Elucidating the Inhibitory Effect of Resveratrol and Its Structural Analogs on Selected Nucleotide-Related Enzymes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Protein Structure Preparation
2.2. Ligand Preparation
2.3. Ligand-Protein Docking
2.4. MM-GBSA Calculation
2.5. Molecular Dynamics Simulation
2.6. DNA Oligo Substrates Extension Assay
3. Results
3.1. Inhibitory Effect of Resveratrol on DNA Polymerase
3.2. Inhibitory Effect on HIV-1 Reverse Transcriptase
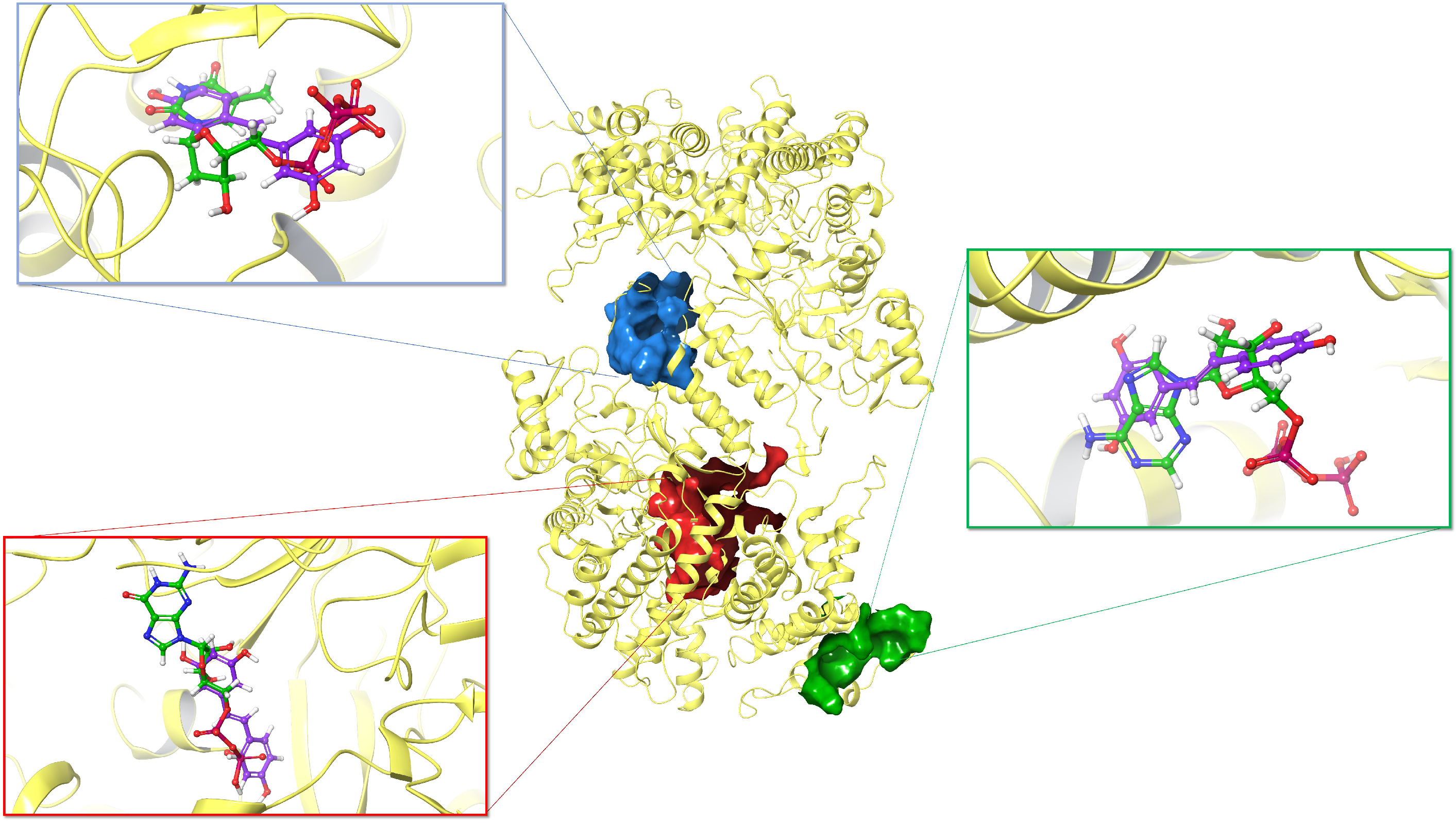
3.3. Inhibitory Effect on Ribonucleotide Reductase
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| MD simulation | molecular dynamics simulation |
| PDB | Protein Data Bank |
| RMSD | rootmean-square deviation |
| M8 | 3,3,4,4,5,5-hexahydroxystilbene |
| DRG | trans-3,5-dihydroxy-4-methoxystilbene |
| 4,4-DHS | trans-4,4-dihydroxystilbene |
| HPSB | 3-hydroxypterostilbene |
| 3,4,5-THS | trans-3,4,5-trihydroxystilbene |
| dCTP | deoxycytidine triphosphate |
| dATP | deoxyadenosine triphosphate |
| ddATP | 2’,3-dideoxyadenosine triphosphate |
| RNR | ribonucleotide reductase |
| Ki | inhibition constant for the inhibitor |
References
- Schneider, Y.; Vincent, F.; Duranton, B.; Badolo, L.; Gossé, F.; Bergmann, C.; Seiler, N.; Raul, F. Anti-proliferative effect of resveratrol, a natural component of grapes and wine, on human colonic cancer cells. Cancer Lett. 2000, 158, 85–91. [Google Scholar] [CrossRef]
- Chin, Y.T.; Hsieh, M.T.; Yang, S.H.; Tsai, P.W.; Wang, S.H.; Wang, C.C.; Lee, Y.S.; Cheng, G.Y.; Huang, W.C.; London, D.; et al. Anti-proliferative and gene expression actions of resveratrol in breast cancer cells in vitro. Oncotarget 2014, 5, 12891. [Google Scholar] [CrossRef] [Green Version]
- Kundu, J.K.; Surh, Y.J. Cancer chemopreventive and therapeutic potential of resveratrol: Mechanistic perspectives. Cancer Lett. 2008, 269, 243–261. [Google Scholar] [CrossRef]
- Gupta, S.C.; Kannappan, R.; Reuter, S.; Kim, J.H.; Aggarwal, B.B. Chemosensitization of tumors by resveratrol. Ann. N. Y. Acad. Sci. 2011, 1215, 150. [Google Scholar] [CrossRef] [Green Version]
- Hsieh, T.-c. (Ed.) Resveratrol: State-of-the-Art Science and Health Applications-Actionable Targets and Mechanisms Of Resveratrol; World Scientific: Singapore, 2018. [Google Scholar]
- Wang, Y.; Catana, F.; Yang, Y.; Roderick, R.; van Breemen, R.B. An LC-MS method for analyzing total resveratrol in grape juice, cranberry juice, and in wine. J. Agric. Food Chem. 2002, 50, 431–435. [Google Scholar] [CrossRef]
- Campagna, M.; Rivas, C. Antiviral activity of resveratrol. Biochem. Soc. Trans. 2010. [Google Scholar] [CrossRef] [PubMed]
- Han, G.; Xia, J.; Gao, J.; Inagaki, Y.; Tang, W.; Kokudo, N. Anti-tumor effects and cellular mechanisms of resveratrol. Drug Discov. Ther. 2015, 9, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Docherty, J.J.; Fu, M.M.H.; Stiffler, B.S.; Limperos, R.J.; Pokabla, C.M.; DeLucia, A.L. Resveratrol inhibition of herpes simplex virus replication. Antivir. Res. 1999, 43, 145–155. [Google Scholar] [CrossRef]
