Ginsenoside Rg3 Prevents Oncogenic Long Noncoding RNA ATXN8OS from Inhibiting Tumor-Suppressive microRNA-424-5p in Breast Cancer Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Rg3 Treatment and Transfection
2.3. Rg3-Quantitative Reverse-Transcription Polymerase Chain Reaction (qRT-PCR)
2.4. Data Mining
2.5. Cell Proliferation and Apoptosis Assay
2.6. Western Blot Analysis
2.7. Statistical Analyses
3. Results
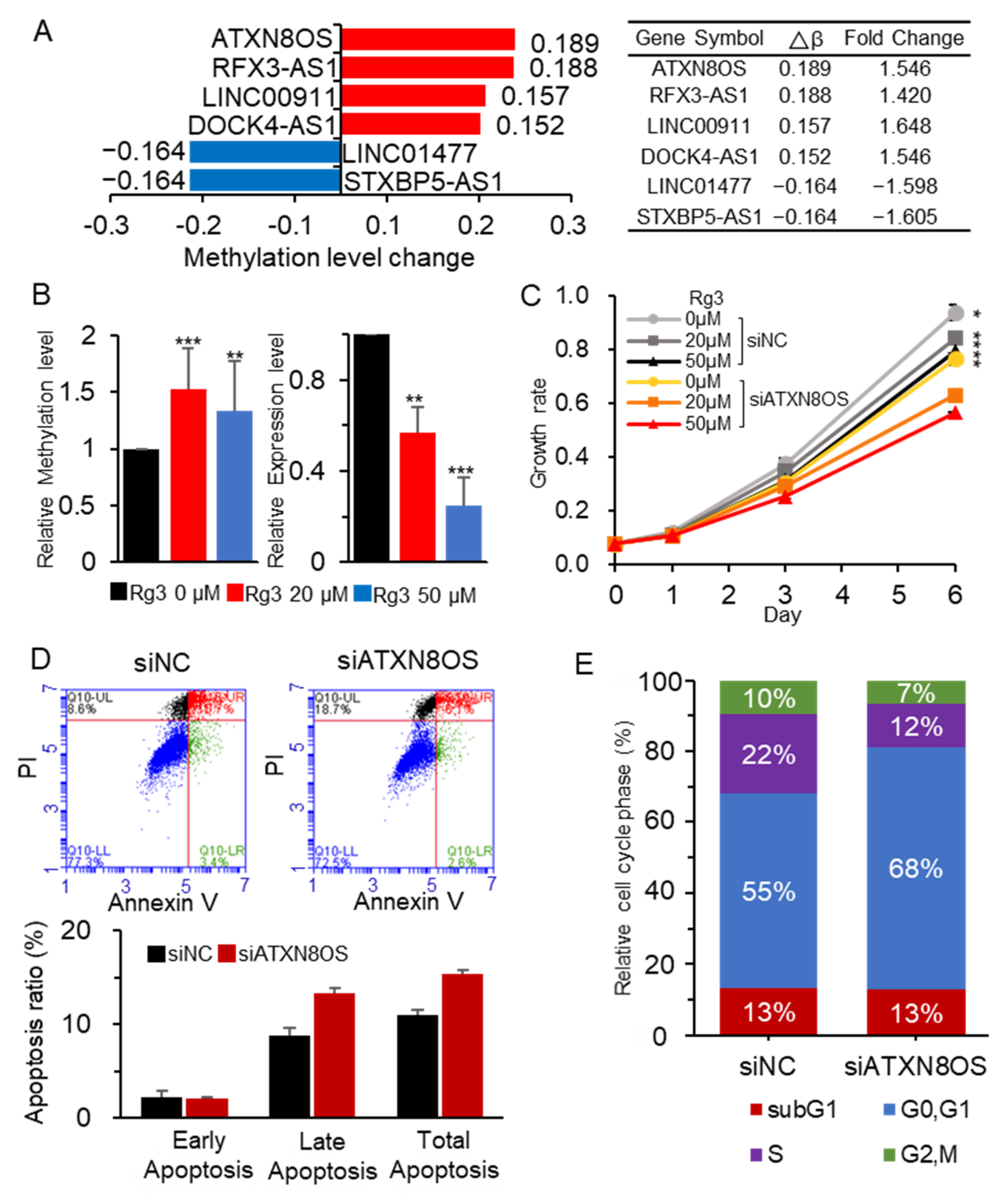
3.1. Rg3 Induces Hypermethylation and Downregulation of ATXN8OS
3.2. ATXN8OS Stimulates Cancer-Cell Proliferation via Sponging miR-424-5p
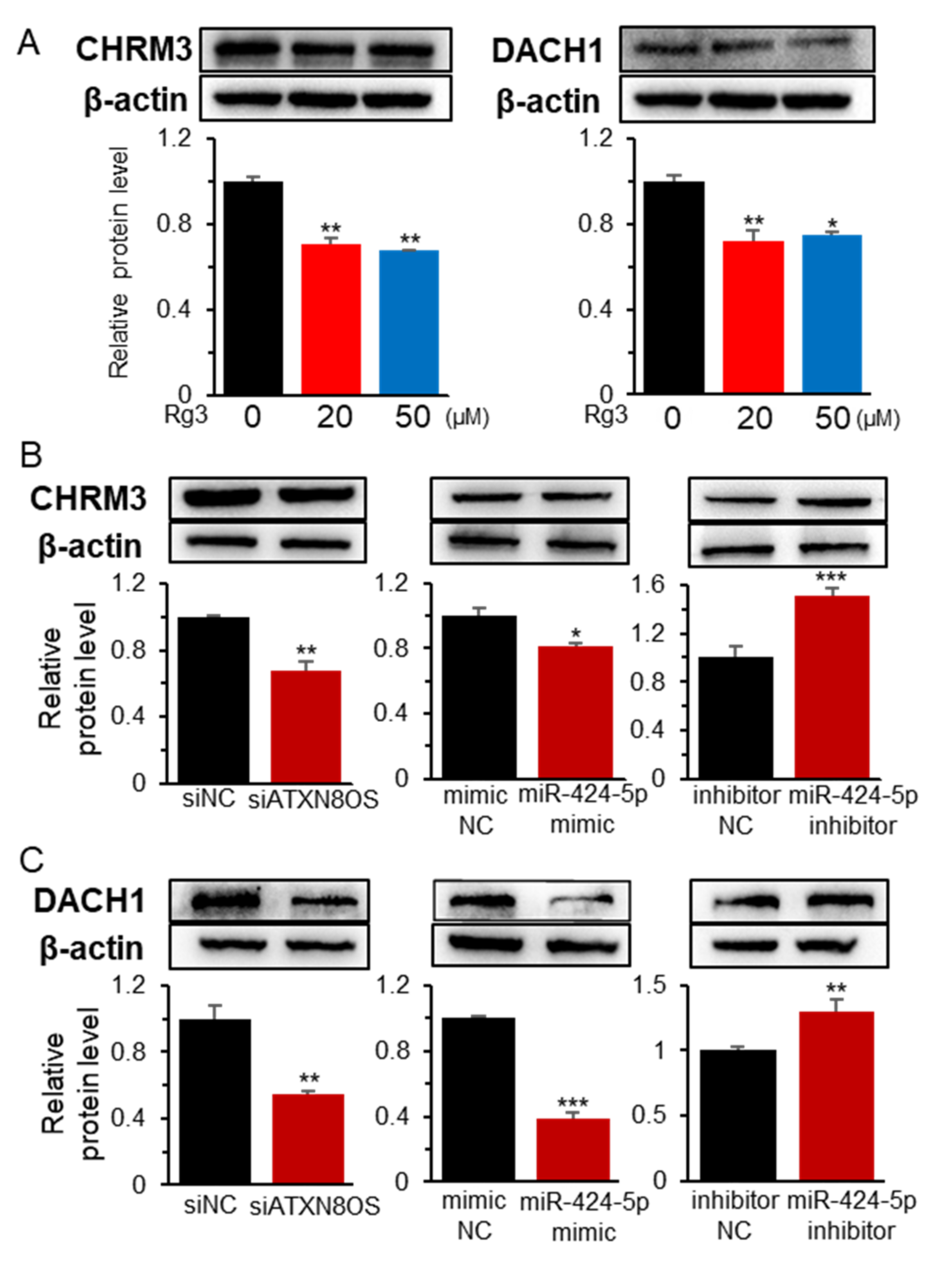
3.3. MiR-424-5p Target Genes are Regulated by ATXN8OS
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nakhjavani, M.; Hardingham, J.E.; Palethorpe, H.M.; Tomita, Y.; Smith, E.; Price, T.J.; Townsend, A.R. Ginsenoside Rg3: Potential molecular targets and therapeutic indication in metastatic breast cancer. Medicines 2019, 6, 17. [Google Scholar] [CrossRef]
- Sun, M.; Ye, Y.; Xiao, L.; Duan, X.; Zhang, Y.; Zhang, H. Anticancer effects of ginsenoside Rg3 (Review). Int. J. Mol. Med. 2017, 39, 507–518. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.M.; Kim, D.H.; Park, J.H.; Na, H.K.; Surh, Y.J. Ginsenoside Rg3 Induces Apoptosis of Human Breast Cancer (MDA-MB-231) Cells. Eur. J. Cancer Prev. 2013, 18, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.-M.; Kim, D.-H.; Park, J.-H.; Surh, Y.-J.; Na, H.-K. Ginsenoside Rg3 inhibits constitutive activation of NF-κB signaling in human breast cancer (MDA-MB-231) cells: ERK and Akt as potential upstream targets. Eur. J. Cancer Prev. 2014, 19, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Jin, Y.; Feng, T.; Wang, H.; Liu, D.; Zhou, Z.; Yan, Q.; Yang, H.; Yang, J.; Yang, J.; et al. Ginsenoside Rg3 Inhibits the Growth of Osteosarcoma and Attenuates Metastasis through the Wnt/β-Catenin and EMT Signaling Pathway. Evid. Based Complement Altern. Med. 2020, 2020, 6065124. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Qu, G. Inhibition of the hypoxia-induced factor-1α and vascular endothelial growth factor expression through ginsenoside Rg3 in human gastric cancer cells. J. Cancer Res. Ther. 2019, 15, 1642–1646. [Google Scholar] [CrossRef]
