Hormone-Independent Mouse Mammary Adenocarcinomas with Different Metastatic Potential Exhibit Different Metabolic Signatures
Abstract
:1. Introduction
2. Materials and Methods
2.1. Syngeneic Tumour Model and Procedures Carried Out in Mice
2.2. Tumour Extracts
2.3. NMR Spectroscopy
2.4. Statistics Analysis and Other Spectral Analysis
3. Results
3.1. 1H-NMR Spectra of Polar and Lipophilic Extracts of Breast Tumours
3.2. Metabolic Differences between 59-2-HI and C7-2-HI Tumour Lines
3.3. Metabolic Differences among Tumours of The Same Line
3.4. Intra-Tumoural Metabolic Differences
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- The World Health Organization Breast Cancer. Available online: http://www.who.int/cancer/prevention/diagnosis-screening/breast-cancer/en/ (accessed on 15 July 2020).
- Koboldt, D.C.; Fulton, R.S.; McLellan, M.D.; Schmidt, H.; Kalicki-Veizer, J.; McMichael, J.F.; Fulton, L.L.; Dooling, D.J.; Ding, L.; Mardis, E.R.; et al. Comprehensive molecular portraits of human breast tumours. Nature 2012, 490, 61–70. [Google Scholar] [CrossRef] [Green Version]
- Reed, A.E.M.; Croft, P.K.; Kutasovic, J.R.; Saunus, J.M.; Lakhani, S.R. Recent advances in breast cancer research impacting clinical diagnostic practice. J. Pathol. 2019, 247, 552–562. [Google Scholar] [CrossRef] [Green Version]
- Haukaas, T.H.; Euceda, L.R.; Giskeødegård, G.F.; Lamichhane, S.; Krohn, M.; Jernström, S.; Aure, M.R.; Lingjærde, O.C.; Schlichting, E.; Garred, Ø.; et al. Metabolic clusters of breast cancer in relation to gene- and protein expression subtypes. Cancer Metab. 2016, 4, 12. [Google Scholar] [CrossRef] [Green Version]
- Sormendi, S.; Wielockx, B. Hypoxia Pathway Proteins as Central Mediators of Metabolism in the Tumor Cells and Their Microenvironment. Front. Immunol. 2018, 9, 40. [Google Scholar] [CrossRef]
- Zardavas, D.; Irrthum, A.; Swanton, C.; Piccart, M. Clinical management of breast cancer heterogeneity. Nat. Rev. Clin. Oncol. 2015, 12, 381–394. [Google Scholar] [CrossRef]
- Rivenbark, A.G.; O’Connor, S.M.; Coleman, W.B. Molecular and Cellular Heterogeneity in Breast Cancer. Am. J. Pathol. 2013, 183, 1113–1124. [Google Scholar] [CrossRef] [Green Version]
- Skibinski, A.; Kuperwasser, C. The origin of breast tumor heterogeneity. Oncogene 2015, 34, 5309–5316. [Google Scholar] [CrossRef] [Green Version]
- Gowda, G.N.; Zhang, S.; Gu, H.; Asiago, V.; Shanaiah, N.; Raftery, D. Metabolomics-based methods for early disease diagnostics. Expert Rev. Mol. Diagn. 2008, 8, 617–633. [Google Scholar] [CrossRef] [Green Version]
- Bathen, T.F.; Jensen, L.R.; Sitter, B.; Fjösne, H.E.; Halgunset, J.; Axelson, D.E.; Gribbestad, I.S.; Lundgren, S. MR-determined metabolic phenotype of breast cancer in prediction of lymphatic spread, grade, and hormone status. Breast Cancer Res. Treat. 2006, 104, 181–189. [Google Scholar] [CrossRef]
- Cao, M.D.; Lamichhane, S.; Lundgren, S.; Bofin, A.; Fjøsne, H.; Giskeødegård, G.F.; Bathen, T.F. Metabolic characterization of triple negative breast cancer. BMC Cancer 2014, 14, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Tang, X.; Lin, C.-C.; Spasojevic, I.; Iversen, E.S.; Chi, J.-T.; Marks, J.R. A joint analysis of metabolomics and genetics of breast cancer. Breast Cancer Res. 2014, 16, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Brockmöller, S.F.; Bucher, E.; Müller, B.M.; Budczies, J.; Hilvo, M.; Griffin, J.L.; Orešič, M.; Kallioniemi, O.; Iljin, K.; Loibl, S.; et al. Integration of Metabolomics and Expression of Glycerol-3-phosphate Acyltransferase (GPAM) in Breast Cancer—Link to Patient Survival, Hormone Receptor Status, and Metabolic Profiling. J. Proteome Res. 2012, 11, 850–860. [Google Scholar] [CrossRef] [PubMed]