- Kulkarni, S.S.; Cantó, C. The molecular targets of resveratrol. Biochim. Et Biophys. Acta (BBA) Mol. Basis Dis. 2015, 1852, 1114–1123. [Google Scholar] [CrossRef] [Green Version]
- Chan, C.N.; Trinité, B.; Levy, D.N. Potent inhibition of HIV-1 replication in resting CD4 T cells by resveratrol and pterostilbene. Antimicrob. Agents Chemother. 2017, 61, e00408-17. [Google Scholar] [CrossRef] [Green Version]
- Locatelli, G.A.; Savio, M.; Forti, L.; Shevelev, I.; Ramadan, K.; Stivala, L.A.; Vannini, V.; Hübscher, U.; Spadari, S.; Maga, G. Inhibition of mammalian DNA polymerases by resveratrol: Mechanism and structural determinants. Biochem. J. 2005, 389, 259–268. [Google Scholar] [CrossRef] [PubMed]
- Fontecave, M.; Lepoivre, M.; Elleingand, E.; Gerez, C.; Guittet, O. Resveratrol, a remarkable inhibitor of ribonucleotide reductase. FEBS Lett. 1998, 421, 277–279. [Google Scholar] [CrossRef] [Green Version]
- Berdis, A.J. Inhibiting DNA polymerases as a therapeutic intervention against cancer. Front. Mol. Biosci. 2017, 4, 78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lange, S.S.; Takata, K.i.; Wood, R.D. DNA polymerases and cancer. Nat. Rev. Cancer 2011, 11, 96–110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maynard, S.; Fang, E.F.; Scheibye-Knudsen, M.; Croteau, D.L.; Bohr, V.A. DNA damage, DNA repair, aging, and neurodegeneration. Cold Spring Harb. Perspect. Med. 2015, 5, a025130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szekeres, T.; Fritzer-Szekeres, M.; Saiko, P.; Jäger, W. Resveratrol and resveratrol analogues—structure—activity relationship. Pharm. Res. 2010, 27, 1042–1048. [Google Scholar] [CrossRef]
- Ovesna, Z.; Horvathova-Kozics, K. Structure-activity relationship of trans-resveratrol and its analogues. Neoplasma 2005, 52, 450. [Google Scholar]
- Barja, G. Free radicals and aging. TRENDS Neurosci. 2004, 27, 595–600. [Google Scholar] [CrossRef]
- Valko, M.; Rhodes, C.; Moncol, J.; Izakovic, M.; Mazur, M. Free radicals, metals and antioxidants in oxidative stress-induced cancer. Chem. Biol. Interact. 2006, 160, 1–40. [Google Scholar] [CrossRef]
- Smith, M.A.; Zhu, X.; Tabaton, M.; Liu, G.; McKeel, D.W., Jr.; Cohen, M.L.; Wang, X.; Siedlak, S.L.; Dwyer, B.E.; Hayashi, T.; et al. Increased iron and free radical generation in preclinical Alzheimer disease and mild cognitive impairment. J. Alzheimer’s Dis. 2010, 19, 363–372. [Google Scholar] [CrossRef] [Green Version]