- Phi, L.T.H.; Wijaya, Y.T.; Sari, I.N.; Kim, K.S.; Yang, Y.G.; Lee, M.W.; Kwon, H.Y. 20(R)-Ginsenoside Rg3 Influences Cancer Stem Cell Properties and the Epithelial-Mesenchymal Transition in Colorectal Cancer via the SNAIL Signaling Axis. Onco Targets Ther. 2019, 12, 10885–10895. [Google Scholar] [CrossRef]
- Ham, J.; Lee, S.; Lee, H.; Jeong, D.; Park, S.; Kim, S.J. Genome-Wide Methylation Analysis Identifies NOX4 and KDM5A as Key Regulators in Inhibiting Breast Cancer Cell Proliferation by Ginsenoside Rg3. Am. J. Chin. Med. 2018, 46, 1333–1355. [Google Scholar] [CrossRef]
- Teng, S.; Wang, Y.; Li, P.; Liu, J.; Wei, A.; Wang, H.; Meng, X.; Pan, D.; Zhang, X. Effects of R type and S type ginsenoside Rg3 on DNA methylation in human hepatocarcinoma cells. Mol. Med. Rep. 2017, 15, 2029–2038. [Google Scholar] [CrossRef]
- Cheng, Z.; Xing, D. Ginsenoside Rg3 inhibits growth and epithelial-mesenchymal transition of human oral squamous carcinoma cells by down-regulating miR-221. Eur. J. Pharmacol. 2019, 853, 353–363. [Google Scholar] [CrossRef]
- Lee, A.; Yun, E.; Chang, W.; Kim, J. Ginsenoside Rg3 protects against iE-DAP–induced endothelial-to-mesenchymal transition by regulating the miR-139-5p–NF-κB axis. J. Ginseng Res. 2020, 44, 300–307. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Lu, J.; Ye, Z.; Han, X.; Zheng, X.; Hou, H.; Chen, W.; Li, X.; Zhao, L. 20(S)-Rg3 blocked epithelial-mesenchymal transition through DNMT3A/miR-145/FSCN1 in ovarian cancer. Oncotarget 2017, 8, 53375–53386. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Chen, H.; He, F.; You, Y.; Feng, Z.; Chen, W.; Li, X.; Zhao, L. Ginsenoside 20(S)-Rg3 upregulates HIF-1α-targeting miR-519a-5p to inhibit the Warburg effect in ovarian cancer cells. Clin. Exp. Pharmacol. Physiol. 2020, 47, 1455–1463. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.; Huang, J.; Xu, T.; Ye, Z.; Jin, F.; Li, N.; Lv, B. MicroRNA-181b blocks gensenoside Rg3-mediated tumor suppression of gallbladder carcinoma by promoting autophagy flux via CREBRF/CREB3 pathway. Am. J. Transl. Res. 2019, 11, 5776–5787. [Google Scholar]
