Determining the Molecular Background of Endometrial Receptivity in Adenomyosis
Abstract
1. Introduction
2. Materials and Methods
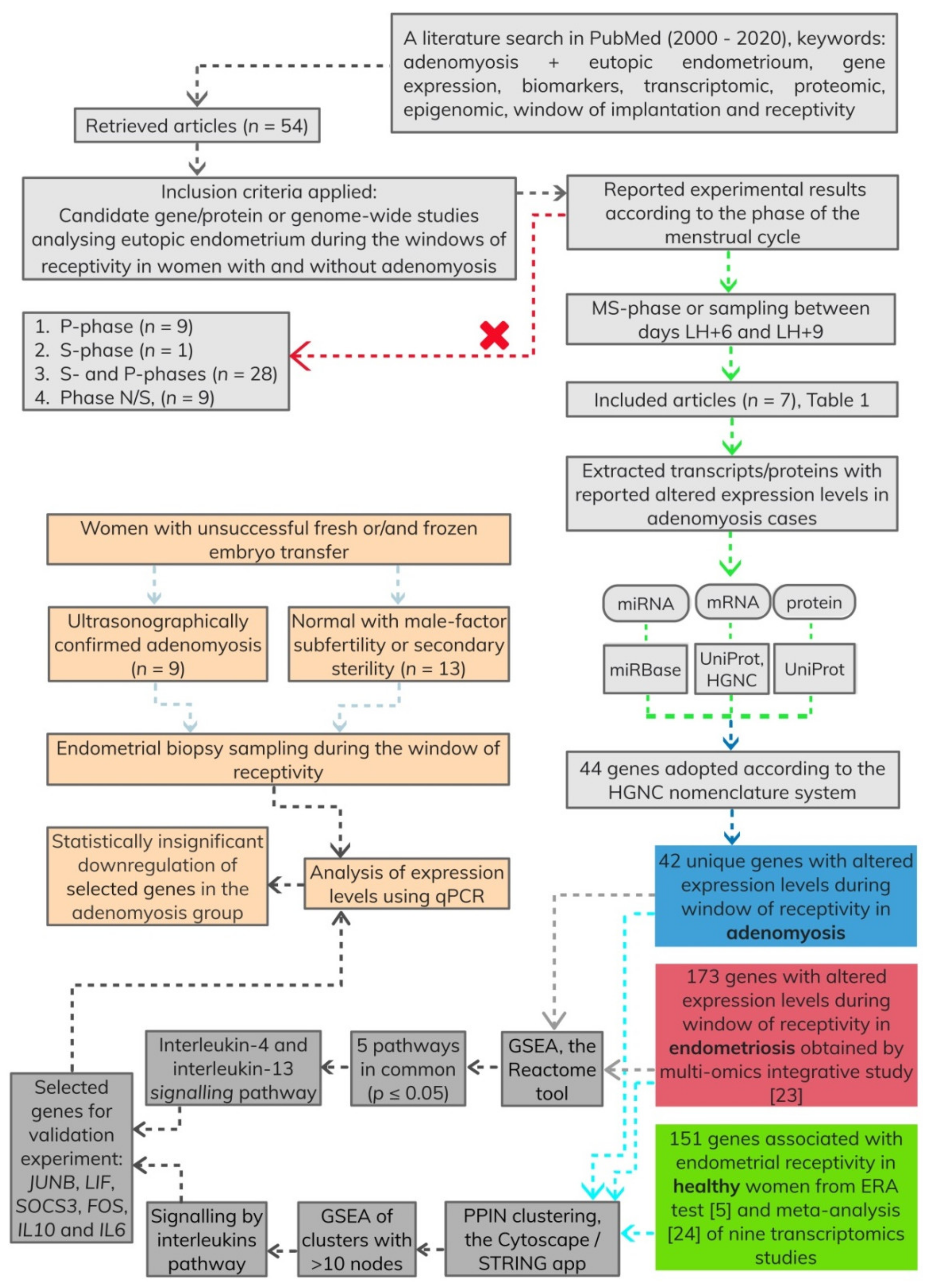
2.1. Identification of Relevant Studies for Development of Gene List
2.2. GSEA Using Retrieved Genes Associated with Adenomyosis, Endometriosis and Healthy Endometrium
2.3. GSEA Using Retrieved Genes Associated with Adenomyosis and Endometriosis Endometrium
2.4. Participating Women
2.5. Clinical Data
2.6. Endometrial Biopsy Sample Collection
2.7. Total RNA Isolation
2.8. Gene Expression Analysis by Quantitative PCR (qPCR)
2.9. Statistical Analysis
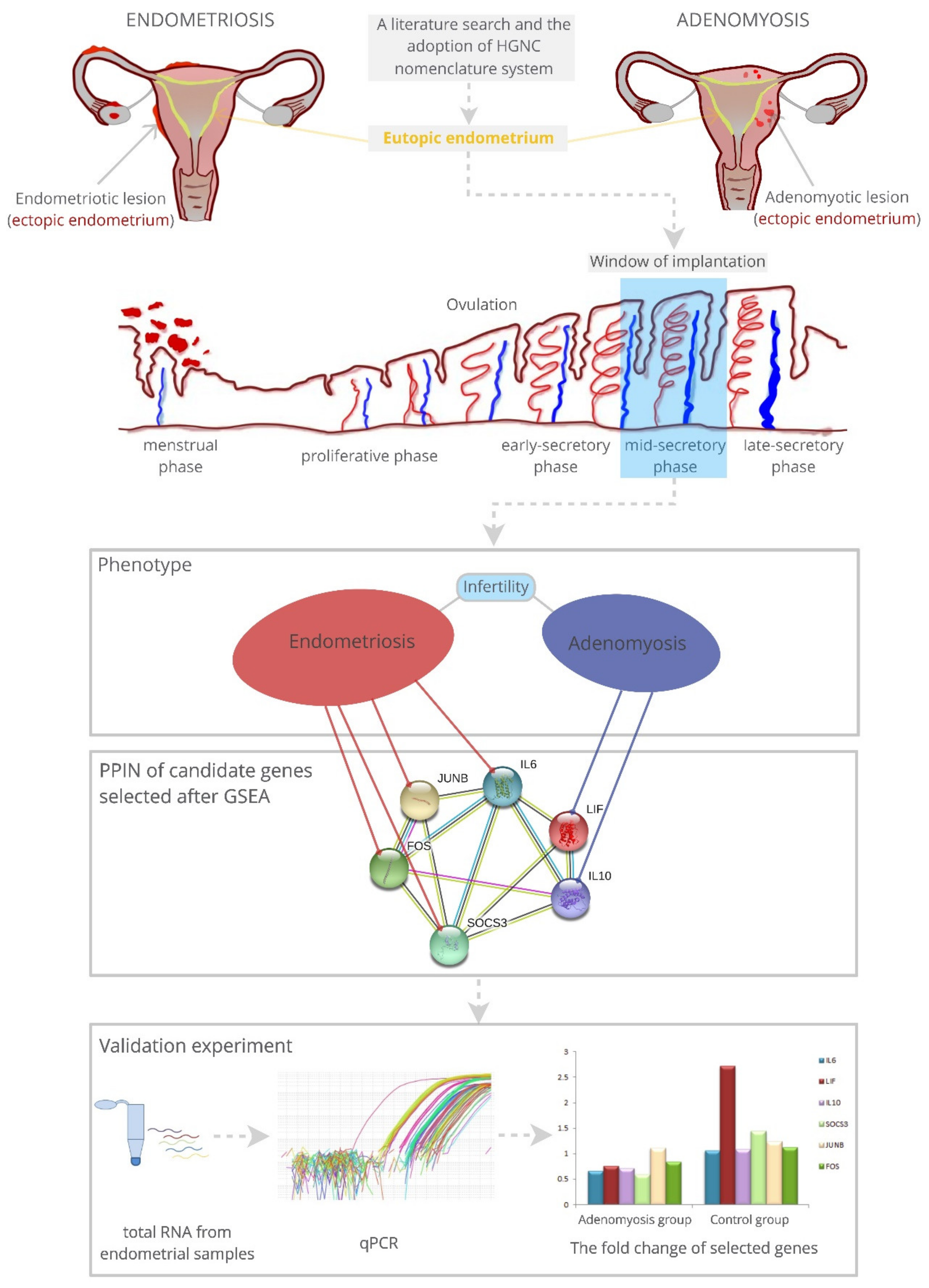
3. Results
3.1. Developed Gene Lists
3.2. Enriched Pathways after PPIN Clustering
3.3. Overlapping Enriched Pathways between Adenomyosis and Endometriosis Gene Lists
3.4. Prioritisation of Candidate Genes for Validation Experiment
3.5. Patients
3.6. Expression Patterns of Candidate Genes
4. Discussion
4.1. Integration of Reported Signatures and Highlighted Enriched Pathways
4.2. The Role of Selected Genes in Reproductive Biology
4.3. Endometrial Tissue Variability
4.4. Limitations of the Study
4.5. Future Directions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Altmäe, S.; Reimand, J.; Hovatta, O.; Zhang, P.; Kere, J.; Laisk-Podar, T.; Saare, M.; Peters, M.; Vilo, J.; Stavreus-Evers, A.; et al. Research Resource: Interactome of Human Embryo Implantation: Identification of Gene Expression Pathways, Regulation, and Integrated Regulatory Networks. Mol. Endocrinol. 2011, 26, 203–217. [Google Scholar] [CrossRef]
- Lessey, B.A.; Young, S.L. What Exactly is Endometrial Receptivity? Fertil. Steril. 2019, 111, 611–617. [Google Scholar] [CrossRef]
- Edgell, T.; Rombauts, L.J.; Salamonsen, L.A. Assessing Receptivity in the Endometrium: The Need for a Rapid, Non-Invasive Test. Reprod. Biomed. Online 2013, 27, 486–496. [Google Scholar] [CrossRef][Green Version]
- Bashiri, A.; Halper, K.I.; Orvieto, R. Recurrent Implantation Failure-Update Overview on Etiology, Diagnosis, Treatment and Future Directions. Reprod. Boil. Endocrinol. 2018, 16, 121. [Google Scholar] [CrossRef]
- Díaz-Gimeno, P.; Horcajadas, J.A.; Martinez-Conejero, J.A.; Esteban, F.J.; Alamá, P.; Pellicer, A.; Simón, C. A Genomic Diagnostic Tool for Human Endometrial Receptivity Based on the Transcriptomic Signature. Fertil. Steril. 2011, 95, 50–60. [Google Scholar] [CrossRef] [PubMed]
- Guerriero, S.; Dueholm, M.; Leone, F.P.G.; Valentin, L.; Rasmussen, C.; Votino, A.; Van Schoubroeck, D.; Landolfo, C.; Installé, A.J.; Guerriero, S.; et al. Terms, Definitions and Measurements to Describe Sonographic Features of Myometrium and Uterine Masses: A Consensus Opinion from the Morphological Uterus Sonographic Assessment (MUSA) Group. Ultrasound Obstet. Gynecol. 2015, 46, 284–298. [Google Scholar] [CrossRef]
- Bazot, M.; Darai, E. Role of Transvaginal Sonography and Magnetic Resonance Imaging in the Diagnosis of Uterine Adenomyosis. Fertil. Steril. 2018, 109, 389–397. [Google Scholar] [CrossRef]
- Puente, J.M.; Fabris, A.; Patel, J.; Patel, A.; Cerrillo, M.; Requena, A.; Garcia-Velasco, J.A. Adenomyosis in Infertile Women: Prevalence and the Role of 3D Ultrasound as a Marker of Severity of the Disease. Reprod. Boil. Endocrinol. 2016, 14, 60. [Google Scholar] [CrossRef] [PubMed]
- Abu Hashim, H.; Elaraby, S.; Fouda, A.A.; Rakhawy, M.E. The Prevalence of Adenomyosis in an Infertile Population: A Cross-Sectional Study. Reprod. Biomed. Online 2020, 40, 842–850. [Google Scholar] [CrossRef]
- Salim, R.; Riris, S.; Saab, W.; Abramov, B.; Khadum, I.; Serhal, P. Adenomyosis Reduces Pregnancy Rates in Infertile Women Undergoing IVF. Reprod. Biomed. Online 2012, 25, 273–277. [Google Scholar] [CrossRef] [PubMed]