- Park, D.; Jeong, H.; Lee, M.N.; Koh, A.; Kwon, O.; Yang, Y.R.; Noh, J.; Suh, P.G.; Park, H.; Ryu, S.H. Resveratrol induces autophagy by directly inhibiting mTOR through ATP competition. Sci. Rep. 2016, 6, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoon, S.H.; Kim, Y.S.; Ghim, S.Y.; Song, B.H.; Bae, Y.S. Inhibition of protein kinase CKII activity by resveratrol, a natural compound in red wine and grapes. Life Sci. 2002, 71, 2145–2152. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, C.L.; Chang, L.S.; Zhang, P.; Hao, H.; Zhu, L.; Lan Toomey, N.; Lee, M.Y. Molecular cloning of the cDNA for the catalytic subunit of human DNA polymerase δ. Nucleic Acids Res. 1992, 20, 735–745. [Google Scholar] [CrossRef] [PubMed]
- Chung, D.W.; Zhang, J.; Tan, C.K.; Davie, E.W.; So, A.G.; Downey, K.M. Primary structure of the catalytic subunit of human DNA polymerase delta and chromosomal location of the gene. Proc. Natl. Acad. Sci. USA 1991, 88, 11197–11201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swan, M.K.; Johnson, R.E.; Prakash, L.; Prakash, S.; Aggarwal, A.K. Structural basis of high-fidelity DNA synthesis by yeast DNA polymerase δ. Nat. Struct. Mol. Biol. 2009, 16, 979. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sastry, G.M.; Adzhigirey, M.; Day, T.; Annabhimoju, R.; Sherman, W. Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aided Mol. Des. 2013, 27, 221–234. [Google Scholar] [CrossRef]
- Shelley, J.C.; Cholleti, A.; Frye, L.L.; Greenwood, J.R.; Timlin, M.R.; Uchimaya, M. Epik: A software program for pK a prediction and protonation state generation for drug-like molecules. J. Comput. Aided Mol. Des. 2007, 21, 681–691. [Google Scholar] [CrossRef]
- Roos, K.; Wu, C.; Damm, W.; Reboul, M.; Stevenson, J.M.; Lu, C.; Dahlgren, M.K.; Mondal, S.; Chen, W.; Wang, L.; et al. OPLS3e: Extending force field coverage for drug-like small molecules. J. Chem. Theory Comput. 2019, 15, 1863–1874. [Google Scholar] [CrossRef]
- Fang, J.G.; Lu, M.; Chen, Z.H.; Zhu, H.H.; Li, Y.; Yang, L.; Wu, L.M.; Liu, Z.L. Antioxidant effects of resveratrol and its analogues against the free-radical-induced peroxidation of linoleic acid in micelles. Chem. Eur. J. 2002, 8, 4191–4198. [Google Scholar] [CrossRef]
- Hayes, J.M.; Archontis, G. MM-GB (PB) SA calculations of protein-ligand binding free energies. Mol. Dyn. Stud. Synth. Biol. Macromol. 2012, 171–190. [Google Scholar] [CrossRef] [Green Version]
- Vanommeslaeghe, K.; MacKerell, A.D., Jr. Automation of the CHARMM General Force Field (CGenFF) I: Bond perception and atom typing. J. Chem. Inf. Model. 2012, 52, 3144–3154. [Google Scholar] [CrossRef] [PubMed]
- Vanommeslaeghe, K.; Raman, E.P.; MacKerell, A.D., Jr. Automation of the CHARMM General Force Field (CGenFF) II: Assignment of bonded parameters and partial atomic charges. J. Chem. Inf. Model. 2012, 52, 3155–3168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vanommeslaeghe, K.; Hatcher, E.; Acharya, C.; Kundu, S.; Zhong, S.; Shim, J.; Darian, E.; Guvench, O.; Lopes, P.; Vorobyov, I.; et al. CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 2010, 31, 671–690. [Google Scholar] [CrossRef] [Green Version]