- Schmitt, A.M.; Chang, H.Y. Long Noncoding RNAs in Cancer Pathways. Cancer Cell. 2016, 29, 452–463. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Su, H.; Zou, C.; Liang, X.; Fei, Z. Ginsenoside Rg3 suppresses the growth of gemcitabine-resistant pancreatic cancer cells by upregulating lncRNA-CASC2 and activating PTEN signaling. J. Biochem. Mol. Toxicol. 2020, 34, e22480. [Google Scholar] [CrossRef]
- Ham, J.; Jeong, D.; Park, S.; Kim, H.W.; Kim, H.; Kim, S.J. Ginsenoside Rg3 and Korean Red Ginseng extract epigenetically regulate the tumor-related long noncoding RNAs RFX3-AS1 and STXBP5-AS1. J. Ginseng Res. 2019, 43, 625–634. [Google Scholar] [CrossRef]
- Li, J.; Qi, Y. Ginsenoside Rg3 inhibits cell growth, migration and invasion in Caco-2 cells by downregulation of lncRNA CCAT1. Exp. Mol. Pathol. 2019, 106, 131–138. [Google Scholar] [CrossRef]
- Tay, Y.; Rinn, J.; Pandolfi, P.P. The multilayered complexity of ceRNA crosstalk and competition. Nature 2014, 505, 344–352. [Google Scholar] [CrossRef]
- Yan, L.; Yang, S.; Yue, C.X.; Wei, X.Y.; Peng, W.; Dong, Z.Y.; Xu, H.N.; Chen, S.L.; Wang, W.R.; Chen, C.J.; et al. Long noncoding RNA H19 acts as a miR-340-3p sponge to promote epithelial-mesenchymal transition by regulating YWHAZ expression in paclitaxel-resistant breast cancer cells. Environ. Toxicol. 2020, 35, 1015–1028. [Google Scholar] [CrossRef]
- Kim, S.J.; Kelly, W.K.; Fu, A.; Haines, K.; Hoffman, A.; Zheng, T.; Zhu, Y. Genome-wide methylation analysis identifies involvement of TNF-α mediated cancer pathways in prostate cancer. Cancer Lett. 2011, 302, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Lee, H.; Bae, H.; Choi, E.H.; Kim, S.J. Epigenetic silencing of miR-19a-3p by cold atmospheric plasma contributes to proliferation inhibition of the MCF-7 breast cancer cell. Sci. Rep. 2016, 6, 30005. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.; Kim, B.; Kang, H.S.; Jeong, G.; Bae, H.; Lee, H.; Lee, S.; Kim, S.J. SCTR regulates cell cycle-related genes toward anti-proliferation in normal breast cells while having pro-proliferation activity in breast cancer cells. Int. J. Oncol. 2015, 47, 1923–1931. [Google Scholar] [CrossRef] [PubMed]