- Vercellini, P.; Consonni, D.; Dridi, D.; Bracco, B.; Frattaruolo, M.P.; Somigliana, E. Uterine Adenomyosis and in vitro Fertilization Outcome: A Systematic Review and Meta-Analysis. Hum. Reprod. 2014, 29, 964–977. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Sun, X.; Yang, X.; Zhang, J.; Xue, Q.; Cai, B.; Zhou, Y. Leukemia Inhibitory Factor is Dysregulated in the Endometrium and Uterine Flushing Fluid of Patients with Adenomyosis during Implantation Window. Fertil. Steril. 2010, 94, 85–89. [Google Scholar] [CrossRef]
- Yen, C.-F.; Liao, S.-K.; Huang, S.J.; Tabak, S.; Arcuri, F.; Lee, C.-L.; Arici, A.; Petraglia, F.; Wang, H.-S.; Kayisli, U.A. Decreased Endometrial Expression of Leukemia Inhibitory Factor Receptor Disrupts the STAT3 Signaling in Adenomyosis During the Implantation Window. Reprod. Sci. 2016, 24, 1176–1186. [Google Scholar] [CrossRef]
- Kunz, G.; Beil, D.; Huppert, P.; Noe, M.; Kissler, S.; Leyendecker, G. Adenomyosis in Endometriosis—Prevalence and Impact on Fertility. Evidence from Magnetic Resonance Imaging. Hum. Reprod. 2005, 20, 2309–2316. [Google Scholar] [CrossRef]
- Meuleman, C.; Vandenabeele, B.; Fieuws, S.; Spiessens, C.; Timmerman, D.; D’Hooghe, T.M. High Prevalence of Endometriosis in Infertile Women with Normal Ovulation and Normospermic Partners. Fertil. Steril. 2009, 92, 68–74. [Google Scholar] [CrossRef]
- Kao, L.C.; Germeyer, A.; Tulac, S.; Lobo, S.; Yang, J.P.; Taylor, R.N.; Osteen, K.; Lessey, B.A.; Giudice, L.C. Expression Profiling of Endometrium from Women with Endometriosis Reveals Candidate Genes for Disease-Based Implantation Failure and Infertility. Endocrinology 2003, 144, 2870–2881. [Google Scholar] [CrossRef]
- Burney, R.O.; Talbi, S.; Hamilton, A.E.; Vo, K.C.; Nyegaard, M.; Nezhat, C.R.; Lessey, B.A.; Giudice, L.C. Gene Expression Analysis of Endometrium Reveals Progesterone Resistance and Candidate Susceptibility Genes in Women with Endometriosis. Endocrinology 2007, 148, 3814–3826. [Google Scholar] [CrossRef]
- Tamaresis, J.S.; Irwin, J.C.; Goldfien, G.A.; Rabban, J.T.; Burney, R.O.; Nezhat, C.; DePaolo, L.V.; Giudice, L. Molecular Classification of Endometriosis and Disease Stage Using High-Dimensional Genomic Data. Endocrinology 2014, 155, 4986–4999. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Fu, J.; Xiao, L.; Yang, S.; Song, Y.; Zhang, X.; Feng, X.; Sun, H.; Xu, W.; Huang, W. miR-196a Overexpression Activates the MEK/ERK Signal and Represses the Progesterone Receptor and Decidualization in Eutopic Endometrium from Women with Endometriosis. Hum. Reprod. 2016, 31, 2598–2608. [Google Scholar] [CrossRef]
- Houshdaran, S.; Nezhat, C.R.; Vo, K.C.; Zelenko, Z.; Irwin, J.C.; Giudice, L.C. Aberrant Endometrial DNA Methylome and Associated Gene Expression in Women with Endometriosis. Boil. Reprod. 2016, 95, 93. [Google Scholar] [CrossRef]
- Barabasi, A.-L.; Gulbahce, N.; Loscalzo, J. Network Medicine: A Network-Based Approach to Human Disease. Nat. Rev. Genet. 2010, 12, 56–68. [Google Scholar] [CrossRef] [PubMed]
- García-Campos, M.A.; Espinal-Enríquez, J.; Hernández-Lemus, E. Pathway Analysis: State of the Art. Front. Physiol. 2015, 6, 383. [Google Scholar] [CrossRef]
- Prašnikar, E.; Knez, J.; Kovačič, B.; Kunej, T. Molecular signature of eutopic endometrium in endometriosis based on the multi-omics integrative synthesis. J. Assist. Reprod. Genet. 2020, 37, 1–19. [Google Scholar] [CrossRef]
- Altmäe, S.; Koel, M.; Võsa, U.; Adler, P.; Suhorutšenko, M.; Laisk-Podar, T.; Kukushkina, V.; Saare, M.; Velthut-Meikas, A.; Krjutškov, K.; et al. Meta-Signature of Human Endometrial Receptivity: A Meta-Analysis and Validation Study of Transcriptomic Biomarkers. Sci. Rep. 2017, 7, 10077. [Google Scholar] [CrossRef]