- Moestue, S.A.; Borgan, E.; Huuse, E.M.; Lindholm, E.M.; Sitter, B.; Børresen-Dale, A.-L.; Engebraaten, O.; Mælandsmo, G.M.; Gribbestad, I.S. Distinct choline metabolic profiles are associated with differences in gene expression for basal-like and luminal-like breast cancer xenograft models. BMC Cancer 2010, 10, 433. [Google Scholar] [CrossRef] [Green Version]
- Grinde, M.T.; Moestue, S.A.; Borgan, E.; Risa, Ø.; Engebraaten, O.; Gribbestad, I.S. 13C High-resolution-magic angle spinning MRS reveals differences in glucose metabolism between two breast cancer xenograft models with different gene expression patterns. NMR Biomed. 2011, 24, 1243–1252. [Google Scholar] [CrossRef] [Green Version]
- Seierstad, T.; Røe, K.; Sitter, B.; Halgunset, J.; Flatmark, K.; Ree, A.H.; Olsen, D.; Gribbestad, I.S.; Bathen, T.F. Principal component analysis for the comparison of metabolic profiles from human rectal cancer biopsies and colorectal xenografts using high-resolution magic angle spinning 1H magnetic resonance spectroscopy. Mol. Cancer 2008, 7, 33. [Google Scholar] [CrossRef] [Green Version]
- Aure, M.R.; Vitelli, V.; Jernström, S.; Kumar, S.; Krohn, M.; Due, E.U.; Haukaas, T.H.; Leivonen, S.; Vollan, H.K.M.; Lüders, T.; et al. Integrative clustering reveals a novel split in the luminal A subtype of breast cancer with impact on outcome. Breast Cancer Res. 2017, 19, 44. [Google Scholar] [CrossRef] [Green Version]
- Borgan, E.; Sitter, B.; Lingjaerde, O.C.; Johnsen, H.; Lundgren, S.; Bathen, T.F.; Sorlie, T.; Borresen-Dale, A.L.; Gribbestad, I.S. Merging transcriptomics and metabolomics--advances in breast cancer profiling. BMC Cancer 2010, 10, 628. [Google Scholar] [CrossRef] [Green Version]
- Sun, X.; Yu, Q. Intra-tumor heterogeneity of cancer cells and its implications for cancer treatment. Acta Pharmacol. Sin. 2015, 36, 1219–1227. [Google Scholar] [CrossRef]
- Dias, A.S.; Almeida, C.R.; Helguero, L.A.; Duarte, I.F. Metabolic crosstalk in the breast cancer microenvironment. Eur. J. Cancer 2019, 121, 154–171. [Google Scholar] [CrossRef]
- Opstad, K.S.; Wright, A.J.; Bell, B.A.; Griffiths, J.R.; Howe, F.A. Correlations between in vivo 1H MRS and ex vivo 1H HRMAS metabolite measurements in adult human gliomas. J. Magn. Reson. Imaging 2010, 31, 289–297. [Google Scholar] [CrossRef]
- Barré, F.P.Y.; Claes, B.S.R.; Dewez, F.; Peutz-Kootstra, C.; Munch-Petersen, H.F.; Grønbæk, K.; Lund, A.H.; Heeren, R.M.A.; Côme, C.; Cillero-Pastor, B. Specific Lipid and Metabolic Profiles of R-CHOP-Resistant Diffuse Large B-Cell Lymphoma Elucidated by Matrix-Assisted Laser Desorption Ionization Mass Spectrometry Imaging and in vivo Imaging. Anal. Chem. 2018, 90, 14198–14206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cimino, J.; Calligaris, D.; Far, J.; Debois, D.; Blacher, S.; Sounni, N.; Noel, A.; De Pauw, E. Towards Lipidomics of Low-Abundant Species for Exploring Tumor Heterogeneity Guided by High-Resolution Mass Spectrometry Imaging. Int. J. Mol. Sci. 2013, 14, 24560–24580. [Google Scholar] [CrossRef] [Green Version]
- Willems, S.M.; van Remoortere, A.; van Zeijl, R.; Deelder, A.M.; McDonnell, L.A.; Hogendoorn, P.C. Imaging mass spectrometry of myxoid sarcomas identifies proteins and lipids specific to tumour type and grade, and reveals biochemical intratumour heterogeneity. J. Pathol. 2010, 222, 400–409. [Google Scholar] [CrossRef]
- Coquery, N.; Stupar, V.; Farion, R.; Maunoir-Regimbal, S.; Barbier, E.L.; Rémy, C.; Fauvelle, F. The three glioma rat models C6, F98 and RG2 exhibit different metabolic profiles: In vivo 1H MRS and ex vivo 1H HRMAS combined with multivariate statistics. Metabolomics 2015, 11, 1834–1847. [Google Scholar] [CrossRef]