- Deng, Z.; Cai, H.; Lin, L.; Zhu, L.; Wu, W.; Yang, S.; Cai, J.; Tan, J. lncRNA ATXN8OS promotes breast cancer by sequestering miR-204. Mol. Med. Rep. 2019, 20, 1057–1064. [Google Scholar] [CrossRef]
- Tian, M.; Li, L.N.; Zheng, R.R.; Yang, L.; Wang, Z.T. Advances on hormone-like activity of Panax ginseng and ginsenosides. Chin. J. Nat. Med. 2020, 18, 526–535. [Google Scholar] [CrossRef]
- Cai, S.; Cheng, X.; Liu, Y.; Lin, Z.; Zeng, W.; Yang, C.; Liu, L.; Chukwuebuka, O.A.; Li, W. EYA1 promotes tumor angiogenesis by activating the PI3K pathway in colorectal cancer. Exp. Cell Res. 2018, 367, 37–46. [Google Scholar] [CrossRef]
- Wang, N.; Yao, M.; Xu, J.; Quan, Y.; Zhang, K.; Yang, R.; Gao, W.Q. Autocrine Activation of CHRM3 Promotes Prostate Cancer Growth and Castration Resistance via CaM/CaMKK-Mediated Phosphorylation of Akt. Clin. Cancer Res. 2015, 21, 4676–4685. [Google Scholar] [CrossRef]
- Hu, X.; Zhang, L.; Li, Y.; Ma, X.; Dai, W.; Gao, X.; Rao, X.; Fu, G.; Wang, R.; Pan, M.; et al. Organoid modelling identifies that DACH1 functions as a tumour promoter in colorectal cancer by modulating BMP signalling. EBioMedicine 2020, 56, 102800. [Google Scholar] [CrossRef]
- Xu, H.; Yu, S.; Yuan, X.; Xiong, J.; Kuang, D.; Pestell, R.G.; Wu, K. DACH1 suppresses breast cancer as a negative regulator of CD44. Sci. Rep. 2017, 7, 4361. [Google Scholar] [CrossRef]
- Wu, K.; Li, Z.; Cai, S.; Tian, L.; Chen, K.; Wang, J.; Hu, J.; Sun, Y.; Li, X.; Ertel, A.; et al. EYA1 phosphatase function is essential to drive breast cancer cell proliferation through cyclin D1. Cancer Res. 2013, 73, 4488–4499. [Google Scholar] [CrossRef]
- Samukawa, M.; Hirano, M.; Saigoh, K.; Kawai, S.; Hamada, Y.; Takahashi, D.; Nakamura, Y.; Kusunoki, S. PSP-Phenotype in SCA8: Case Report and Systemic Review. Cerebellum 2019, 18, 76–84. [Google Scholar] [CrossRef] [PubMed]
- Li, X.X.; Wang, L.J.; Hou, J.; Liu, H.Y.; Wang, R.; Wang, C.; Xie, W.H. Identification of Long Noncoding RNAs as Predictors of Survival in Triple-Negative Breast Cancer Based on Network Analysis. BioMed Res. Int. 2020, 2020, 8970340. [Google Scholar] [CrossRef] [PubMed]