- Braschi, B.; Denny, P.; Gray, K.; Jones, T.E.; Seal, R.; Tweedie, S.; Yates, B.; Bruford, E.A. Genenames.org: The HGNC and VGNC Resources in 2019. Nucleic Acids Res. 2018, 47, D786–D792. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–Protein Association Networks with Increased Coverage, Supporting Functional Discovery in Genome-Wide Experimental Datasets. Nucleic Acids Res. 2018, 47, D607–D613. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Feng, X.; Stein, L. A Human Functional Protein Interaction Network and its Application to Cancer Data Analysis. Genome Boil. 2010, 11, R53. [Google Scholar] [CrossRef] [PubMed]
- Fabregat, A.; Sidiropoulos, K.; Garapati, P.V.; Gillespie, M.E.; Hausmann, K.; Haw, R.A.; Jassal, B.; Jupe, S.; Korninger, F.; McKay, S.; et al. The Reactome pathway Knowledgebase. Nucleic Acids Res. 2015, 44, D481–D487. [Google Scholar] [CrossRef] [PubMed]
- Naftalin, J.; Hoo, W.; Pateman, K.; Mavrelos, D.; Jurkovic, D.; Holland, T.K. How Common is Adenomyosis? A Prospective Study of Prevalence Using Transvaginal Ultrasound in a Gynaecology Clinic. Hum. Reprod. 2012, 27, 3432–3439. [Google Scholar] [CrossRef]
- WHO. Laboratory Manual for the Examination and Processing of Human Semen, 5th ed.; Cooper, T.G., Ed.; WHO Press: Geneva, Switzerland, 2010. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Fischer, C.P.; Kayisili, U.; Taylor, H.S. HOXA10 Expression is Decreased in Endometrium of Women with Adenomyosis. Fertil. Steril. 2011, 95, 1133–1136. [Google Scholar] [CrossRef]
- Xiao, Y.; Li, T.; Xia, E.; Yang, X.; Sun, X.; Zhou, Y. Expression of Integrin β3 and Osteopontin in the Eutopic Endometrium of Adenomyosis during the Implantation Window. Eur. J. Obstet. Gynecol. Reprod. Boil. 2013, 170, 419–422. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Huang, C.; Jiang, R.; Du, Y.; Zhou, J.; Jiang, Y.; Yan, Q.; Xing, J.; Hou, X.; Zhou, J.; et al. Decreased Endometrial IL-10 Impairs Endometrial Receptivity by Downregulating HOXA10 Expression in Women with Adenomyosis. BioMed Res. Int. 2018, 2018, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Yan, Q.; Yan, G.; Zhang, C.; Wang, Z.; Huang, C.; Wang, J.; Zhou, J.; Liu, Y.; Ding, L.; Zhang, Q.; et al. miR-21 Reverses Impaired Decidualization Through Modulation of KLF12 and NR4A1 Expression in Human Endometrial Stromal Cells. Boil. Reprod. 2019, 100, 1395–1405. [Google Scholar] [CrossRef]
- Martinez-Conejero, J.A.; Morgan, M.; Montesinos, M.; Fortuño, S.; Meseguer, M.; Simón, C.; Horcajadas, J.A.; Pellicer, A. Adenomyosis does not Affect Implantation, but is Associated with Miscarriage in Patients Undergoing Oocyte Donation. Fertil. Steril. 2011, 96, 943–950. [Google Scholar] [CrossRef]
- Stephens, A.N.; Hannan, N.J.; Rainczuk, A.; Meehan, K.L.; Chen, J.; Nicholls, P.K.; Rombauts, L.J.F.; Stanton, P.G.; Robertson, D.M.; Salamonsen, L.A. Post-Translational Modifications and Protein-Specific Isoforms in Endometriosis Revealed by 2D DIGE. J. Proteome Res. 2010, 9, 2438–2449. [Google Scholar] [CrossRef]
- Okada, H.; Tsuzuki, T.; Murata, H. Decidualization of the Human Endometrium. Reprod. Med. Boil. 2018, 17, 220–227. [Google Scholar] [CrossRef]
- Campo, S.; Campo, V.; Benagiano, G. Adenomyosis and Infertility. Reprod. Biomed. Online 2012, 24, 35–46. [Google Scholar] [CrossRef]
- Chaouat, G.; Dubanchet, S.; Ledee, N. Cytokines: Important for implantation? J. Assist. Reprod. Genet. 2007, 24, 491–505. [Google Scholar] [CrossRef]
- Flynn, L.; Byrne, B.; Carton, J.; O’Farrelly, C.; Kelehan, P.; O’Herlihy, C. Menstrual Cycle Dependent Fluctuations in NK and T-Lymphocyte Subsets from Non-Pregnant Human Endometrium. Am. J. Reprod. Immunol. 2000, 43, 209–217. [Google Scholar] [CrossRef] [PubMed]
- Van Nieuwenhoven, A.V.; Heineman, M.; Faas, M. The Immunology of Successful Pregnancy. Hum. Reprod. Updat. 2003, 9, 347–357. [Google Scholar] [CrossRef] [PubMed]
- Enciso, M.; Carrascosa, J.P.; Sarasa, J.; A Martínez-Ortiz, P.; Munné, S.; A Horcajadas, J.; Aizpurua, J. Development of a New Comprehensive and Reliable Endometrial Receptivity Map (ER Map/ER Grade) Based on RT-qPCR Gene Expression Analysis. Hum. Reprod. 2018, 33, 220–228. [Google Scholar] [CrossRef] [PubMed]