- Zhang, Q.; Lou, Y.; Yang, J.; Wang, J.; Feng, J.; Zhao, Y.; Wang, L.; Huang, X.; Fu, Q.; Ye, M.; et al. Integrated multiomic analysis reveals comprehensive tumour heterogeneity and novel immunophenotypic classification in hepatocellular carcinomas. Gut 2019, 68, 2019–2031. [Google Scholar] [CrossRef]
- Hensley, C.T.; Faubert, B.; Yuan, Q.; Lev-Cohain, N.; Jin, E.; Kim, J.; Jiang, L.; Ko, B.; Skelton, R.; Loudat, L.; et al. Metabolic Heterogeneity in Human Lung Tumors. Cell 2016, 164, 681–694. [Google Scholar] [CrossRef] [Green Version]
- Okegawa, T.; Morimoto, M.; Nishizawa, S.; Kitazawa, S.; Honda, K.; Araki, H.; Tamura, T.; Ando, A.; Satomi, Y.; Nutahara, K.; et al. Intratumor Heterogeneity in Primary Kidney Cancer Revealed by Metabolic Profiling of Multiple Spatially Separated Samples within Tumors. EBioMedicine 2017, 19, 31–38. [Google Scholar] [CrossRef] [Green Version]
- Park, V.Y.; Yoon, D.; Koo, J.S.; Kim, E.-K.; Kim, S., II; Choi, J.S.; Park, S.; Park, H.S.; Kim, S.; Kim, M.J. Intratumoral Agreement of High-Resolution Magic Angle Spinning Magnetic Resonance Spectroscopic Profiles in the Metabolic Characterization of Breast Cancer. Medicine (Baltimore) 2016, 95, e3398. [Google Scholar] [CrossRef]
- Gogiashvili, M.; Horsch, S.; Marchan, R.; Gianmoena, K.; Cadenas, C.; Tanner, B.; Naumann, S.; Ersova, D.; Lippek, F.; Rahnenführer, J.; et al. Impact of intratumoral heterogeneity of breast cancer tissue on quantitative metabolomics using high-resolution magic angle spinning 1 H NMR spectroscopy. NMR Biomed. 2018, 31, e3862. [Google Scholar] [CrossRef]
- Mao, X.; He, J.; Li, T.; Lu, Z.; Sun, J.; Meng, Y.; Abliz, Z.; Chen, J. Application of imaging mass spectrometry for the molecular diagnosis of human breast tumors. Sci. Rep. 2016, 6, 1–12. [Google Scholar] [CrossRef]
- Helguero, L.A.; Viegas, M.; Asaithamby, A.; Shyamala, G.; Lanari, C.; Molinolo, A.A. Progesterone Receptor Expression in Medroxyprogesterone Acetate-Induced Murine Mammary Carcinomas and Response to Endocrine Treatment. Breast Cancer Res. Treat. 2003, 79, 379–390. [Google Scholar] [CrossRef]
- Lanari, C.; Lamb, C.A.; Fabris, V.T.; Helguero, L.A.; Soldati, R.; Bottino, M.C.; Giulianelli, S.; Cerliani, J.P.; Wargon, V.; Molinolo, A. The MPA mouse breast cancer model: Evidence for a role of progesterone receptors in breast cancer. Endocr. Relat. Cancer 2009, 16, 333–350. [Google Scholar] [CrossRef] [Green Version]
- Wu, H.; Southam, A.D.; Hines, A.; Viant, M.R. High-throughput tissue extraction protocol for NMR- and MS-based metabolomics. Anal. Biochem. 2008, 372, 204–212. [Google Scholar] [CrossRef]
- Sterin, M.; Cohen, J.S.; Ringel, I. Hormone Sensitivity is Reflected in the Phospholipid Profiles of Breast Cancer Cell Lines. Breast Cancer Res. Treat. 2004, 87, 1–11. [Google Scholar] [CrossRef]
- Oostendorp, M.; Engelke, U.F.H.; Willemsen, M.A.A.P.; Wevers, R.A. Diagnosing Inborn Errors of Lipid Metabolism with Proton Nuclear Magnetic Resonance Spectroscopy. Clin. Chem. 2006, 52, 1395–1405. [Google Scholar] [CrossRef] [Green Version]
- Mika, A.; Kaczynski, Z.; Stepnowski, P.; Kaczor, M.; Proczko-Stepaniak, M.; Kaska, L.; Sledzinski, T. Potential Application of 1H NMR for Routine Serum Lipidome Analysis –Evaluation of Effects of Bariatric Surgery. Sci. Rep. 2017, 7, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Canadian Institutes of Health Research; Canada Foundation for Innovation; The M.I.C. (TMIC) Human Metabolome Database. Available online: www.hmdb.ca (accessed on 15 July 2020).