- Cen, D.; Huang, H.; Yang, L.; Guo, K.; Zhang, J. Long noncoding RNA STXBP5-AS1 inhibits cell proliferation, migration, and invasion through inhibiting the PI3K/AKT signaling pathway in gastric cancer cells. Onco Targets Ther. 2019, 12, 1929–1936. [Google Scholar] [CrossRef]
- Dastmalchi, N.; Hosseinpourfeizi, M.A.; Khojasteh, S.M.B.; Baradaran, B.; Safaralizadeh, R. Tumor suppressive activity of miR-424-5p in breast cancer cells through targeting PD-L1 and modulating PTEN/PI3K/AKT/mTOR signaling pathway. Life Sci. 2020, 259, 118239. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.; Zhang, Y.; Xia, Y.; Zhao, H.; Liu, A.; Chen, Y. LncRNA MEG3 targeting miR-424-5p via MAPK signaling pathway mediates neuronal apoptosis in ischemic stroke. Aging (Albany N.Y.). 2020, 12, 3156–3174. [Google Scholar] [CrossRef]
- Liu, X.; Fu, Y.; Zhang, G.; Zhang, D.; Liang, N.; Li, F.; Li, C.; Sui, C.; Jiang, J.; Lu, H.; et al. miR-424-5p Promotes Anoikis Resistance and Lung Metastasis by Inactivating Hippo Signaling in Thyroid Cancer. Mol. Ther. Oncolytics. 2019, 15, 248–260. [Google Scholar] [CrossRef]
- Vimalraj, S.; Saravanan, S.; Raghunandhakumar, S.; Anuradha, D. Melatonin regulates tumor angiogenesis via miR-424-5p/VEGFA signaling pathway in osteosarcoma. Life Sci. 2020, 256, 118011. [Google Scholar] [CrossRef]
- Wu, J.; Yang, B.; Zhang, Y.; Feng, X.; He, B.; Xie, H.; Zhou, L.; Wu, J.; Zheng, S. miR-424-5p represses the metastasis and invasion of intrahepatic cholangiocarcinoma by targeting ARK5. Int. J. Biol. Sci. 2019, 15, 1591–1599. [Google Scholar] [CrossRef]
- Yue, X.; Wang, Z. Long Intergenic Non-Coding RNA LINC00922 Aggravates the Malignant Phenotype of Breast Cancer by Regulating the microRNA-424-5p/BDNF Axis. Cancer Manag. Res. 2020, 12, 7539–7552. [Google Scholar] [CrossRef]
- Shen, X.; Li, Y.; He, F.; Kong, J. LncRNA CDKN2B-AS1 Promotes Cell Viability, Migration, and Invasion of Hepatocellular Carcinoma via Sponging miR-424-5p. Cancer Manag. Res. 2020, 12, 6807–6819. [Google Scholar] [CrossRef]
