. Natl. Acad. Sci. USA 2002, 99, 7536–7541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hale, C.; Kleppe, K.; Terns, M.P.; Terns, M.P. Prokaryotic silencing (psi)RNAs in Pyrococcus furiosus. RNA 2008, 14, 2572–2579. [Google Scholar] [CrossRef] [Green Version]
- Lillestøl, R.K.; Shah, S.A.; Brügger, K.; Redder, P.; Phan, H.; Christiansen, J.; Garrett, R.A. CRISPR families of the crenarchaeal genus Sulfolobus: Bidirectional transcription and dynamic properties. Mol. Microbiol. 2009, 72, 259–272. [Google Scholar] [CrossRef]
- Tang, T.-H.; Polacek, N.; Żywicki, M.; Huber, H.; Brügger, K.; Garrett, R.; Bachellerie, J.P.; Hüttenhofer, A. Identification of novel non-coding RNAs as potential antisense regulators in the archaeon Sulfolobus solfataricus. Mol. Microbiol. 2004, 55, 469–481. [Google Scholar] [CrossRef]
- Juranek, S.; Eban, T.; Altuvia, Y.; Brown, M.; Morozov, P.; Tuschl, T.; Margalit, H. A genome-wide view of the expression and processing patterns of Thermus thermophilus HB8 CRISPR RNAs. RNA 2012, 18, 783–794. [Google Scholar] [CrossRef] [Green Version]
- Richter, C.; Chang, J.T.; Fineran, P.C. Function and regulation of clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR associated (Cas) systems. Viruses 2012, 4, 2291–2311. [Google Scholar] [CrossRef]
- Crawley, A.B.; Henriksen, E.D.; Stout, E.; Brandt, K.; Barrangou, R. Characterizing the activity of abundant, diverse and active CRISPR-Cas systems in lactobacilli. Sci. Rep. 2018, 8, 11544. [Google Scholar] [CrossRef] [Green Version]
- Young, J.C.; Dill, B.D.; Pan, C.; Hettich, R.L.; Banfield, J.F.; Shah, M.; Fremaux, C.; Horvath, P.; Barrangou, R.; Verberkmoes, N.C. Phage-Induced Expression of CRISPR-Associated Proteins Is Revealed by Shotgun Proteomics in Streptococcus thermophilus. PLoS ONE 2012, 7, e38077. [Google Scholar] [CrossRef]
- Agari, Y.; Sakamoto, K.; Tamakoshi, M.; Oshima, T.; Kuramitsu, S.; Shinkai, A. Transcription Profile of Thermus thermophilus CRISPR Systems after Phage Infection. J. Mol. Boil. 2010, 395, 270–281. [Google Scholar] [CrossRef] [PubMed]
- Vestergaard, G.; Garrett, R.A.; Shah, S.A. CRISPR adaptive immune systems of Archaea. RNA Boil. 2014, 11, 156–167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, T.; Li, Y.; Wang, X.; Ye, Q.; Li, H.; Liang, Y.; She, Q.; Peng, N. Transcriptional regulator-mediated activation of adaptation genes triggers CRISPR de novo spacer acquisition. Nucleic Acids Res. 2015, 43, 1044–1055. [Google Scholar] [CrossRef]
- He, F.; Vestergaard, G.; Peng, W.; She, Q.; Peng, X. CRISPR-Cas type I-A Cascade complex couples viral infection surveillance to host transcriptional regulation in the dependence of Csa3b. Nucleic Acids Res. 2016, 45, 1902–1913. [Google Scholar] [CrossRef] [Green Version]
- Nickel, L.; Weidenbach, K.; Jäger, D.; Backofen, R.; Lange, S.J.; Heidrich, N.; Schmitz, R.A. Two CRISPR-Cas systems inMethanosarcina mazeistrain Gö1 display common processing features despite belonging to different types I and III. RNA Boil. 2013, 10, 779–791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weidenbach, K.; Nickel, L.; Neve, H.; Alkhnbashi, O.S.; Künzel, S.; Kupczok, A.; Bauersachs, T.; Cassidy, L.; Tholey, A.; Backofen, R.; et al. Methanosarcina Spherical Virus, a Novel Archaeal Lytic Virus Targeting Methanosarcina Strains. J. Virol. 2017, 91, e00955-17. [Google Scholar] [CrossRef] [Green Version]