- Veselkov, K.A.; Lindon, J.C.; Ebbels, T.M.D.; Crockford, D.; Volynkin, V.V.; Holmes, E.; Davies, D.B.; Nicholson, J.K. Recursive Segment-Wise Peak Alignment of Biological 1 H NMR Spectra for Improved Metabolic Biomarker Recovery. Anal. Chem. 2009, 81, 56–66. [Google Scholar] [CrossRef]
- Trygg, J.; Holmes, E.; Lundstedt, T. Chemometrics in Metabonomics. J. Proteome Res. 2007, 6, 469–479. [Google Scholar] [CrossRef]
- Bridge, P.D.; Sawilowsky, S.S. Increasing Physicians’ Awareness of the Impact of Statistics on Research Outcomes. J. Clin. Epidemiol. 1999, 52, 229–235. [Google Scholar] [CrossRef]
- Berben, L.; Sereika, S.M.; Engberg, S. Effect size estimation: Methods and examples. Int. J. Nurs. Stud. 2012, 49, 1039–1047. [Google Scholar] [CrossRef]
- Ranstam, J. Multiple p-values and Bonferroni correction. Osteoarthr. Cartil. 2016, 24, 763–764. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cloarec, O.; Dumas, M.-E.; Craig, A.; Barton, R.H.; Trygg, J.; Hudson, J.; Blancher, C.; Gauguier, D.; Lindon, J.C.; Holmes, E.; et al. Statistical Total Correlation Spectroscopy: An Exploratory Approach for Latent Biomarker Identification from Metabolic 1 H NMR Data Sets. Anal. Chem. 2005, 77, 1282–1289. [Google Scholar] [CrossRef]
- Mosconi, E.; Fontanella, M.; Sima, D.M.; Van Huffel, S.; Fiorini, S.; Sbarbati, A.; Marzola, P. Investigation of adipose tissues in Zucker rats using in vivo and ex vivo magnetic resonance spectroscopy. J. Lipid Res. 2011, 52, 330–336. [Google Scholar] [CrossRef] [Green Version]
- Sitter, B.; Sonnewald, U.; Spraul, M.; Fjösne, H.E.; Gribbestad, I.S. High-resolution magic angle spinning MRS of breast cancer tissue. NMR Biomed. 2002, 15, 327–337. [Google Scholar] [CrossRef] [PubMed]
- Nittoli, A.C.; Costantini, S.; Sorice, A.; Capone, F.; Ciarcia, R.; Marzocco, S.; Budillon, A.; Severino, L. Effects of α-zearalenol on the metabolome of two breast cancer cell lines by 1H-NMR approach. Metabolomics 2018, 14, 33. [Google Scholar] [CrossRef] [PubMed]
- Araújo, R.; Bispo, D.; Helguero, L.A.; Gil, A.M. Metabolomic studies of breast cancer in murine models: A review. Biochim. Biophys. Acta Mol. Basis Dis. 2020, 1866, 165713. [Google Scholar] [CrossRef]