- Vogel, C.; Marcotte, E.M. Insights into Regulation of Protein Abundance from Proteomics and Transcriptomis Analyses. Nat. Rev. Genet. 2013, 13, 227–232. [Google Scholar] [CrossRef] [PubMed]
- Shuya, L.L.; Menkhorst, E.; Yap, J.; Li, P.; Lane, N.; Dimitriadis, E. Leukemia Inhibitory Factor Enhances Endometrial Stromal Cell Decidualization in Humans and Mice. PLoS ONE 2011, 6, e25288. [Google Scholar] [CrossRef] [PubMed]
- Chung, T.-W.; Park, M.-J.; Kim, H.S.; Choi, H.-J.; Ha, K.-T. Integrin αVβ3 and αVβ5 are Required for Leukemia Inhibitory Factor-Mediated the Adhesion of Trophoblast Cells to the Endometrial Cells. Biochem. Biophys. Res. Commun. 2016, 469, 936–940. [Google Scholar] [CrossRef]
- Serafini, P.; Silva, I.D.; Smith, G.D.; Motta, E.L.; Rocha, A.; Baracat, E.C. Endometrial Claudin-4 and Leukemia Inhibitory Factor are Associated with Assisted Reproduction Outcome. Reprod. Boil. Endocrinol. 2009, 7, 30. [Google Scholar] [CrossRef]
- Franasiak, J.M.; Holoch, K.J.; Yuan, L.; Schammel, D.P.; Young, S.L.; Lessey, B.A. Prospective Assessment of Midsecretory Endometrial Leukemia Inhibitor Factor Expression Versus ανβ3 Testing in Women with Unexplained Infertility. Fertil. Steril. 2014, 101, 1724–1731. [Google Scholar] [CrossRef]
- Kühn, R.; Löhler, J.; Rennick, D.; Rajewsky, K.; Muller, W. Interleukin-10-Deficient Mice Develop Chronic Enterocolitis. Cell 1993, 75, 263–274. [Google Scholar] [CrossRef]
- Thaxton, J.E.; Sharma, S. Interleukin-10: A Multi-Faceted Agent of Pregnancy. Am. J. Reprod. Immunol. 2010, 63, 482–491. [Google Scholar] [CrossRef]
- Diehl, S.A.; Rincoón, M. The Two Faces of IL-6 on Th1/Th2 Differentiation. Mol. Immunol. 2002, 39, 531–536. [Google Scholar] [CrossRef]
- Von Wolff, M.; Thaler, C.; Strowitzki, T.; Broome, J.; Stolz, W.; Tabibzadeh, S. Regulated Expression of Cytokines in Human Endometrium Throughout the Menstrual Cycle: Dysregulation in Habitual Abortion. Mol. Hum. Reprod. 2000, 6, 627–634. [Google Scholar] [CrossRef] [PubMed]
- Zhihong, N.; Yun, F.; Pinggui, Z.; Sulian, Z.; Zhang, A. Cytokine Profiling in the Eutopic Endometrium of Adenomyosis During the Implantation Window After Ovarian Stimulation. Reprod. Sci. 2015, 23, 124–133. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.-H.; Wu, M.-Y.; Chang, D.-Y.; Chang, C.-H.; Yang, Y.-S.; Ho, H.-N. Increased Interleukin-6 Messenger RNA Expression in Macrophage-Cocultured Endometrial Stromal Cells in Adenomyosis. Am. J. Reprod. Immunol. 2006, 55, 181–187. [Google Scholar] [CrossRef]
- Ponce, C.; Torres, M.; Galleguillos, C.; Sovino, H.; Boric, M.A.; Fuentes, A.; Johnson, M.C. Nuclear Factor κB Pathway and Interleukin-6 are Affected in Eutopic Endometrium of Women with Endometriosis. Reproduction 2009, 137, 727–737. [Google Scholar] [CrossRef]
- Jochum, W.; Passegué, E.; Wagner, E.F. AP-1 in Mouse Development and Tumorigenesis. Oncogene 2001, 20, 2401–2412. [Google Scholar] [CrossRef]
- Raimundo, N.; Vanharanta, S.; Aaltonen, L.A.; Hovatta, I.; Suomalainen, A. Downregulation of SRF–FOS–JUNB Pathway in Fumarate Hydratase Deficiency and in Uterine Leiomyomas. Oncogene 2009, 28, 1261–1273. [Google Scholar] [CrossRef]
- Li, B.; Tournier, C.; Davis, R.J.; Flavell, R.A. Regulation of IL-4 Expression by the Transcription Factor JunB during T Helper Cell Differentiation. EMBO J. 1999, 18, 420–432. [Google Scholar] [CrossRef]
- Absenger, Y.; Hess-Stumpp, H.; Kreft, B.; Krätzschmar, J.; Haendler, B.; Schütze, N.; Regidor, P.; Winterhager, E. Cyr61, a Deregulated Gene in Endometriosis. Mol. Hum. Reprod. 2004, 10, 399–407. [Google Scholar] [CrossRef][Green Version]
- Pan, H.; Sheng, J.-Z.; Tang, L.; Zhu, R.; Zhou, T.-H.; Huang, H. Increased Expression of C-Fos Protein Associated with Increased Matrix Metalloproteinase-9 Protein Expression in the Endometrium of Endometriotic Patients. Fertil. Steril. 2008, 90, 1000–1007. [Google Scholar] [CrossRef]