- Nickel, L.; Ulbricht, A.; Alkhnbashi, O.S.; Förstner, K.U.; Cassidy, L.; Weidenbach, K.; Backofen, R.; Schmitz, R.A. Cross-cleavage activity of Cas6b in crRNA processing of two different CRISPR-Cas systems in Methanosarcina mazei Gö1. RNA Boil. 2018, 16, 492–503. [Google Scholar] [CrossRef] [Green Version]
- Deppenmeier, U.; Johann, A.; Hartsch, T.; Merkl, R.; Schmitz, R.A.; Martinez-Arias, R.; Henne, A.; Wiezer, A.; Bäumer, S.; Jacobi, C.; et al. The genome of Methanosarcina mazei: Evidence for lateral gene transfer between bacteria and archaea. J. Mol. Microbiol. Biotechnol. 2002, 4, 453–461. [Google Scholar]
- Ehlers, C.; Weidenbach, K.; Veit, K.; Deppenmeier, U.; Metcalf, W.W.; Schmitz, R.A. Development of genetic methods and construction of a chromosomal glnK1 mutant in Methanosarcina mazei strain Gö1. Mol. Genet. Genom. 2005, 273, 290–298. [Google Scholar] [CrossRef]
- Hanahan, D. Studies on transformation of Escherichia coli with plasmids. J. Mol. Boil. 1983, 166, 557–580. [Google Scholar] [CrossRef]
- Miller, V.L.; Mekalanos, J.J. A novel suicide vector and its use in construction of insertion mutations: Osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 1988, 170, 2575–2583. [Google Scholar] [CrossRef] [Green Version]
- Inoue, H.; Nojima, H.; Okayama, H. High efficiency transformation of Escherichia coli with plasmids. Gene 1990, 96, 23–28. [Google Scholar] [CrossRef]
- Guss, A.M.; Rother, M.; Zhang, J.K.; Kulkkarni, G.; Metcalf, W.W. New methods for tightly regulated gene expression and highly efficient chromosomal integration of cloned genes for Methanosarcina species. Archaea 2008, 2, 193–203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Metcalf, W.W.; Zhang, J.K.; Apolinario, E.; Sowers, K.R.; Wolfe, R.S. A genetic system for Archaea of the genus Methanosarcina: Liposome-mediated transformation and construction of shuttle vectors. Proc. Natl. Acad. Sci. USA 1997, 94, 2626–2631. [Google Scholar] [CrossRef] [Green Version]
- Weidenbach, K.; Ehlers, C.; Kock, J.; Schmitz, R.A.; Schmitz, R.A. NrpRII mediates contacts between NrpRI and general transcription factors in the archaeon Methanosarcina mazei Gö1. FEBS J. 2010, 277, 4398–4411. [Google Scholar] [CrossRef] [PubMed]
- Jäger, M.; Sharma, C.M.; Thomsen, J.; Ehlers, C.; Vogel, J.; Schmitz, R.A. Deep sequencing analysis of the Methanosarcina mazei Go1 transcriptome in response to nitrogen availability. Proc. Natl. Acad. Sci. USA 2009, 106, 21878–21882. [Google Scholar] [CrossRef] [Green Version]
- Weidenbach, K.; Glöer, J.; Ehlers, C.; Sandman, K.; Reeve, J.N.; Schmitz, R.A.; Schmitz, R.A. Deletion of the archaeal histone inMethanosarcina mazeiGö1 results in reduced growth and genomic transcription. Mol. Microbiol. 2008, 67, 662–671. [Google Scholar] [CrossRef]
- Veit, K.; Ehlers, C.; Schmitz, R.A. Effects of nitrogen and carbon sources on transcription of soluble methyltransferases in Methanosarcina mazei strain Gö1. J. Bacteriol. 2005, 187, 6147–6154. [Google Scholar] [CrossRef] [Green Version]