- Coum, A.; Ouldamer, L.; Noury, F.; Barantin, L.; Saint-Hilaire, A.; Vilde, A.; Bougnoux, P.; Gambarota, G. In vivo MR spectroscopy of human breast tissue: Quantification of fatty acid composition at a clinical field strength (3 T). Magn. Reson. Mater. Phys. Biol. Med. 2016, 29, 1–4. [Google Scholar] [CrossRef]
- Merchant, T.E.; Kasimos, J.N.; Vroom, T.; de Bree, E.; Iwata, J.L.; de Graaf, P.W.; Glonek, T. Malignant breast tumor phospholipid profiles using 31P magnetic resonance. Cancer Lett. 2002, 176, 159–167. [Google Scholar] [CrossRef]
- Morse, D.L.; Carroll, D.; Day, S.; Gray, H.; Sadarangani, P.; Murthi, S.; Job, C.; Baggett, B.; Raghunand, N.; Gillies, R.J. Characterization of breast cancers and therapy response by MRS and quantitative gene expression profiling in the choline pathway. NMR Biomed. 2009, 22, 114–127. [Google Scholar] [CrossRef] [Green Version]
- Paul, A.; Kumar, S.; Raj, A.; Sonkar, A.A.; Jain, S.; Singhai, A.; Roy, R. Alteration in lipid composition differentiates breast cancer tissues: A 1H HRMAS NMR metabolomic study. Metabolomics 2018, 14, 119. [Google Scholar] [CrossRef]
- Dória, M.L.; Cotrim, C.Z.; Simões, C.; Macedo, B.; Domingues, P.; Domingues, M.R.; Helguero, L.A. Lipidomic analysis of phospholipids from human mammary epithelial and breast cancer cell lines. J. Cell. Physiol. 2013, 228, 457–468. [Google Scholar] [CrossRef] [PubMed]
- Dória, M.L.; Cotrim, Z.; MacEdo, B.; Simões, C.; Domingues, P.; Helguero, L.; Domingues, M.R. Lipidomic approach to identify patterns in phospholipid profiles and define class differences in mammary epithelial and breast cancer cells. Breast Cancer Res. Treat. 2012, 133, 635–648. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Li, X.; Xiang, L.; Song, Y.; Liu, Y.; Jiang, Y.; Cai, Z. Metabolomics and lipidomics study unveils the impact of polybrominated diphenyl ether-47 on breast cancer mice. J. Hazard. Mater. 2020, 390, 121451. [Google Scholar] [CrossRef]
- Huang, Y.C.; Chung, H.H.; Dutkiewicz, E.P.; Chen, C.L.; Hsieh, H.Y.; Chen, B.R.; Wang, M.Y.; Hsu, C.C. Predicting Breast Cancer by Paper Spray Ion Mobility Spectrometry Mass Spectrometry and Machine Learning. Anal. Chem. 2020, 92, 1653–1657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simões, R.V.; Serganova, I.S.; Kruchevsky, N.; Leftin, A.; Shestov, A.A.; Thaler, H.T.; Sukenick, G.; Locasale, J.W.; Blasberg, R.G.; Koutcher, J.A.; et al. Metabolic Plasticity of Metastatic Breast Cancer Cells: Adaptation to Changes in the Microenvironment. Neoplasia 2015, 17, 671–684. [Google Scholar] [CrossRef] [Green Version]