- Gutiérrez, I.S.; Lin, F.Y.; Vanommeslaeghe, K.; Lemkul, J.A.; Armacost, K.A.; Brooks, C.L., III; MacKerell, A.D., Jr. Parametrization of halogen bonds in the CHARMM general force field: Improved treatment of ligand–protein interactions. Bioorg. Med. Chem. 2016, 24, 4812–4825. [Google Scholar] [CrossRef] [Green Version]
- Yu, W.; He, X.; Vanommeslaeghe, K.; MacKerell, A.D., Jr. Extension of the CHARMM general force field to sulfonyl-containing compounds and its utility in biomolecular simulations. J. Comput. Chem. 2012, 33, 2451–2468. [Google Scholar] [CrossRef] [Green Version]
- Baranovskiy, A.G.; Babayeva, N.D.; Suwa, Y.; Gu, J.; Pavlov, Y.I.; Tahirov, T.H. Structural basis for inhibition of DNA replication by aphidicolin. Nucleic Acids Res. 2014, 42, 14013–14021. [Google Scholar] [CrossRef] [Green Version]
- Olsson, M.H.; Søndergaard, C.R.; Rostkowski, M.; Jensen, J.H. PROPKA3: Consistent treatment of internal and surface residues in empirical p K a predictions. J. Chem. Theory Comput. 2011, 7, 525–537. [Google Scholar] [CrossRef]
- Ono, K.; Nakane, H. Mechanisms of inhibition of various cellular DNA and RNA polymerases by several flavonoids. J. Biochem. 1990, 108, 609–613. [Google Scholar] [CrossRef]
- Mizushina, Y.; Ishidoh, T.; Kamisuki, S.; Nakazawa, S.; Takemura, M.; Sugawara, F.; Yoshida, H.; Sakaguchi, K. Flavonoid glycoside: A new inhibitor of eukaryotic DNA polymerase α and a new carrier for inhibitor-affinity chromatography. Biochem. Biophys. Res. Commun. 2003, 301, 480–487. [Google Scholar] [CrossRef]
- Shiomi, K.; Kuriyama, I.; Yoshida, H.; Mizushina, Y. Inhibitory effects of myricetin on mammalian DNA polymerase, topoisomerase and human cancer cell proliferation. Food Chem. 2013, 139, 910–918. [Google Scholar] [CrossRef] [PubMed]
- Das, K.; Martinez, S.E.; Arnold, E. Structural insights into HIV reverse transcriptase mutations Q151M and Q151M complex that confer multinucleoside drug resistance. Antimicrob. Agents Chemother. 2017, 61, e00224-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kolberg, M.; Strand, K.R.; Graff, P.; Andersson, K.K. Structure, function, and mechanism of ribonucleotide reductases. Biochim. Biophys. Acta (BBA) Proteins Proteom. 2004, 1699, 1–34. [Google Scholar] [CrossRef]
- Aye, Y.; Li, M.; Long, M.; Weiss, R. Ribonucleotide reductase and cancer: Biological mechanisms and targeted therapies. Oncogene 2015, 34, 2011–2021. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, M.F.; Alam, I.; Huff, S.E.; Pink, J.; Flanagan, S.A.; Shewach, D.; Misko, T.A.; Oleinick, N.L.; Harte, W.E.; Viswanathan, R.; et al. Potent competitive inhibition of human ribonucleotide reductase by a nonnucleoside small molecule. Proc. Natl. Acad. Sci. USA 2017, 114, 8241–8246. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, M.F.; Dealwis, C.G. The structural basis for the allosteric regulation of ribonucleotide reductase. In Progress in Molecular Biology and Translational Science; Elsevier: Amsterdam, The Netherlands, 2013; Volume 117, pp. 389–410. [Google Scholar] [CrossRef] [Green Version]