- Dugar, G.; Herbig, A.; Förstner, K.U.; Heidrich, N.; Reinhardt, R.; Nieselt, K.; Sharma, C.M. High-Resolution Transcriptome Maps Reveal Strain-Specific Regulatory Features of Multiple Campylobacter jejuni Isolates. PLoS Genet. 2013, 9, e1003495. [Google Scholar] [CrossRef]
- Prasse, D.; Thomsen, J.; De Santis, R.; Muntel, J.; Becher, D.; Schmitz, R.A.; Schmitz, R.A. First description of small proteins encoded by spRNAs in Methanosarcina mazei strain Gö1. Biochimie 2015, 117, 138–148. [Google Scholar] [CrossRef]
- Förstner, K.U.; Vogel, J.; Sharma, C.M. READemption—A tool for the computational analysis of deep-sequencing-based transcriptome data. Bioinformatics 2014, 30, 3421–3423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoffmann, S.; Otto, C.; Kurtz, S.; Sharma, C.M.; Khaitovich, P.; Vogel, J.; Stadler, P.F.; Hackermüller, J. Fast Mapping of Short Sequences with Mismatches, Insertions and Deletions Using Index Structures. PLoS Comput. Boil. 2009, 5, e1000502. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 002832. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Studier, F.W. Protein production by auto-induction in high-density shaking cultures. Protein Expr. Purif. 2005, 41, 207–234. [Google Scholar] [CrossRef] [PubMed]
- Laemmli, U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Rabilloud, T.; Carpentier, G.; Tarroux, P. Improvement and simplification of low-background silver staining of proteins by using sodium dithionite. Electrophoresis 1988, 9, 288–291. [Google Scholar] [CrossRef]
- Kisiela, M.; Faust, A.; Ebert, B.; Maser, E.; Scheidig, A.J. Crystal structure and catalytic characterization of the dehydrogenase/reductase SDR family member 4 ( DHRS 4) from Caenorhabditis elegans. FEBS J. 2017, 285, 275–293. [Google Scholar] [CrossRef] [Green Version]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1989. [Google Scholar]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.; Sternberg, M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [Green Version]
- Makarova, K.S.; Anantharaman, V.; Grishin, N.V.; Koonin, E.V.; Aravind, L. CARF and WYL domains: Ligand-binding regulators of prokaryotic defense systems. Front. Genet. 2014, 5, 102. [Google Scholar] [CrossRef]
- Martinez-Hackert, E.; Stock, A.M. Structural relationships in the OmpR family of winged-helix transcription factors 1 1Edited by M. Gottesman. J. Mol. Boil. 1997, 269, 301–312. [Google Scholar] [CrossRef]
- Teichmann, M.; Dumay-Odelot, H.; Fribourg, S. Structural and functional aspects of winged-helix domains at the core of transcription initiation complexes. Transcription 2012, 3, 2–7. [Google Scholar] [CrossRef] [PubMed]
- Lintner, N.G.; Frankel, K.A.; Tsutakawa, S.E.; Alsbury, D.L.; Copié, V.; Young, M.J.; Tainer, J.A.; Lawrence, C.M. The Structure of the CRISPR-Associated Protein Csa3 Provides Insight into the Regulation of the CRISPR/Cas System. J. Mol. Boil. 2011, 405, 939–955. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holm, L.; Park, J. DaliLite workbench for protein structure comparison. Bioinformatics 2000, 16, 566–567. [Google Scholar] [CrossRef] [PubMed]
- Armougom, F.; Moretti, S.; Poirot, O.; Audic, S.; Dumas, P.; Schaeli, B.; Keduas, V.; Notredame, C. Expresso: Automatic incorporation of structural information in multiple sequence alignments using 3D-Coffee. Nucleic Acids Res. 2006, 34, W604–W608. [Google Scholar] [CrossRef]