- Williams, N.C.; O’Neill, L.A.J. A Role for the Krebs Cycle Intermediate Citrate in Metabolic Reprogramming in Innate Immunity and Inflammation. Front. Immunol. 2018, 9, 141. [Google Scholar] [CrossRef] [Green Version]
- Icard, P.; Poulain, L.; Lincet, H. Understanding the central role of citrate in the metabolism of cancer cells. Biochim. Biophys. Acta Rev. Cancer 2012, 1825, 111–116. [Google Scholar] [CrossRef]
- Iacobazzi, V.; Infantino, V. Citrate—New functions for an old metabolite. Biol. Chem. 2014, 395, 387–399. [Google Scholar] [CrossRef]
- Caneba, C.A.; Bellance, N.; Yang, L.; Pabst, L.; Nagrath, D. Pyruvate uptake is increased in highly invasive ovarian cancer cells under anoikis conditions for anaplerosis, mitochondrial function, and migration. Am. J. Physiol. Metab. 2012, 303, E1036–E1052. [Google Scholar] [CrossRef] [Green Version]
- Davis, R.T.; Blake, K.; Ma, D.; Gabra, M.B.I.; Hernandez, G.A.; Phung, A.T.; Yang, Y.; Maurer, D.; Lefebvre, A.E.Y.T.; Alshetaiwi, H.; et al. Transcriptional diversity and bioenergetic shift in human breast cancer metastasis revealed by single-cell RNA sequencing. Nat. Cell Biol. 2020, 22, 310–320. [Google Scholar] [CrossRef]
- Sancak, Y.; Peterson, T.R.; Shaul, Y.D.; Lindquist, R.A.; Thoreen, C.C.; Bar-Peled, L.; Sabatini, D.M. The Rag GTPases Bind Raptor and Mediate Amino Acid Signaling to mTORC1. Science 2008, 320, 1496–1501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Csibi, A.; Fendt, S.-M.; Li, C.; Poulogiannis, G.; Choo, A.Y.; Chapski, D.J.; Jeong, S.M.; Dempsey, J.M.; Parkhitko, A.; Morrison, T.; et al. The mTORC1 Pathway Stimulates Glutamine Metabolism and Cell Proliferation by Repressing SIRT4. Cell 2013, 153, 840–854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Selwan, E.M.; Edinger, A.L. Branched chain amino acid metabolism and cancer: The importance of keeping things in context. Transl. Cancer Res. 2017, 6, S578–S584. [Google Scholar] [CrossRef]