- Heredia, A.; Davis, C.; Amin, M.N.; Le, N.M.; Wainberg, M.A.; Oliveira, M.; Deeks, S.G.; Wang, L.X.; Redfield, R.R. Targeting host nucleotide biosynthesis with resveratrol inhibits emtricitabine (FTC)-resistant HIV-1. AIDS 2014, 28, 317. [Google Scholar] [CrossRef] [Green Version]
- He, Y.; Yan, C.; Fang, J.; Inouye, C.; Tjian, R.; Ivanov, I.; Nogales, E. Near-atomic resolution visualization of human transcription promoter opening. Nature 2016, 533, 359–365. [Google Scholar] [CrossRef] [Green Version]
- Baell, J.B. Feeling nature’s PAINS: Natural products, natural product drugs, and pan assay interference compounds (PAINS). J. Nat. Prod. 2016, 79, 616–628. [Google Scholar] [CrossRef]
- Ali, D.; Chen, L.; Kowal, J.M.; Okla, M.; Manikandan, M.; AlShehri, M.; AlMana, Y.; AlObaidan, R.; AlOtaibi, N.; Hamam, R.; et al. Resveratrol inhibits adipocyte differentiation and cellular senescence of human bone marrow stromal stem cells. Bone 2020, 115252. [Google Scholar] [CrossRef]
- Kiskova, T.; Kubatka, P.; Büsselberg, D.; Kassayova, M. The Plant-Derived Compound Resveratrol in Brain Cancer: A Review. Biomolecules 2020, 10, 161. [Google Scholar] [CrossRef] [Green Version]
- Hostenbach, S.; D’Haeseleer, M.; Kooijman, R.; De Keyser, J. Modulation of Cytokine-Induced Astrocytic Endothelin-1 Production as a Possible New Approach to the Treatment of Multiple Sclerosis. Front. Pharmacol. 2019, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caon, I.; Bartolini, B.; Moretto, P.; Parnigoni, A.; Caravà, E.; Vitale, D.L.; Alaniz, L.; Viola, M.; Karousou, E.; De Luca, G.; et al. Sirtuin 1 reduces hyaluronan synthase 2 expression by inhibiting nuclear translocation of NF-kB and expression of the long-non coding RNA HAS2-AS1. J. Biol. Chem. 2020, 295, 3485–3496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsieh, T.C.; Wu, J.M. Tumor PD-L1 Induction by Resveratrol/Piceatannol May Function as a Search, Enhance, and Engage (“SEE”) Signal to Facilitate the Elimination of “Cold, Non-Responsive” Low PD-L1-Expressing Tumors by PD-L1 Blockade. Int. J. Mol. Sci. 2019, 20, 5969. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xia, N.; Daiber, A.; Förstermann, U.; Li, H. Antioxidant effects of resveratrol in the cardiovascular system. Br. J. Pharmacol. 2017, 174, 1633–1646. [Google Scholar] [CrossRef] [Green Version]
- Xie, L.; Ge, X.; Tan, H.; Xie, L.; Zhang, Y.; Hart, T.; Yang, X.; Bourne, P.E. Towards structural systems pharmacology to study complex diseases and personalized medicine. PLoS Comput. Biol. 2014, 10, e1003554. [Google Scholar] [CrossRef] [Green Version]
- Fulda, S. Resveratrol and derivatives for the prevention and treatment of cancer. Drug Discov. Today 2010, 15, 757–765. [Google Scholar] [CrossRef]