- Morsch, D.M.; Carneiro, M.M.; Lecke, S.B.; Araújo, F.C.; Camargos, A.F.; Reis, F.M.; Spritzer, P.M. C-Fos Gene and Protein Expression in Pelvic Endometriosis: A Local Marker of Estrogen Action. J. Mol. Histol. 2009, 40, 53–58. [Google Scholar] [CrossRef] [PubMed]
- Lang, R.; Pauleau, A.-L.; Parganas, E.; Takahashi, Y.; Mages, J.; Ihle, J.N.; Rutschman, R.; Murray, P.J. SOCS3 Regulates the Plasticity of gp130 Signaling. Nat. Immunol. 2003, 4, 546–550. [Google Scholar] [CrossRef] [PubMed]
- Dong, Q.; Fan, R.; Zhao, S.; Wang, Y. Over-expression of SOCS-3 Gene Promotes IL-10 Production by JEG-3 Trophoblast Cells. Placenta 2009, 30, 11–14. [Google Scholar] [CrossRef]
- Braunschweig, A.; Poehlmann, T.G.; Busch, S.; Schleussner, E.; Markert, U.R. Signal Transducer and Activator of Transcription 3 (STAT3) and Suppressor of Cytokine Signaling (SOCS3) Balance Controls Cytotoxicity and IL-10 Expression in Decidual-Like Natural Killer Cell Line NK-92. Am. J. Reprod. Immunol. 2011, 66, 329–335. [Google Scholar] [CrossRef] [PubMed]
- Suhorutshenko, M.; Kukushkina, V.; Velthut-Meikas, A.; Altmäe, S.; Peters, M.; Mägi, R.; Krjutškov, K.; Koel, M.; Codoñer, F.M.; Martinez-Blanch, J.F.; et al. Endometrial Receptivity Revisited: Endometrial Transcriptome Adjusted for Tissue Cellular Heterogeneity. Hum. Reprod. 2018, 33, 2074–2086. [Google Scholar] [CrossRef]



| Study Design of Retrieved Studies | Biotype of Assessed Biological Entity in Source Reference | Biological Entities Assessed in Source Reference | The HGNC Gene Symbol | Approved Gene Name | Reported Up-(↑) or Down-(↓) Regulation in Adenomyosis (A) Compared to Control (C) Group, (Statistical Significance) | Indication and Procedure for Endometrial Tissue Samples Collection | Inclusion Criteria and Composition of A. Number (n) of Participants | Inclusion Criteria and Composition of C. n of Participants. | Source Reference |
| Candidate gene/protein | miRNA | miR-21 | MIR21 | microRNA 21 | ~50% ↓ (p < 0.001) | Endometrial biopsy between days 19 and 23 of the menstrual cycle | Confirmed 2 of 5 sonographic features of A. Aged 31.4 ± 0.7 years. n = 12 | ≥1 normal pregnancy and/or delivery. Aged 30.4 ± 0.6 years. n = 12 | [36] |
| protein | KLF12 | KLF12 | Kruppel like factor 12 | ~2-fold ↑ (p < 0.001) | |||||
| protein | NR4A1 | NR4A1 | Nuclear receptor subfamily 4 group A member 1 | ~50% ↓ (p < 0.001) | |||||
| protein | IL-10 | IL10 | Interleukin 10 | 40% ↓ (p < 0.001) | Endometrial biopsy in LH+7 using an endometrial curette | >2 of 5 sonographic features of A, clinical symptoms (secondary and progressive dysmenorrhea, menorrhagia and menostaxis), and clinical signs (homogenous enlargement of local uplift of the uterus, firmness and tenderness). Aged 40 years or younger. n = 23 | Tubal factor infertility. Aged 40 years or younger. n = 23 | [35] | |
| protein | HOXA10 | HOXA10 | Homeobox A10 | ~50% ↓ (p < 0.001) | |||||
| Protein phosphorylation | p(Y705)-STAT3 | STAT3 | Signal transducer and activator of transcription 3 | ~50% ↓ (p < 0.001) | |||||
| mRNA, protein | LIFR | LIFR | LIF receptor subunit alpha | 0.3-fold ↓ mRNA (p < 0.01). Protein ↓ according to H-score: - glandular cells 43.6 in A vs. 92.3 in C (p < 0.001). - stromal cells: 27.3 in A vs. 57.7 in C (p < 0.001). | Laparoscopically assisted vaginal hysterectomy (LAVH) or myomectomy (part of C samples) between days 19 and 23 of the menstrual cycle. | Diagnosed A preoperatively with clinical symptoms and ultrasonogram. A confirmed postoperatively by pathologist. Aged 39.8 ± 0.8 years. - Diffuse A, n = 5. - Focal A, n = 4. | - Intramural or subserosal leiomyoma, n = 16. - Submucosal leiomyoma, n = 3. Aged 41.2 ± 0.5 years. | [13] | |
| mRNA, protein | LIF | LIF | LIF interleukin 6 family cytokine | 0.15-fold ↓ mRNA (p < 0.01). protein ↓ according to the H-score: - glandular cells 61.5 in A vs. 231.8 in C (p < 0.001). - stromal cells: 40.7 in A vs. 135.4 in C (p < 0.001). | |||||