- Díez-Villaseñor, C.; Guzmán, N.M.; Almendros, C.; García-Martínez, J.; Mojica, F.J. CRISPR-spacer integration reporter plasmids reveal distinct genuine acquisition specificities among CRISPR-Cas I-E variants of Escherichia coli. RNA Boil. 2013, 10, 792–802. [Google Scholar] [CrossRef] [Green Version]
- Erdmann, S.; Garrett, R.A. Selective and hyperactive uptake of foreign DNA by adaptive immune systems of an archaeon via two distinct mechanisms. Mol. Microbiol. 2012, 85, 1044–1056. [Google Scholar] [CrossRef]
- Yosef, I.; Goren, M.G.; Qimron, U. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 2012, 40, 5569–5576. [Google Scholar] [CrossRef] [Green Version]
- Nuñez, J.K.; Lee, A.S.Y.; Engelman, A.; Doudna, J.A. Integrase-mediated spacer acquisition during CRISPR–Cas adaptive immunity. Nature 2015, 519, 193–198. [Google Scholar] [CrossRef] [Green Version]
- Crooks, G.E.; Hon, G.; Chandonia, J.-M.; Brenner, S.E. WebLogo: A Sequence Logo Generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [Green Version]
- Makarova, K.S.; Wolf, Y.I.; Koonin, E.V. The basic building blocks and evolution of CRISPR-Cas systems. Biochem. Soc. Trans. 2013, 41, 1392–1400. [Google Scholar] [CrossRef]
- Krupovic, M.; Makarova, K.S.; Forterre, P.; Prangishvili, D.; Koonin, E.V. Casposons: A new superfamily of self-synthesizing DNA transposons at the origin of prokaryotic CRISPR-Cas immunity. BMC Boil. 2014, 12, 36. [Google Scholar] [CrossRef] [PubMed]
- Pul, Ü.; Wurm, R.; Arslan, Z.; Geißen, R.; Hofmann, N.; Wagner, R. Identification and characterization of E. coli CRISPR-caspromoters and their silencing by H-NS. Mol. Microbiol. 2010, 75, 1495–1512. [Google Scholar] [CrossRef]
- Westra, E.R.; Pul, Ü.; Heidrich, N.; Jore, M.M.; Lundgren, M.; Stratmann, T.; Wurm, R.; Raine, A.; Mescher, M.; Van Heereveld, L.; et al. H-NS-mediated repression of CRISPR-based immunity in Escherichia coli K12 can be relieved by the transcription activator LeuO. Mol. Microbiol. 2010, 77, 1380–1393. [Google Scholar] [CrossRef] [PubMed]
- Medina-Aparicio, L.; Rebollar-Flores, J.E.; Gallego-Hernandez, A.L.; Vazquez, A.; Olvera, L.; Gutierrez-Rios, R.M.; Calva, E.; Hernández-Lucas, I. The CRISPR/Cas Immune System Is an Operon Regulated by LeuO, H-NS, and Leucine-Responsive Regulatory Protein in Salmonella enterica Serovar Typhi. J. Bacteriol. 2011, 193, 2396–2407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, T.; Liu, Z.; Ye, Q.; Pan, S.; Wang, X.; Li, Y.; Peng, W.; Liang, Y.; She, Q.; Peng, N. Coupling transcriptional activation of CRISPR-Cas system and DNA repair genes by Csa3a in Sulfolobus islandicus. Nucleic Acids Res. 2017, 45, 8978–8992. [Google Scholar] [CrossRef] [PubMed]
- Goren, M.G.; Doron, S.; Globus, R.; Amitai, G.; Sorek, R.; Qimron, U. Repeat Size Determination by Two Molecular Rulers in the Type I-E CRISPR Array. Cell Rep. 2016, 16, 2811–2818. [Google Scholar] [CrossRef] [Green Version]
- Nuñez, J.K.; Bai, L.; Harrington, L.B.; Hinder, T.L.; Doudna, J.A. CRISPR Immunological Memory Requires a Host Factor for Specificity. Mol. Cell 2016, 62, 824–833. [Google Scholar] [CrossRef] [Green Version]
- Shah, S.A.; Garrett, R.A. CRISPR/Cas and Cmr modules, mobility and evolution of adaptive immune systems. Res. Microbiol. 2011, 162, 27–38. [Google Scholar] [CrossRef]