- Saha, B.; Pai, G.B.; Subramanian, M.; Gupta, P.; Tyagi, M.; Patro, B.S.; Chattopadhyay, S. Resveratrol analogue, trans-4, 4′-dihydroxystilbene (DHS), inhibits melanoma tumor growth and suppresses its metastatic colonization in lungs. Biomed. Pharmacother. 2018, 107, 1104–1114. [Google Scholar] [CrossRef]
- Mahbub, A.A.; Le Maitre, C.L.; Haywood-Small, S.; Cross, N.A.; Jordan-Mahy, N. Polyphenols enhance the activity of alkylating agents in leukaemia cell lines. Oncotarget 2019, 10, 4570. [Google Scholar] [CrossRef]
- Ren, K.W.; Li, Y.H.; Wu, G.; Ren, J.Z.; Lu, H.B.; Li, Z.M.; Han, X.W. Quercetin nanoparticles display antitumor activity via proliferation inhibition and apoptosis induction in liver cancer cells. Int. J. Oncol. 2017, 50, 1299–1311. [Google Scholar] [CrossRef]
- Turumtay, H.; Midilli, A.; Turumtay, E.A.; Demir, A.; Selvi, E.K.; Budak, E.E.; Er, H.; Kocaimamoglu, F.; Baykal, H.; Belduz, A.O.; et al. Gram (-) microorganisms DNA polymerase inhibition, antibacterial and chemical properties of fruit and leaf extracts of Sorbus acuparia and Sorbus caucasica var. yaltirikii. Biomed. Chromatogr. 2017, 31, e3901. [Google Scholar] [CrossRef]
- Nosrati, M.; Shakeran, Z.; Shakeran, Z. Frangulosid as a novel hepatitis B virus DNA polymerase inhibitor: A virtual screening study. Silico Pharmacol. 2018, 6, 10. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Zhu, M.; Wang, L.; Yu, S. Anti-tumor effects and associated molecular mechanisms of myricetin. Biomed. Pharmacother. 2019, 120, 109506. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Sk, M.F.; Sonawane, A.; Kar, P.; Sadhukhan, S. Plant-derived natural polyphenols as potential antiviral drugs against SARS-CoV-2 via RNA-dependent RNA polymerase (RdRp) inhibition: An in-silico analysis. J. Biomol. Struct. Dyn. 2020, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Ong, K.C.; Khoo, H.E. Biological effects of myricetin. Gen. Pharmacol. Vasc. Syst. 1997, 29, 121–126. [Google Scholar] [CrossRef]
- Mikuła-Pietrasik, J.; Sosińska, P.; Murias, M.; Wierzchowski, M.; Brewińska-Olchowik, M.; Piwocka, K.; Szpurek, D.; Książek, K. High potency of a novel resveratrol derivative, 3, 3′, 4, 4′-tetrahydroxy-trans-stilbene, against ovarian cancer is associated with an oxidative stress-mediated imbalance between DNA damage accumulation and repair. Oxid. Med. Cell. Longev. 2015, 2015. [Google Scholar] [CrossRef] [Green Version]
- McFadden, D. A review of pterostilbene antioxidant activity and disease modification. Oxid. Med. Cell. Longev. 2013, 2013. [Google Scholar] [CrossRef] [Green Version]
- Cheng, T.C.; Lai, C.S.; Chung, M.C.; Kalyanam, N.; Majeed, M.; Ho, C.T.; Ho, Y.S.; Pan, M.H. Potent anti-cancer effect of 3′-hydroxypterostilbene in human colon xenograft tumors. PLoS ONE 2014, 9, e111814. [Google Scholar] [CrossRef] [Green Version]