| mRNA, protein | Integrin β3 | ITGB3 | Integrin subunit beta 3 | mRNA ↓: Median value 6.2 in A vs. 21.5 in C (p < 0.01). protein ↓ according to the H-score: 2.0 in A vs. 2.7 in C (p < 0.05). | A group: hysterectomy for dysmenorrhea, hysteroscopy and diagnostic curettage for history of infertility. C group: testing for tubal patency or hysterectomy for pathological changes of the cervix. Ultrasound monitoring of menstrual cycle for endometrial sampling between days 7 and 9 following ovulation. Histologically confirmed mid-late secretory phase. | Enlarged uterus revealed by MRI and/or high level of serum CA125. Aged 40.1 ± 2.7 years. - Dysmenorrhea, n = 11. - History of infertility, n = 17. | Born at least 1 child and used some form of contraception with: - tubal factor of infertility or - pathological changes of the cervix. Aged 40.9 ± 2.1 years. n = 27 | [34] | |
| mRNA, protein | OPN | SPP1 | Secreted phosphoprotein 1 | mRNA ↓: Median value 12.2 in A vs. 24.2 in C (p < 0.01). protein ↓ according to the H-score: 2.1 in A vs. 2.7 in C (p < 0.05). | |||||
| mRNA, protein | LIF | LIF | LIF interleukin 6 family cytokine | Protein ↓ according to the H-score: 2.0 in A vs. 2.7 in C (p < 0.05). | Laparoscopy for tubal ligation, testing for tubal patency, or women without endometriosis who had hysterectomy for pathological changes of the cervix. Endometrial sampling by curettage and dating determined by histology. | Preoperative A diagnosis on clinical presentation, enlarged uterus determined by TVUS and/or high values of CA125. A confirmed by two histopathologists. Aged 40.3 ± 2.7 years. Total n = 28 with: - infertility. n = 21 or/and - dysmenorrhea. n = 11. | Fertile with: - tubal ligation - pathologic changes of the cervix Aged 40.9 ± 2.6 years. n = 27 | [12] | |
| protein | HOXA10 | HOXA10 | Homeobox A10 | ↓ according to the H-score: 1.4 in A vs. 2.1 in C (p < 0.001). | A group: hysterectomy for hypermenorrhea or adnexal mass. A confirmed by histopathology. C group: hysterectomy for pelvic organ prolapse or benign adnexal mass. Histologically confirmed MS-phase of the menstrual cycle | Aged between 25 to 52 years old. n = 19 | Fertile with: - pelvic organ prolapse or, - benign adnexal mass. Aged between 25 to 52 years old. Total n = 12 | [33] | |
| Genome-wide | mRNA | TMPRSS11B | TMPRSS11B | Transmembrane serine protease 11B | 215.5 fold change (FC) ↑, (pfp* = 0.0421) | Systematic TVUS to detect ovulation. When a follicle of 17–18 mm was detected, LH peak was determined by urinary test. Endometrial biopsies sampling in LH+7 using Pipelle catheter. | A diagnosed by MRI and TVUS - Never pregnant, n = 2 - Two term pregnancies, n = 1 - A single pregnancy, n = 1 - Two early miscarriages, n = 1 - A miscarriage, n = 1 | Young women with regular menses, no uterine or endocrine anomalies, proven fertility (previous spontaneous pregnancy at term). Normal results in a uterine ultrasonography. n = 6 | [37] |
| mRNA | CHD5 | CHD5 | Chromodomain helicase DNA binding protein 5 | 31.8 FC ↑ (0.00000) Confirmed by qPCR validation | |||||
| mRNA | SST | SST | Somatostatin | 10.6 FC ↑ (0.0350) Confirmed by qPCR validation | |||||
| mRNA | SPBC25 | SPC25 | SPC25 component of NDC80 kinetochore complex | 9.8 FC ↑ (0.0367) | |||||
| mRNA | FLJ20105 | ERCC6L | ERCC excision repair 6 like, spindle assembly checkpoint helicase | 9.1 FC ↑ (0.0496) | |||||
| mRNA | AKR1B10 | AKR1B10 | Aldo-keto reductase family 1 member B10 | 8.7 FC ↑ (0.0317) Confirmed by qPCR validation | |||||
| mRNA | CDKN3 | CDKN3 | Cyclin dependent kinase inhibitor 3 | 8.0 FC ↑ (0.0406) | |||||
| mRNA | ATP1A2 | ATP1A2 | ATPase Na+/K+ transporting subunit alpha 2 | 6.2 FC ↑ (0.04069 Confirmed by qPCR validation | |||||
| mRNA | MB | MB | Myoglobin | 5.8 FC ↑ (0.0262) | |||||