- Cluntun, A.A.; Lukey, M.J.; Cerione, R.A.; Locasale, J.W. Glutamine Metabolism in Cancer: Understanding the Heterogeneity. Trends Cancer 2017, 3, 169–180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kung, H.-N.; Marks, J.R.; Chi, J.-T. Glutamine Synthetase Is a Genetic Determinant of Cell Type–Specific Glutamine Independence in Breast Epithelia. PLoS Genet. 2011, 7, e1002229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glunde, K.; Bhujwalla, Z.M.; Ronen, S.M. Choline metabolism in malignant transformation. Nat. Rev. Cancer 2011, 11, 835–848. [Google Scholar] [CrossRef] [Green Version]
- Sukocheva, O.; Wadham, C. Role of sphingolipids in oestrogen signalling in breast cancer cells: An update. J. Endocrinol. 2014, 220, R25–R35. [Google Scholar] [CrossRef] [Green Version]
- Rocha, C.M.; Barros, A.S.; Gil, A.M.; Goodfellow, B.J.; Humpfer, E.; Spraul, M.; Carreira, I.M.; Melo, J.B.; Bernardo, J.; Gomes, A.; et al. Metabolic Profiling of Human Lung Cancer Tissue by 1 H High Resolution Magic Angle Spinning (HRMAS) NMR Spectroscopy. J. Proteome Res. 2010, 9, 319–332. [Google Scholar] [CrossRef]
- Kim, H.J.; Lee, S.; Oh, S.C. The Inositide Signaling Pathway as a Target for Treating Gastric Cancer and Colorectal Cancer. Front. Physiol. 2016, 7, 168. [Google Scholar] [CrossRef]


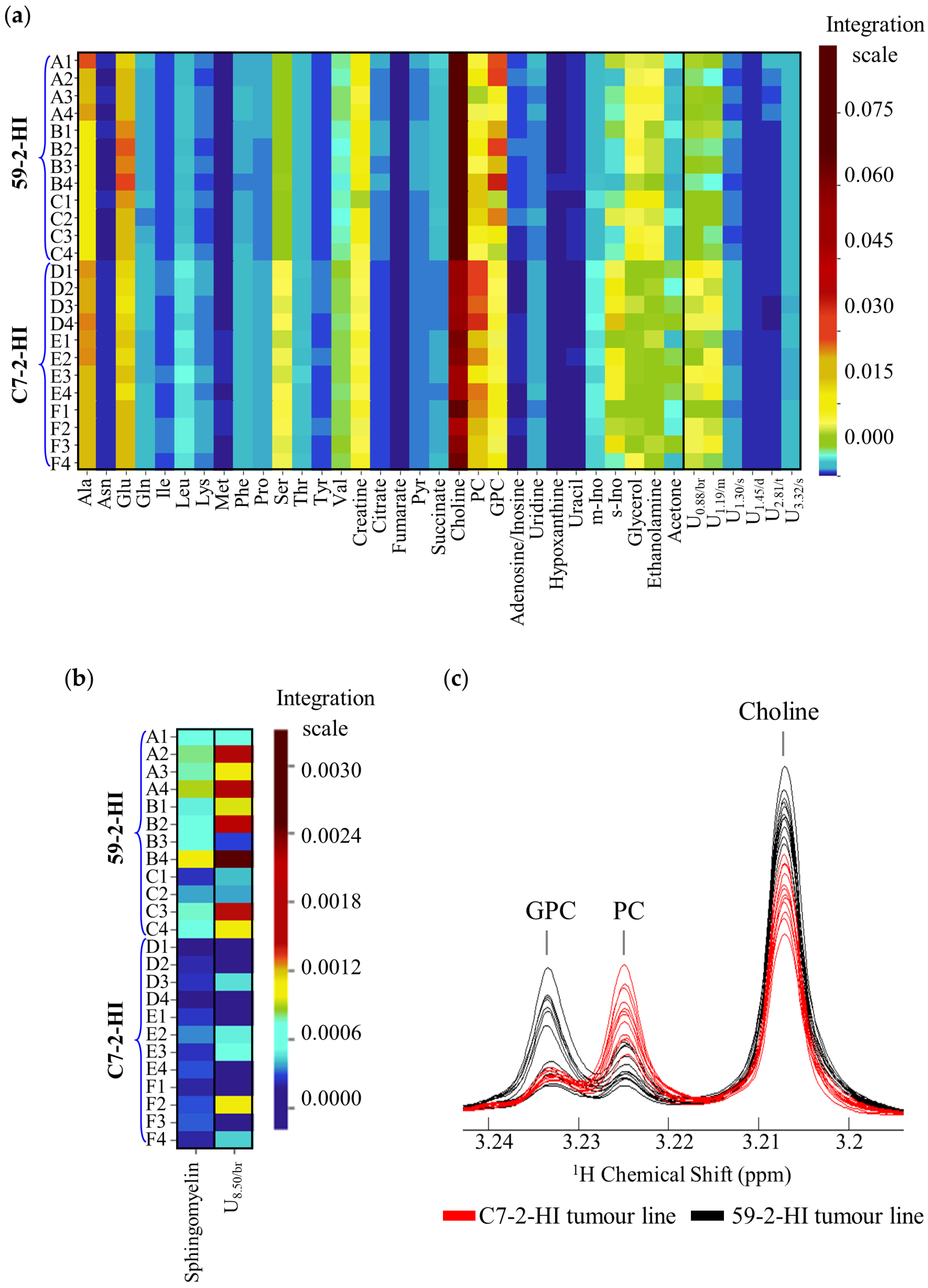
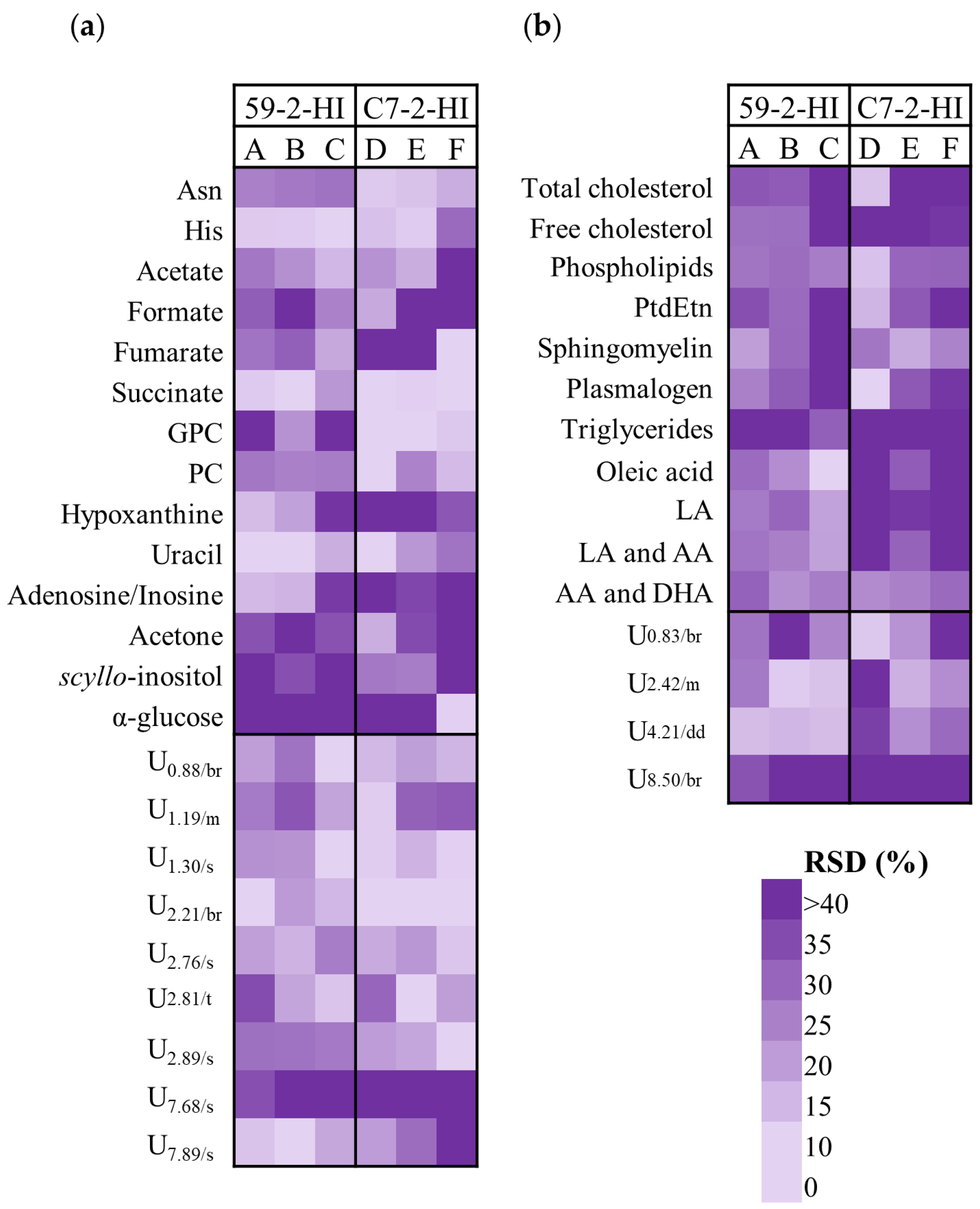
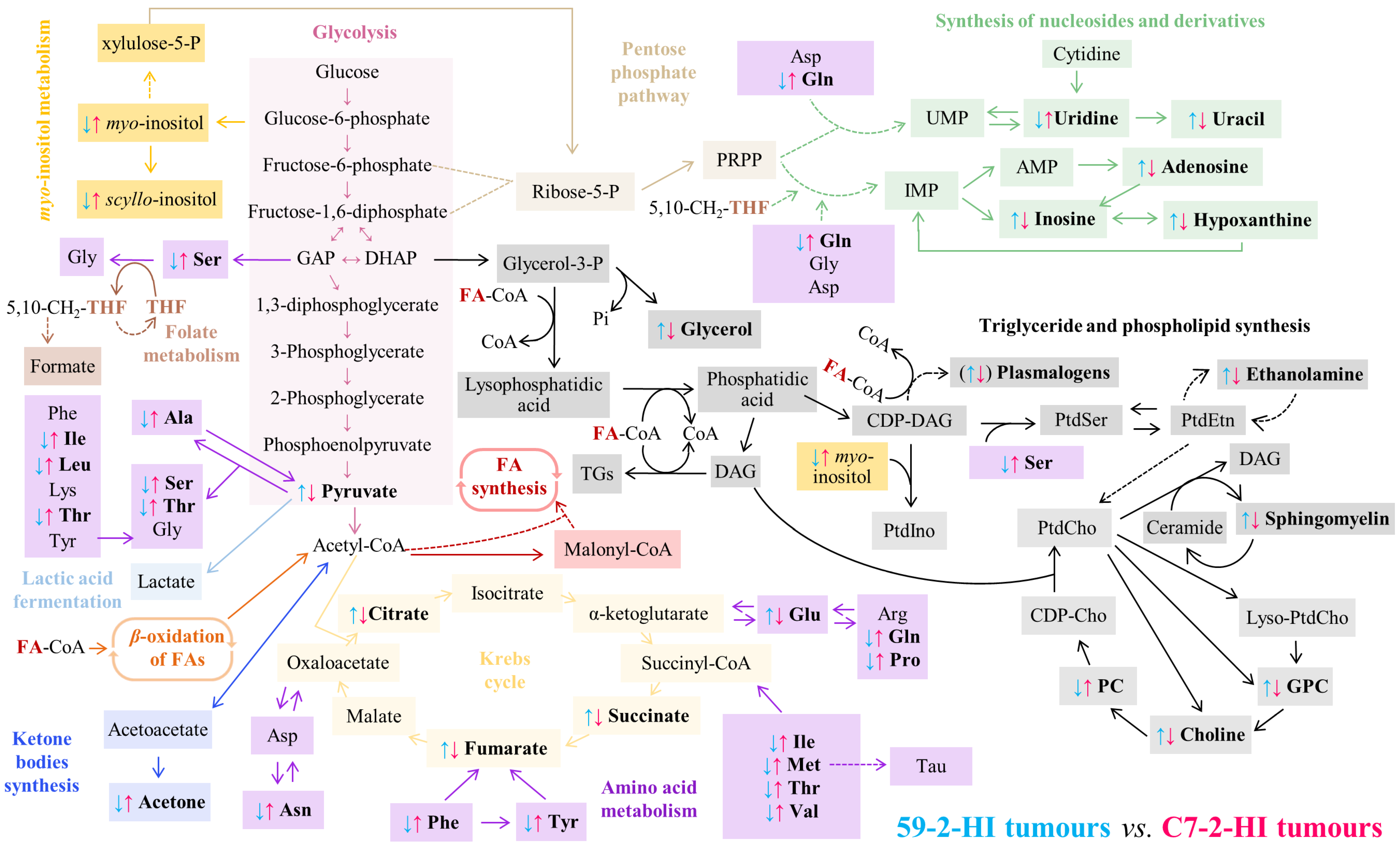
| δ1H/ppm a (Multiplicity) | Metabolite | 52-2-HI (A, B, C) vs. C7-2-HI (D, E, F) | |
|---|---|---|---|
| Effect Size (ES) b | p-Value c | ||
| Polar compounds | |||
| 1.48 (d) | Alanine | –1.78 ± 0.94 | 3.23 × 10−3 d |
| 2.84 (dd) | Asparagine | –1.35 ± 0.89 | 1.11 × 10−2 d |
| 2.36 (m) | Glutamate | 1.67 ± 0.93 | 1.50 × 10−3 d |
| 2.46 (m) | Glutamine | –1.54 ± 0.91 | 3.89 × 10−3 d |
| 1.02 (d) | Isoleucine | –2.76 ± 1.12 | 8.64 × 10−5 |
| 0.97 (d) | Leucine | –4.12 ± 1.41 | 4.15 × 10−5 |
| 1.92 (m) | Lysine | –3.07 ± 1.18 | 3.23 × 10−5 |
| 2.65 (t) | Methionine | –1.57 ± 0.92 | 1.82 × 10−3 d |
| 7.33 (m) | Phenylalanine | –3.37 ± 1.25 | 4.15 × 10−5 |
| 2.03 (m) | Proline | –2.77 ± 1.12 | 1.10 × 10−4 |
| 3.95 (dd) | Serine | –5.66 ± 1.79 | 3.23 × 10−5 |
| 4.25 (dd) | Threonine | –3.41 ± 1.25 | 3.23 × 10−5 |
| 6.91 (d) | Tyrosine | –2.04 ± 0.99 | 2.76 × 10−4 |
| 1.05 (d) | Valine | –2.92 ± 1.15 | 6.78 × 10−5 |