- Bradde, S.; Mora, T.; Walczak, A.M. Cost and benefits of clustered regularly interspaced short palindromic repeats spacer acquisition. Philos. Trans. R. Soc. B Boil. Sci. 2019, 374, 20180095. [Google Scholar] [CrossRef] [Green Version]
- Krupovic, M.; Shmakov, S.; Makarova, K.S.; Forterre, P.; Koonin, E.V. Recent Mobility of Casposons, Self-Synthesizing Transposons at the Origin of the CRISPR-Cas Immunity. Genome Boil. Evol. 2016, 8, 375–386. [Google Scholar] [CrossRef]
- Casacuberta, E.; González, J. The impact of transposable elements in environmental adaptation. Mol. Ecol. 2013, 22, 1503–1517. [Google Scholar] [CrossRef] [PubMed]
- Hickman, A.B.; Kailasan, S.; Genzor, P.; Haase, A.D.; Dyda, F. Casposase structure and the mechanistic link between DNA transposition and spacer acquisition by CRISPR-Cas. eLife 2020, 9, 9. [Google Scholar] [CrossRef] [PubMed]
- Babu, M.; Beloglazova, N.; Flick, R.; Graham, C.; Skarina, T.; Nocek, B.; Gagarinova, A.; Pogoutse, O.; Brown, G.; Binkowski, A.; et al. A dual function of the CRISPR-Cas system in bacterial antivirus immunity and DNA repair. Mol. Microbiol. 2010, 79, 484–502. [Google Scholar] [CrossRef] [PubMed] [Green Version]





| ORF | Gene/Protein | log2 Fold Change | Padj | |
|---|---|---|---|---|
| Upregulated | ||||
| Energy metabolism | MM_1439 | Methylcobalamin:coenzyme M methyltransferase MtbA2 | 1.51 | 5.69 × 10−9 |
| MM_1438 | Monomethylamine corrinoid protein MtmC1 | 1.55 | 5.69 × 10−9 | |
| MM_1437 | Monomethylamine:corrinoid methyltransferase MtmB1 | 1.86 | 3.54 × 10−8 | |
| MM_1436 | Monomethylamine:corrinoid methyltransferase MtmB1 (C-terminal domain) | 2.26 | 2.26 × 10−13 | |
| MM_1435 | Monomethylamine permease MtmP | 2.17 | 1.05 × 10−13 | |
| MM_1434 | Monomethylamine permease MtmP (C-terminal domain) | 2.27 | 5.36 × 10−12 | |
| MM_1433 | Efflux RND transporter permease subunit | 2.47 | 1.72 × 10−16 | |
| MM_1694 | Dimethylamine:corrinoid methyltransferase MtbB1 (C-terminal domain) | 1.54 | 1.76 × 10−5 | |
| MM_1693 | Dimethylamine:corrinoid methyltransferase MtbB1 | 2.00 | 8.15 × 10−10 | |
| MM_0388 | Heterodisulfate reductase, subunit HdrC | 1.54 | 2.05 × 10−9 | |
| MM_0389 | Heterodisulfate reductase, subunit HdrB | 1.56 | 5.50 × 10−10 | |
| MM_0753 | 7-cyano-7-deazaguanine synthase queC | 1.56 | 1.23 × 10−5 | |
| MM_1055 | Trimethylamine corrionid protein | 1.78 | 0.0005 | |
| MM_2084 | CO dehydrogenase/acetyl-CoA synthase, gamma subunit | 1.83 | 7.47 × 10−12 | |
| MM_3334 | Monomethylamine corrinoid protein MtmC2 | 1.83 | 3.26 × 10−10 | |
| MM_3335 | Monomethylamine:corrinoid methyltransferase MtmB2 | 2.08 | 1.44 × 10−11 | |
| MM_0465 | Cobalamin biosynthesis protein CbiM | 1.95 | 5.94 × 10−15 | |
| MM_0337 | Tryptophan synthase, beta chain | 2.25 | 2.01 × 10−20 | |
| MM_1738 | Heme biosynthesis protein | 2.25 | 2.39 × 10−9 | |
| MM_1737 | Heme biosynthesis protein | 3.25 | 8.66 × 10−25 | |
| MM_1733 | Metalloproteinase | 3.04 | 1.18 × 10−8 | |
| MM_1734 | Conserved protein | 3.37 | 1.43 × 10−14 | |
| MM_2692 | NAD(P)/FAD-dependent oxidoreductase | 6.91 | 2.13 × 10−66 | |
| Membrane associated | MM_1800 | queuosine precursor transporter | 1.70 | 3.81 × 10−5 |
| MM_1799 | Conserved protein | 1.77 | 8.73 × 10−9 | |