| mRNA | KCNA4 | KCNA4 | Potassium voltage-gated channel subfamily A member 4 | 5.5 FC ↑ (0.0255) | |||||
| mRNA | MMP20 | MMP20 | Matrix metallopeptidase 20 | 5.1 FC ↑ (0.0468) | |||||
| mRNA | FNDC1 | FNDC1 | Fibronectin type III domain containing 1 | 5.0 FC ↑ (0.0464) | |||||
| mRNA | TUBAL3 | TUBAL3 | Tubulin alpha like 3 | 4.9 FC ↑ (0.0425) | |||||
| mRNA | SPINK2 | SPINK2 | Serine peptidase inhibitor Kazal type 2 | 4.8 FC ↑ (0.0406) | |||||
| mRNA | COL11A1 | COL11A1 | Collagen type XI alpha 1 chain | 4.6 FC ↑ (0.0493) | |||||
| mRNA | LOC220115 | / | 4.1 FC ↑ (0.0278) | ||||||
| mRNA | LIPH | LIPH | Lipase H | 3.9 FC ↑ (0.0356) | |||||
| lncRNA | C21orf121 | ZNF295-AS1 | ZNF295 antisense RNA 1 | 3.5 FC ↑ (0.0343) | |||||
| mRNA | PSG6 | PSG6 | Pregnancy specific beta-1-glycoprotein 6 | 3.3 FC ↑ (0.0280) | |||||
| mRNA | C3orf33 | C3orf33 | Chromosome 3 open reading frame 33 | 3.2 FC ↑ (0.0475) | |||||
| mRNA | MDAC1 | TMEM190 | Transmembrane protein 190 | 2.4 FC ↑ (0.0275) | |||||
| mRNA | COL8A1 | COL8A1 | Collagen type VII alpha 1 chain | 2.2 FC ↑ (0.0488) | |||||
| mRNA | LTF | LTF | Lactotransferrin | 1.9 FC ↑ (0.0340) | |||||
| mRNA | SULT1E1 | SULT1E1 | Sulfotransferase family 1E member 1 | 1.8 FC ↑ (0.0410) | |||||
| mRNA | TBX15 | TBX15 | T-box transcription factor 15 | 1.7 FC ↑ (0.0340) | |||||
| mRNA | ATP12A | ATP12A | ATPase H+/K+ transporting non-gastric alpha2 subunit | −5.1 FC ↓ (0.0340) | |||||
| mRNA | LOC401233 | / | −3.8 FC ↓ (0.0000) | ||||||
| mRNA | CLDN4 | CLDN4 | Claudin 4 | −3.7 FC ↓ (0.0488) | |||||
| mRNA | LOC643338 | C15orf62 | Chromosome 15 open reading frame 62 | −3.2 FC ↓ (0.0450) | |||||
| mRNA | SCGB2A2 | SCGB2A2 | Secretoglobin family 2A member 2 | −3.1 FC ↓ (0.0233) | |||||
| mRNA | TCN1 | TCN1 | Transcobalamin 1 | −2.6 FC ↓ (0.0220) | |||||
| mRNA | GPR78 | GPR78 | G protein-coupled receptor 78 | −1.7 ↓ (0.0175) | |||||
| mRNA | CACNA1E | CACNA1E | Calcium voltage-gated channel subunit alpha1 E | −1.6 FC ↓ (0.0471) | |||||
| mRNA | CYP3A7 | CYP3A7 | Cytochrome P450 family 3 subfamily A member 7 | −1.2 FC ↓ (0.0433) |
| Group of Women | Gene List |
|---|---|
| Adenomyosis | AKR1B10, ATP12A, ATP1A2, C15orf62, C3orf33, CACNA1E, CDKN3, CHD5, CLDN4, COL11A1, COL8A1, CYP3A7, ERCC6L, FNDC1, GPR78, HOXA10, IL10, ITGB3, KCNA4, KLF12, LIF, LIFR, LIPH, LTF, MB, MIR21, MMP20, NR4A1, PSG6, SCGB2A2, SPC25, SPINK2, SPP1, SST, STAT3, SULT1E1, TBX15, TCN1, TMEM190, TMPRSS11B, TUBAL3, ZNF295-AS1. |
| Endometriosis | ABCB11, ABCC3, ACKR1, ACO2, ADGRF1, AFF4, AGT, AIMP1, ALPI, AMY1A, AMY2A, AMY2B, ANXA2, ANXA5, AOC1, ATF3, BST2, C1QA, C1QTNF6, CA1, CA12, CASP5, CCBE1, CCL3, CCL3L1, CCL3L3, CCL8, CCN1, CCT8, CDA, CDK5R1, CELF1, COL12A1, CORO1B, CRABP1, CRISP3, CST7, CTSW, CWH43, CXCL2, CYP3A5, DDIT4L, DDX17, DEPP1, DLG5, DNAJC3, DST, EDNRB, EGR1, EGR2, EGR3, EIF1, EIF4A1, EIF4A2, ENPP3, FMN2, FOS, FOSB, GALP, GSN, GUCY1B1, GZMA, HACD1, HOXA9, HPCAL4, HSP90B1, FNA21, IL6, IMMT, JUNB, KCNK2, KRIT1, KRT18, KRT5, KRTAP19-2, LAMA3, LCK, LONRF2, LPP, LRRD1, LTB4R2, LUZP1, MALL, MAP4, MAPK8, MET, MIR135A1, MIR138-1, MIR138-2, MIR1915, MIR194-2, MIR196A1, MIR196A2, MIR219B, MIR22, MIR26B, MIR3196, MIR339, MIR365B, MIR3686, MIR374B, MIR4251, MIR4252, MIR4254, MIR4425, MIR4723, MIR505, MIR542, MIR548AA2, MIR548AP, MIR548T, MIR5585, MIR921, MMP26, MUC7, MYL12A, NCR1, NEAT1, NFAT5, NR4A1, NR4A3, PAX8, PCSK5, PCYOX1, PDHB, PER1, PITX1, PLEK, PLEKHA2, POMZP3, PRDX6, PRIM2, PRRC2C, PTAFR, RAB9BP1, RBBP4, RGS1, RIF1, RIN1, RNF150, RNH1, RSRP1, S100A3, S100A8, SAP30L, SCG2, SCGB2A2, SEMA3C, SERPINB8, SHB, SLA, SLC15A4, SLC1A1, SLC44A2, SMG1, SOCS3, SON, SP3P, TAF6L, TGFB3, THRAP3, TRIM15, TRPM6, TUBA1C, VDAC1P1, VEGFA, VHL, VIM, YBX1, YBX1P2, YWHAE, ZFP36, ZIC2. |