- Zhou, K.; Li, S.; Du, G.; Fan, Y.; Wu, P.; Sun, H.; Zhang, T. LncRNA XIST depletion prevents cancer progression in invasive pituitary neuroendocrine tumor by inhibiting bFGF via upregulation of microRNA-424-5p. Onco Targets Ther. 2019, 12, 7095–7109. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Zhou, Y.; Chen, W.; Chen, L.; Lu, J.; He, F.; Li, X.; Zhao, L. Ginsenoside 20(S)-Rg3 Prevents PKM2-Targeting miR-324-5p from H19 Sponging to Antagonize the Warburg Effect in Ovarian Cancer Cells. Cell. Physiol. Biochem. 2018, 51, 1340–1353. [Google Scholar] [CrossRef] [PubMed]
- Xue, L.P.; Fu, X.L.; Hu, M.; Zhang, L.W.; Li, Y.D.; Peng, Y.L.; Ding, P. Rg1 inhibits high glucose-induced mesenchymal activation and fibrosis via regulating miR-2113/RP11-982M15.8/Zeb1 pathway. Biochem. Biophys. Res. Commun. 2018, 501, 827–832. [Google Scholar] [CrossRef] [PubMed]
- Figliuzzi, M.; Marinari, E.; De Martino, A. MicroRNAs as a selective channel of communication between competing RNAs: A steady-state theory. Biophys. J. 2013, 104, 1203–1213. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, H.; Ji, H.W.; Kim, H.W.; Yun, S.H.; Park, J.E.; Kim, S.J. Ginsenoside Rg3 Prevents Oncogenic Long Noncoding RNA ATXN8OS from Inhibiting Tumor-Suppressive microRNA-424-5p in Breast Cancer Cells. Biomolecules 2021, 11, 118. https://doi.org/10.3390/biom11010118
Kim H, Ji HW, Kim HW, Yun SH, Park JE, Kim SJ. Ginsenoside Rg3 Prevents Oncogenic Long Noncoding RNA ATXN8OS from Inhibiting Tumor-Suppressive microRNA-424-5p in Breast Cancer Cells. Biomolecules. 2021; 11(1):118. https://doi.org/10.3390/biom11010118
Chicago/Turabian StyleKim, Heejoo, Hwee Won Ji, Hyeon Woo Kim, Sung Hwan Yun, Jae Eun Park, and Sun Jung Kim. 2021. "Ginsenoside Rg3 Prevents Oncogenic Long Noncoding RNA ATXN8OS from Inhibiting Tumor-Suppressive microRNA-424-5p in Breast Cancer Cells" Biomolecules 11, no. 1: 118. https://doi.org/10.3390/biom11010118
APA StyleKim, H., Ji, H. W., Kim, H. W., Yun, S. H., Park, J. E., & Kim, S. J. (2021). Ginsenoside Rg3 Prevents Oncogenic Long Noncoding RNA ATXN8OS from Inhibiting Tumor-Suppressive microRNA-424-5p in Breast Cancer Cells. Biomolecules, 11(1), 118. https://doi.org/10.3390/biom11010118