- Dai, Y.; Tan, A.L.C.; Chen, H.; Ong, P.S.; Xiang, X.; Wu, J.; Lin, H.S. Quantification of desoxyrhapontigenin (4-methoxyresveratrol) in rat plasma by LC–MS/MS: Application to pre-clinical pharmacokinetic study. J. Pharm. Biomed. Anal. 2018, 153, 95–101. [Google Scholar] [CrossRef]
- Du, M.; Zhang, Z.; Gao, T. Piceatannol induced apoptosis through up-regulation of microRNA-181a in melanoma cells. Biol. Res. 2017, 50. [Google Scholar] [CrossRef]
- Vendrely, V.; Peuchant, E.; Buscail, E.; Moranvillier, I.; Rousseau, B.; Bedel, A.; Brillac, A.; de Verneuil, H.; Moreau-Gaudry, F.; Dabernat, S. Resveratrol and capsaicin used together as food complements reduce tumor growth and rescue full efficiency of low dose gemcitabine in a pancreatic cancer model. Cancer Lett. 2017, 390, 91–102. [Google Scholar] [CrossRef]
- Yuan, Z.; Luan, G.; Wang, Z.; Hao, X.; Li, J.; Suo, Y.; Li, G.; Wang, H. Flavonoids from Potentilla parvifolia Fisch. and Their Neuroprotective Effects in Human Neuroblastoma SH-SY 5Y Cells in vitro. Chem. Biodivers. 2017, 14, e1600487. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Cruz, E.; Cerezo, A.B.; Cantos-Villar, E.; Richard, T.; Troncoso, A.M.; Garcia-Parrilla, M.C. Inhibition of VEGFR-2 phosphorylation and effects on downstream signaling pathways in cultivated human endothelial cells by stilbenes from Vitis spp. J. Agric. Food Chem. 2019, 67, 3909–3918. [Google Scholar] [CrossRef] [PubMed]
- Almeida, L.; Vaz-da Silva, M.; Falcão, A.; Soares, E.; Costa, R.; Loureiro, A.I.; Fernandes-Lopes, C.; Rocha, J.F.; Nunes, T.; Wright, L.; et al. Pharmacokinetic and safety profile of trans-resveratrol in a rising multiple-dose study in healthy volunteers. Mol. Nutr. Food Res. 2009, 53, S7–S15. [Google Scholar] [CrossRef] [PubMed]
- Lepak, A.; Gutmann, A.; Kulmer, S.T.; Nidetzky, B. Creating a water-soluble resveratrol-based antioxidant by site-selective enzymatic glucosylation. ChemBioChem 2015, 16, 1870–1874. [Google Scholar] [CrossRef]
- Ghiselli, A.; Nardini, M.; Baldi, A.; Scaccini, C. Antioxidant activity of different phenolic fractions separated from an Italian red wine. J. Agric. Food Chem. 1998, 46, 361–367. [Google Scholar] [CrossRef]
- Bachelor, F.; Loman, A.; Snowdon, L. Synthesis of pinosylvin and related heartwood stilbenes. Can. J. Chem. 1970, 48, 1554–1557. [Google Scholar] [CrossRef] [Green Version]
- Mikulski, D.; Górniak, R.; Molski, M. A theoretical study of the structure–radical scavenging activity of trans-resveratrol analogues and cis-resveratrol in gas phase and water environment. Eur. J. Med. Chem. 2010, 45, 1015–1027. [Google Scholar] [CrossRef]
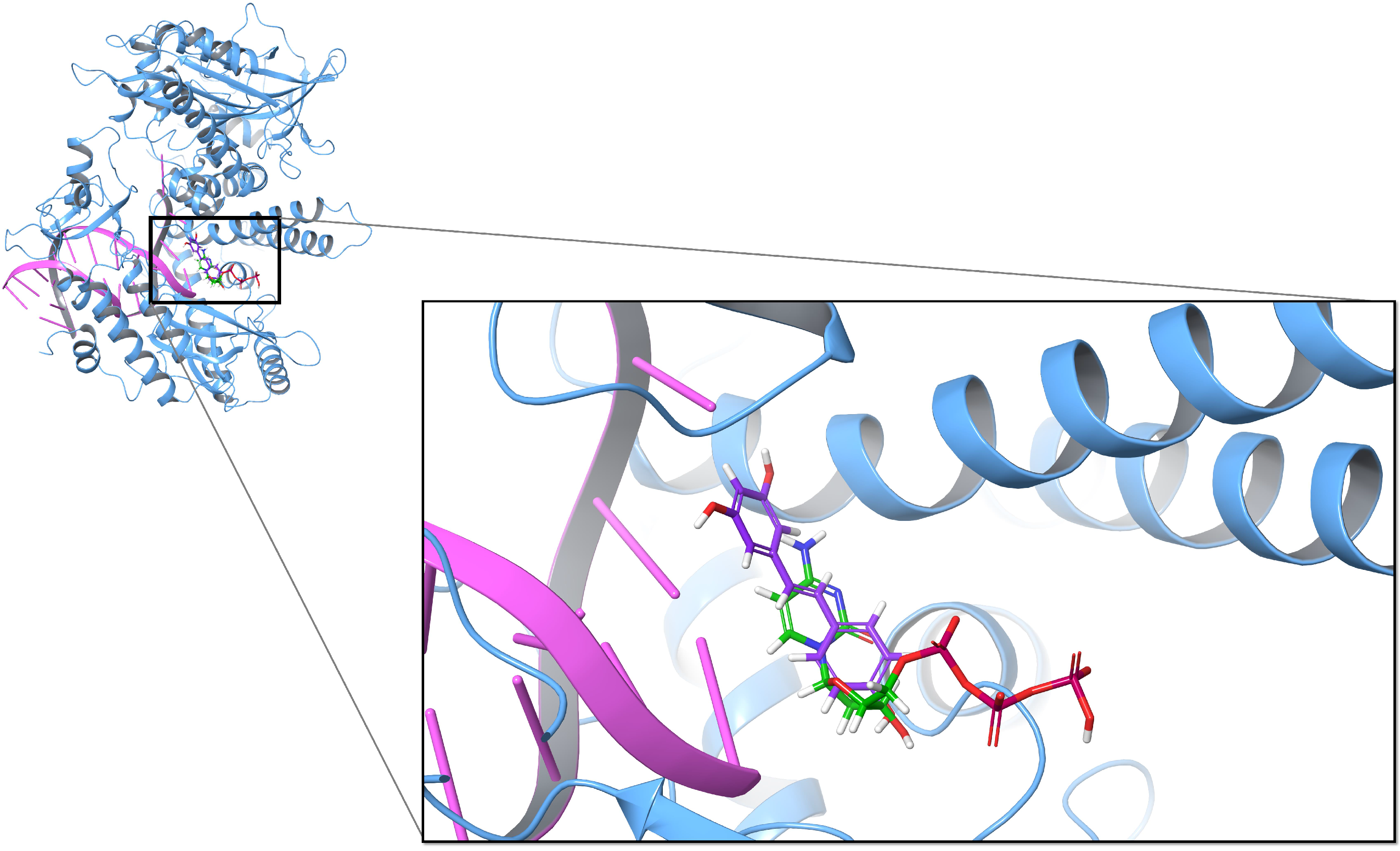
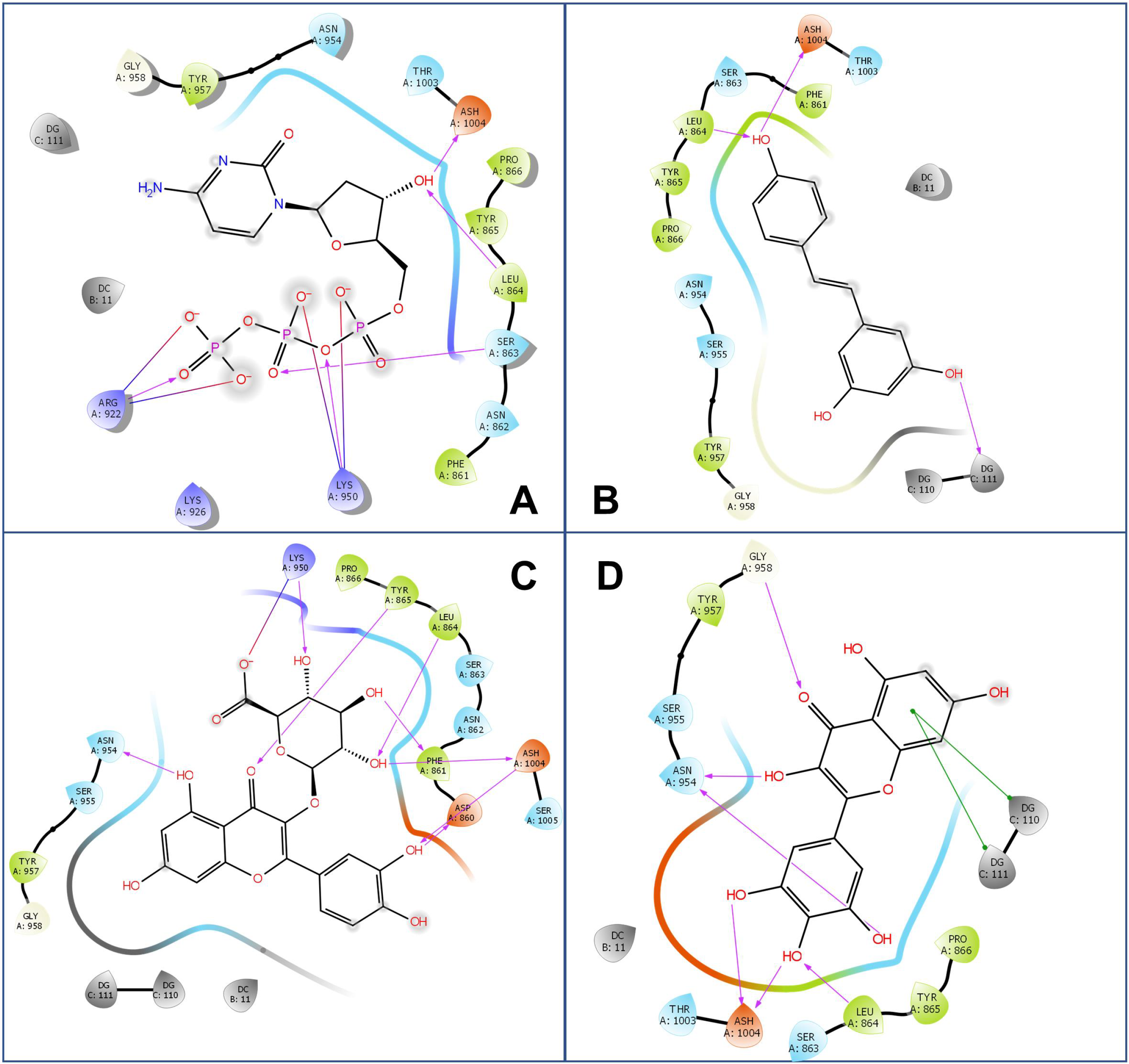
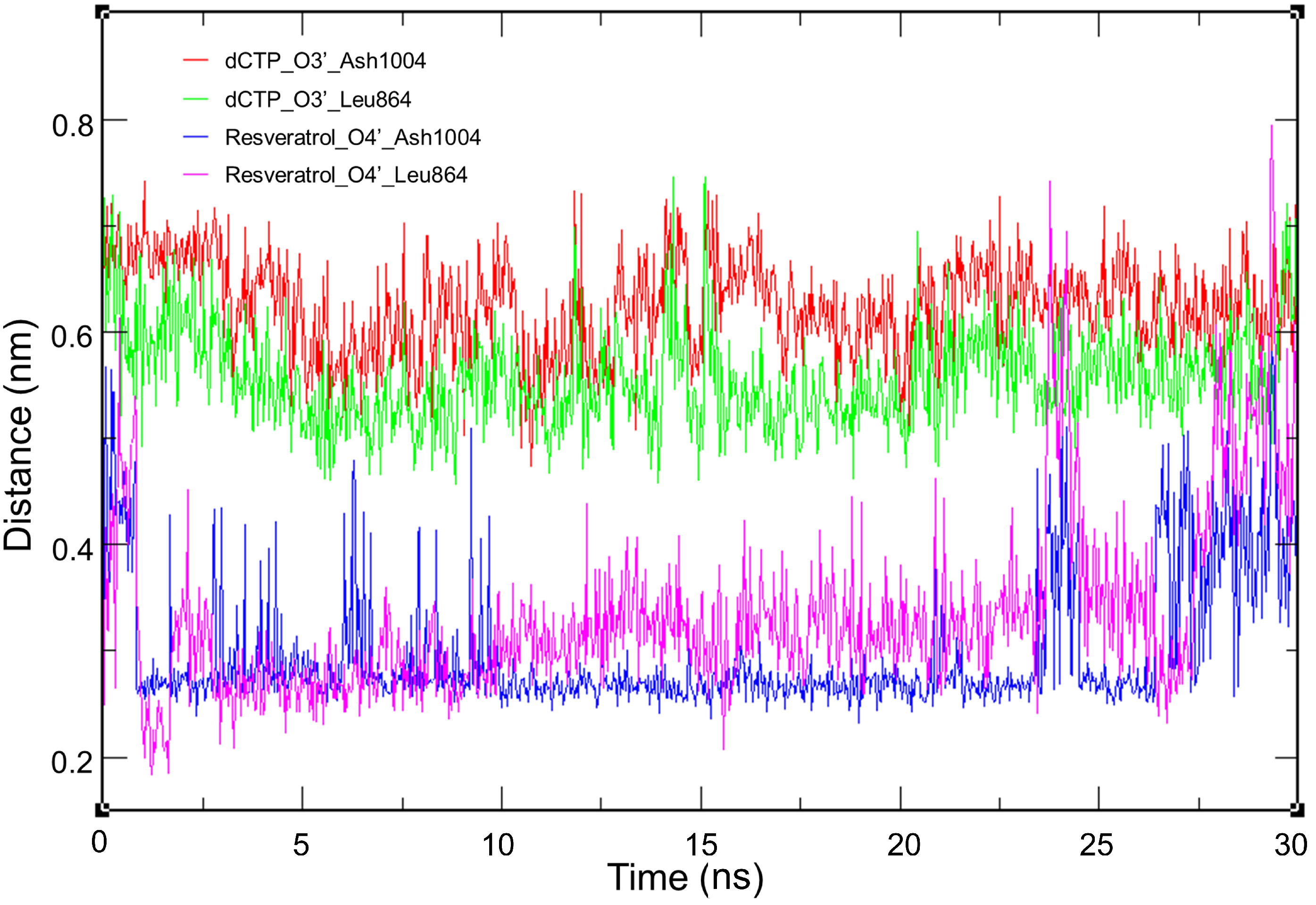
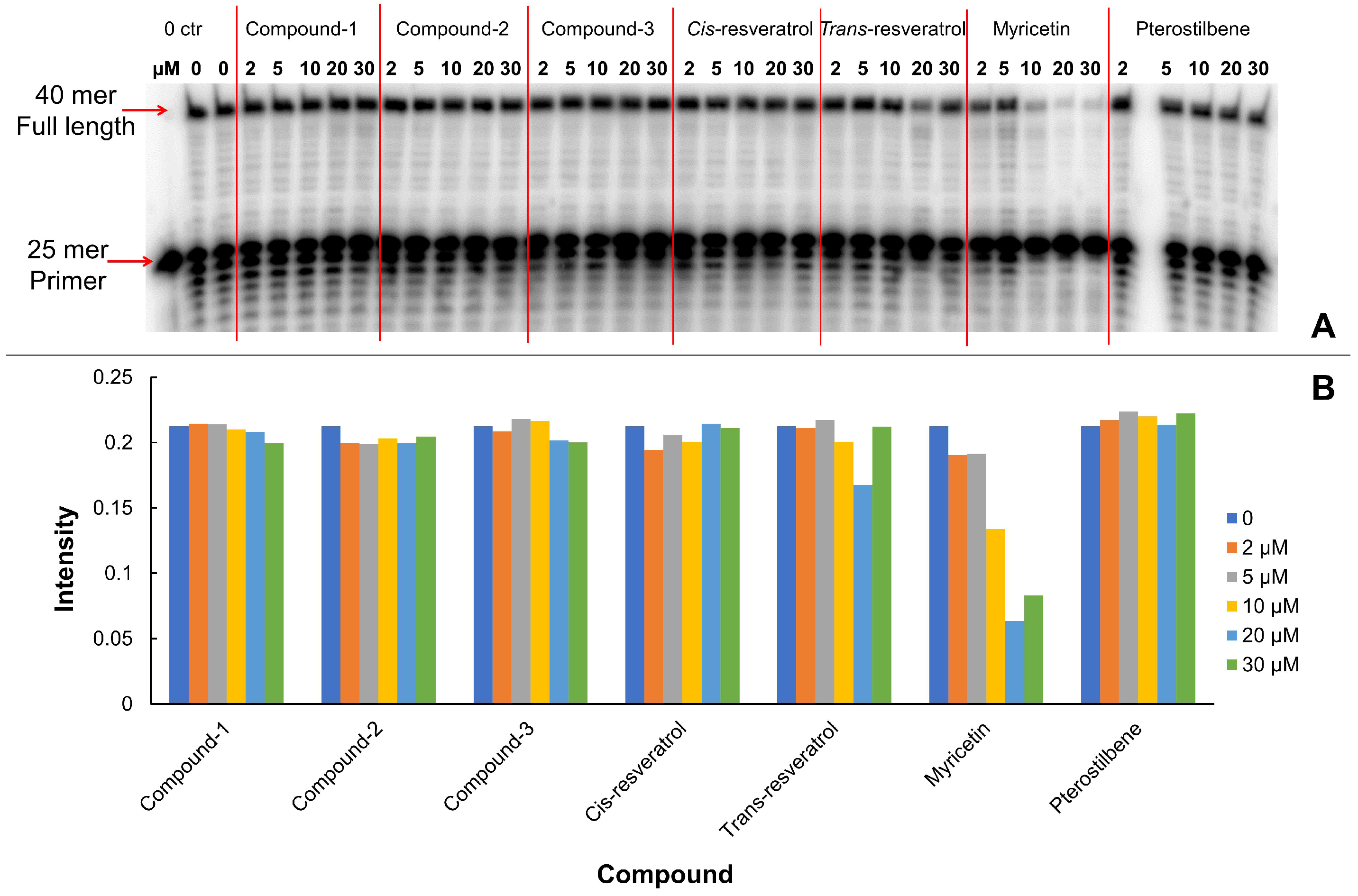
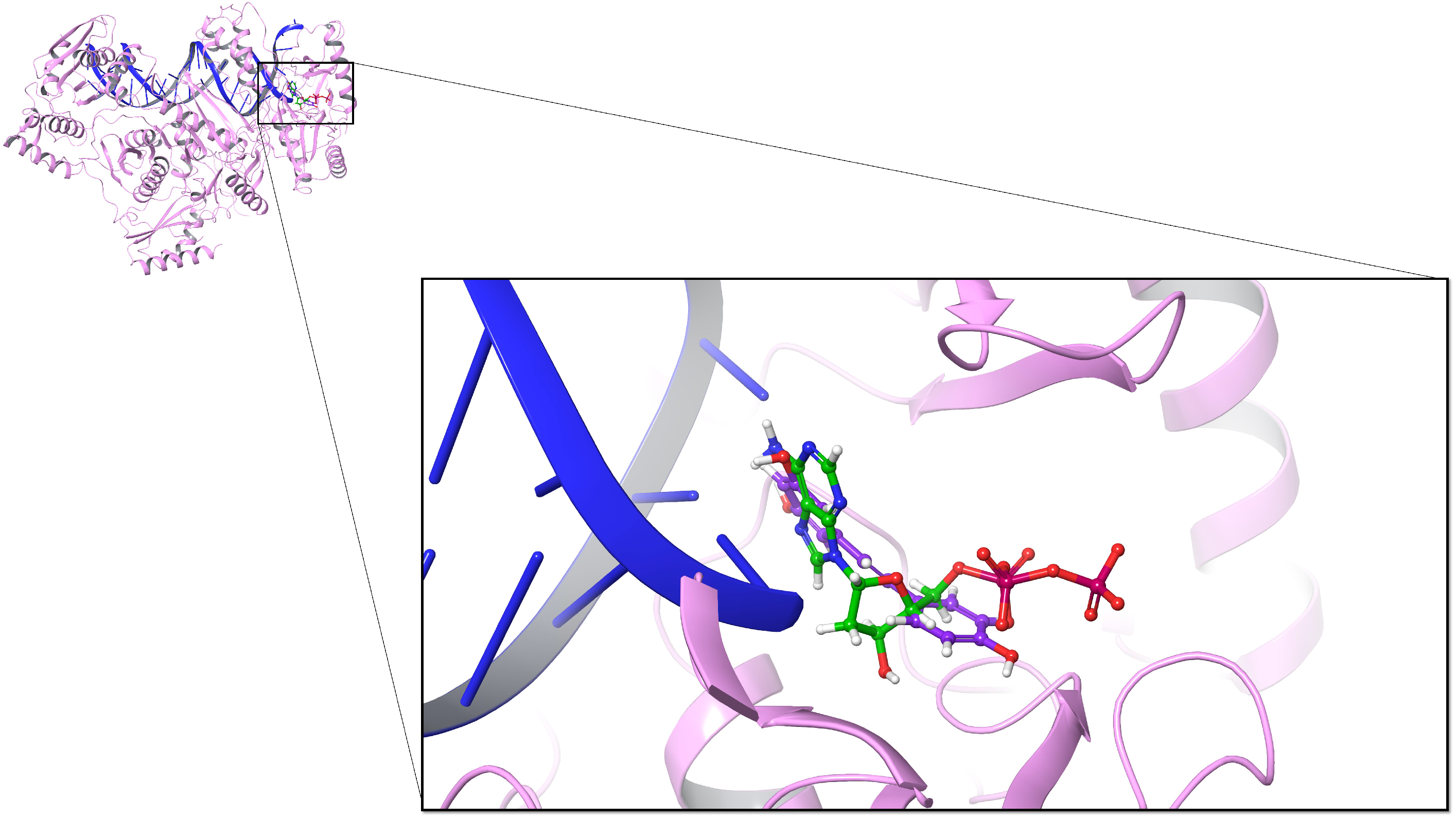
| Compound | MM-GBSA (kcal/mol) | |
|---|---|---|
| 4Q5V | 5TXM | |