| MM_0464 | Cobalt transport protein CbiN | 2.24 | 5.21 × 10−19 | |
| MM_1362 | ABC transporter, periplasmic binding protein | 2.43 | 2.22 × 10−24 | |
| Transposons | MM_0766 | Transposase | 2.51 | 3.16 × 10−22 |
| MM_2686 | Transposase | 2.71 | 2.25 × 10−6 | |
| MM_3249 | Cas1-solo | 3.04 | 1.96 × 10−24 | |
| MM_3250 | Hypothetical protein | 5.09 | 2.33 × 10−81 | |
| Others | MM_2688 | radical SAM protein | 4.42 | 4.20 × 10−22 |
| MM_2693 | archaeosine biosynthesis radical SAM protein RaSEA | 8.65 | 1.02 × 10−12 | |
| Unknown function | MM_1613 | Conserved protein (DUF111 family protein) | 1.53 | 0.004 |
| MM_1731 | Conserved protein | 2.17 | 2.09 × 10−8 | |
| MM_2118 | PEF-CTERM sorting domain-containing protein | 2.20 | 3.37 × 10−17 | |
| MM_3251 | Hypothetical protein | 2.28 | 2.94 × 10−5 | |
| MM_1612 | Conserved protein | 2.40 | 7.11 × 10−10 | |
| MM_2687 | Hypothetical protein | 4.40 | 6.85 × 10−21 | |
| MM_2691 | Hypothetical protein | 6.49 | 6.84 × 10−56 | |
| Downregulated | ||||
| Energy metabolism | MM_3043 | Coenzyme F420 hydrogenase | −1.56 | 3.00 × 10−9 |
| MM_3042 | Coenzyme F420 hydrogenase | −1.68 | 1.34 × 10−11 | |
| MM_2005 | Phosphate-binding protein | −1.90 | 2.67 × 10−7 | |
| MM_0977 | F420-dependent NADP reductase | −1.98 | 9.62 × 10−9 | |
| Membrane associated | MM_2576 | Ferrous iron transport protein B | −1.58 | 1.30 × 10−5 |
| MM_1909 | Glutathione-regulated potassium-efflux system protein | −5.21 | 5.30 × 10−84 | |
| Transposons | MM_2590 | Transposase | −2.34 | 1.63 × 10−8 |
| MM_2699 | Transposase | −3.16 | 2.80 × 10−12 | |
| Transport | MM_0077 | Metallophosphoesterase | −1.80 | 1.48 × 10−10 |
| Unknown function | MM_2575 | Hypothetical protein | −1.53 | 0.006 |
| MM_2602 | Hypothetical protein | −1.59 | 1.50 × 10−5 | |
| MM_0978 | Hypothetical protein | −1.64 | 2.16 × 10−5 | |
| MM_1865 | Conserved protein | −1.66 | 8.15 × 10−10 | |
| MM_3369 | Hypothetical protein | −1.99 | 0.0004 | |
| MM_1864 | Conserved protein | −4.35 | 5.37 × 10−77 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ulbricht, A.; Nickel, L.; Weidenbach, K.; Vargas Gebauer, H.; Kießling, C.; Förstner, K.U.; Schmitz, R.A. The CARF Protein MM_0565 Affects Transcription of the Casposon-Encoded cas1-solo Gene in Methanosarcina mazei Gö1. Biomolecules 2020, 10, 1161. https://doi.org/10.3390/biom10081161
Ulbricht A, Nickel L, Weidenbach K, Vargas Gebauer H, Kießling C, Förstner KU, Schmitz RA. The CARF Protein MM_0565 Affects Transcription of the Casposon-Encoded cas1-solo Gene in Methanosarcina mazei Gö1. Biomolecules. 2020; 10(8):1161. https://doi.org/10.3390/biom10081161
Chicago/Turabian StyleUlbricht, Andrea, Lisa Nickel, Katrin Weidenbach, Herman Vargas Gebauer, Claudia Kießling, Konrad U. Förstner, and Ruth A. Schmitz. 2020. "The CARF Protein MM_0565 Affects Transcription of the Casposon-Encoded cas1-solo Gene in Methanosarcina mazei Gö1" Biomolecules 10, no. 8: 1161. https://doi.org/10.3390/biom10081161
APA StyleUlbricht, A., Nickel, L., Weidenbach, K., Vargas Gebauer, H., Kießling, C., Förstner, K. U., & Schmitz, R. A. (2020). The CARF Protein MM_0565 Affects Transcription of the Casposon-Encoded cas1-solo Gene in Methanosarcina mazei Gö1. Biomolecules, 10(8), 1161. https://doi.org/10.3390/biom10081161