| Healthy | ABCC3, ACADSB, ADAMTS1, ALPL, AMIGO2, ANG, ANO1, ANXA2, ANXA4, AOX1, APOD, ARG2, ARID5B, ASPM, ATP1B1, ATP6V0E2, ATP6V1A, BARD1, BCL6, BUB1B, C1R, C4BPA, CAPN6, CATSPERB, CCNB2, CDA, CDC20, CDK1, CENPE, CEP55, CFD, CLDN4, CLU, COL16A1, COMP, CP, CRABP2, CSRP2, CTNNA2, CXCL13, CXCL14, DDX52, DEFB1, DEPP1, DEPTOR, DKK1, DLGAP5, DPP4, DYNLT3, ECI2, ECM1, EDN3, EDNRB, EFNA1, ENPEP, EPHB3, FANCI, FOSL2, FXYD2, G0S2, GABARAPL1, GADD45A, GALNT12, GALNT4, GAS1, GAST, GBP2, GDF15, GPX3, GREM2, HABP2, HEY2, HLA-DOB, HPSE, ID4, IDO1, IL15, IMPA2, KCNG1, KIF11, KIF20A, KIF4A, KMO, KRT7, LAMB3, LIF, LMCD1, LMOD1, LRRC17, LYPD3, MAOA, MAP2K6, MFAP2, MFAP5, MMP26, MPPED2, MSX1, MT1G, MT1H, MT2A, MTCL1, NDC80, NDRG1, NDRG2, NNMT, NRG2, OLFM1, OLFM4, PAEP, PAQR4, PBK, PENK, PLA1A, PLAAT3, PLAAT4, PMEPA1, POLD4, POSTN, PRC1, PRKCQ, PRR15L, PRUNE2, PTPRR, RASSF2, RETREG1, RNASE4, RPRM, S100A1, S100A4, S100P, SCGB2A2, SERPINA5, SERPING1, SFRP4, SLC1A1, SNX10, SOD2, SORD, SOX17, SPDEF, SPP1, SYNE2, TACC3, TAGLN, TBC1D2, TCN1, THBD, TMSB15A, TOP2A, TRH, TSPAN8. |
| Adenomyosis | Endometriosis | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Reactome Pathway ID | Number of Total Reactions | Annotated Genes | Reaction Count | p-Value | FDR-Value | Annotated Genes | Reaction Count | p-Value | FDR-Value |
| R-HSA-8950505: Gene and protein expression by JAK-STAT signalling after Interleukin-12 stimulation | 36 | IL10 | 1 | 0.043 | 0.197 | CA1, ANXA2 | 2 | 0.043 | 0.450 |
| R-HSA-6783783: Interleukin-10 signalling | 15 | IL10, STAT3, LIF | 15 | 6.26 × 10−4 | 5.38 × 10−2 | IL6, CCL3L1, CCL3L3, PTAFR, CCL3, CXCL2 | 2 | 6.08 × 10−7 | 2.35 × 10−4 |
| R-HSA-6785807: Interleukin-4 and interleukin-13 signalling | 46 | IL10, STAT3, LIF | 19 | 2.60 × 10−3 | 0.086 | SOCS3, IL6, VIM, FOS, JUNB, VEGFA, HSP90B1 | 5 | 1.37 × 10−4 E-04 | 0.015 |
| R-HSA-6798695: Neutrophil degranulation | 10 | TCN1, LIPH, CHD5, CACNA1E, MMP20, LTF | 7 | 0.020 | 0.197 | CDA, AOC1, GSN, ANXA2, SLC44A2, CRISP3, PTAFR, CXCL2, PRDX6, CCT8, DNAJC3, BST2, SLC15A4, SCG2, KCNK2, S100A8 | 9 | 0.014 | 0.360 |
| R-HSA-8849474: PTK6 activation of STAT3 | 9 | STAT3 | 6 | 0.031 | 0.197 | SOCS3 | 3 | 0.007 | 0.281 |
| Clinical Characteristics | Adenomyosis Group (n = 9) | Control Group (n = 13) | p-Value * |
|---|---|---|---|
| Age (years) | 33 [32; 39] | 36 [32; 39] | 0.946 |
| BMI (kg/m2) | 29.4 [17.8; 34.6] | 22.8 [19.6; 24.0] | 0.057 |
| Endometrial thickness (mm) | 7.3 [4.6; 9.7] | 9.2 [7.4; 13.0] | 0.030 |
| Number of previous ART | 2 [1; 3] | 2 [1; 4] | 0.387 |
| Female sterility: | 0.522 | ||
| primary | 5 | 9 | |
| secondary | 4 | 4 |
| Candidate Gene | Adenomyosis Group (n = 9) | Control Group (n = 13) | p-Value |
|---|---|---|---|
| IL6 | 0.664 [0.402; 1.589] | 1.054 [0.533; 2.224] | 0.333 |
| LIF | 0.761 [0.140; 4.317] | 2.717 [0.423; 6.870] | 0.262 |
| IL10 | 0.716 [0.295; 2.080] | 1.081 [0.523; 1.995] | 0.193 |
| SOCS3 | 0.597 [0.223; 2.780] | 1.446 [0.935; 3.283] | 0.301 |
| JUNB | 1.106 [0.419; 1.557] | 1.242 [0.714; 1.953] | 0.526 |
| FOS | 0.844 [0.420; 1.657] | 1.120 [0.509; 1.705] | 0.271 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Prašnikar, E.; Kunej, T.; Repnik, K.; Potočnik, U.; Knez, J.; Kovačič, B. Determining the Molecular Background of Endometrial Receptivity in Adenomyosis. Biomolecules 2020, 10, 1311. https://doi.org/10.3390/biom10091311
Prašnikar E, Kunej T, Repnik K, Potočnik U, Knez J, Kovačič B. Determining the Molecular Background of Endometrial Receptivity in Adenomyosis. Biomolecules. 2020; 10(9):1311. https://doi.org/10.3390/biom10091311
Chicago/Turabian StylePrašnikar, Erika, Tanja Kunej, Katja Repnik, Uroš Potočnik, Jure Knez, and Borut Kovačič. 2020. "Determining the Molecular Background of Endometrial Receptivity in Adenomyosis" Biomolecules 10, no. 9: 1311. https://doi.org/10.3390/biom10091311
APA StylePrašnikar, E., Kunej, T., Repnik, K., Potočnik, U., Knez, J., & Kovačič, B. (2020). Determining the Molecular Background of Endometrial Receptivity in Adenomyosis. Biomolecules, 10(9), 1311. https://doi.org/10.3390/biom10091311