| Substrate | −49.46(dCTP) | −47.88(ddATP) −46.41(dATP) |
| Trans-resveratrol | −49.58 | −55.42 |
| Cis-resveratrol | −48.85 | −42.47 |
| Piceatannol | −50.67 | −41.76 |
| M8 | −59.6 | −40.04 |
| 3,4,5-THS | −63.64 | −38.37 |
| 4,4-DHS | −56.1 | −41.36 |
| Pterostilbene | −46.68 | −40.81 |
| HPSB | −50.51 | −42.28 |
| DRG | −45.91 | −44.08 |
| Pinostilbene | −43.86 | −35.87 |
| Compound-1 | −42.87 | −27.84 |
| Compound-2 | −34.08 | −47.19 |
| Compound-3 | −52.48 | −41.53 |
| Quercetin | −59.08 | −52.96 |
| Myricetin | −61.90 | −49.87 |
| Miquelianin | −73.53 * | −75.71 * |
| Astringin | −56.22 | −60.16 |
| Mulberroside A | −59.33 | −47.25 |
| Hydroxyurea | −20.25 | −10.82 |
| Compound | MM-GBSA (kcal/mol) | ||
|---|---|---|---|
| A Site | S Site | C Site | |
| Substrate | −42.27(ATP) | −76.81(TTP) | −49.42(GDP) |
| Trans-resveratrol | −42.14 | −52.8 | −37.02 |
| Cis-resveratrol | −38.22 | −51.05 | −43.24 |
| Piceatannol | −44.24 | −58.18 | −45.25 |
| M8 | −41.23 | −65.52 | −46.42 |
| 3,4,5-THS | −39.08 | −59.32 | −31.65 |
| 4,4-DHS | −37.21 | −46.51 | −35.58 |
| Pterostilbene | −46.8 | −51.16 | −42.23 |
| HPSB | −34.11 | −56.22 | −38.31 |
| DRG | −43.66 | −58.32 | −40.16 |
| Pinostilbene | −32 | −55 | −50.57 |
| Compound-1 | −23.42 | −46.50 | −38.51 |
| Compound-2 | −24.98 | −49.11 | −40.11 |
| Compound-3 | −24.98 | −59.82 | −29.60 |
| Quercetin | −24.14 | −59.47 | −46.89 |
| Myricetin | −30.91 | −59.43 | −46.59 |
| Miquelianin | −60.20 * | −62.45 | −59.59 * |
| Astringin | −50.80 | −58.74 | −51.77 |
| Mulberroside A | −58.42 | −65.12 | −48.44 |
| Hydroxyurea | −14.73 | −24.24 | −20.80 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, Y.; Hsieh, T.-c.; Wu, J.M.; Wang, X.; Christopher, J.S.; Pham, A.H.; Swaby, J.D.-L.; Lou, L.; Xie, Z.-R. Elucidating the Inhibitory Effect of Resveratrol and Its Structural Analogs on Selected Nucleotide-Related Enzymes. Biomolecules 2020, 10, 1223. https://doi.org/10.3390/biom10091223
Wu Y, Hsieh T-c, Wu JM, Wang X, Christopher JS, Pham AH, Swaby JD-L, Lou L, Xie Z-R. Elucidating the Inhibitory Effect of Resveratrol and Its Structural Analogs on Selected Nucleotide-Related Enzymes. Biomolecules. 2020; 10(9):1223. https://doi.org/10.3390/biom10091223
Chicago/Turabian StyleWu, Yifei, Tze-chen Hsieh, Joseph M. Wu, Xiaoxiao Wang, Joshua S. Christopher, Amanda H. Pham, Justin David-Li Swaby, Lei Lou, and Zhong-Ru Xie. 2020. "Elucidating the Inhibitory Effect of Resveratrol and Its Structural Analogs on Selected Nucleotide-Related Enzymes" Biomolecules 10, no. 9: 1223. https://doi.org/10.3390/biom10091223
APA StyleWu, Y., Hsieh, T.-c., Wu, J. M., Wang, X., Christopher, J. S., Pham, A. H., Swaby, J. D.-L., Lou, L., & Xie, Z.-R. (2020). Elucidating the Inhibitory Effect of Resveratrol and Its Structural Analogs on Selected Nucleotide-Related Enzymes. Biomolecules, 10(9), 1223. https://doi.org/10.3390/biom10091223