| 3.93 (s) | Creatine | 1.18 ± 0.87 | 2.68 × 10−3 d |
| 2.52 (d) | Citrate | 6.59 ± 2.03 | 3.23 × 10−5 |
| 6.52(s) | Fumarate | 1.64 ± 0.93 | 1.22 × 10−3 |
| 2.39 (s) | Pyruvate | 1.78 ± 0.95 | 8.12 × 10−4 |
| 2.41 (s) | Succinate | 3.18 ± 1.21 | 3.23 × 10–5 |
| 3.21 (s) | Choline | 3.68 ± 1.31 | 3.23 × 10−5 |
| 3.22 (s) | Phosphocholine | –2.25 ± 1.02 | 2.20 × 10−4 |
| 2.23 (s) | Glycerophosphocholine | 1.41 ± 0.89 | 2.82 × 10−2 d |
| 8.35 (s) | Adenosine/Inosine | 3.61 ± 1.30 | 3.23 × 10−5 |
| 7.86 (d) | Uridine | –1.27 ± 0.88 | 1.11 × 10−2 d |
| 8.22 (s) | Hypoxanthine | 1.98 ± 0.98 | 5.32 × 10−4 |
| 5.81 (d) | Uracil | 2.38 ± 1.05 | 8.64 × 10−5 |
| 3.36 (s) | scyllo–inositol | –1.96 ± 0.97 | 4.29 × 10−4 |
| 3.63 (t) | myo–inositol e | –3.21 ± 1.21 | 8.64 × 10−5 |
| 3.55 (dd) | Glycerol | 2.48 ± 1.06 | 1.39 × 10−4 |
| 3.15 (t) | Ethanolamine | 2.00 ± 0.98 | 2.20 × 10−4 |
| 2.24 (s) | Acetone | –1.90 ± 0.96 | 6.58 × 10−4 |
| 0.88 (br) | U0.88/br | –1.97 ± 0.98 | 4.29 × 10−4 |
| 1.19 (m) | U1.19/m | –1.85 ± 0.96 | 1.22 × 10−3 |
| 1.30 (s) | U1.30/s | –1.77 ± 0.94 | 4.29 × 10−4 |
| 1.45 (d) | U1.45/d | –3.06 ± 1.18 | 5.31 × 10−5 |
| 2.81 (t) | U2.81/t | 1.28 ± 0.88 | 6.78 × 10−5 |
| 3.12 (s) | U3.12/s | 1.95 ± 0.97 | 9.99 × 10−4 |
| Lipophilic compounds | |||
| 5.68 (m) | Sphingomyelin | 2.54 ± 1.08 | 1.10 × 10−4 |
| 8.50 (br) | U8.50/br | 1.51 ± 0.85 | 2.21 × 10−3 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bispo, D.; Fabris, V.; Lamb, C.A.; Lanari, C.; Helguero, L.A.; Gil, A.M. Hormone-Independent Mouse Mammary Adenocarcinomas with Different Metastatic Potential Exhibit Different Metabolic Signatures. Biomolecules 2020, 10, 1242. https://doi.org/10.3390/biom10091242
Bispo D, Fabris V, Lamb CA, Lanari C, Helguero LA, Gil AM. Hormone-Independent Mouse Mammary Adenocarcinomas with Different Metastatic Potential Exhibit Different Metabolic Signatures. Biomolecules. 2020; 10(9):1242. https://doi.org/10.3390/biom10091242
Chicago/Turabian StyleBispo, Daniela, Victoria Fabris, Caroline A. Lamb, Claudia Lanari, Luisa A. Helguero, and Ana M. Gil. 2020. "Hormone-Independent Mouse Mammary Adenocarcinomas with Different Metastatic Potential Exhibit Different Metabolic Signatures" Biomolecules 10, no. 9: 1242. https://doi.org/10.3390/biom10091242







