Transcriptomic Analysis of Naïve Human Embryonic Stem Cells Cultured in Three-Dimensional PEG Scaffolds
Abstract
:1. Introduction
2. Materials and Methods
2.1. Maintenance of Naïve ESCs in 2-D Culture
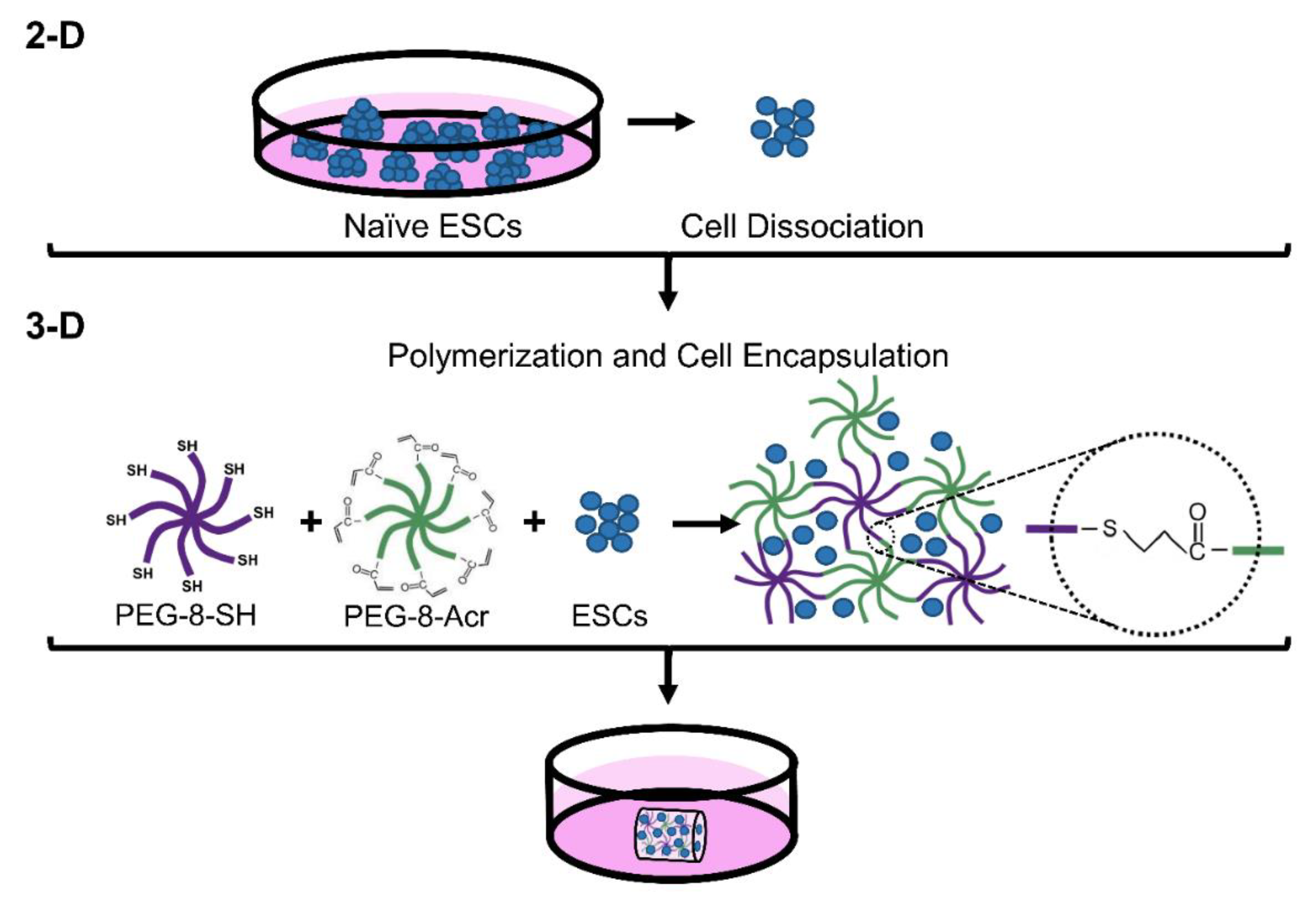
2.2. Self-Assembly of 3-D Scaffolds Via Thiol-Michael Addition Reaction for Encapsulation of Cells
2.3. Cell Proliferation of 3-D Grown ESCs
2.4. Teratoma Formation Assay
2.5. Gene Expression Analysis Using qRT–PCR
2.6. RNA-Sequencing (RNA-seq)
2.7. Bioinformatic Analysis
2.8. Immunohistochemical Analysis
2.9. Statistical Analysis
3. Results
3.1. Proliferation and Characteristics of Pluripotent ESCs Grown in 3-D Self-Assembling Scaffolds
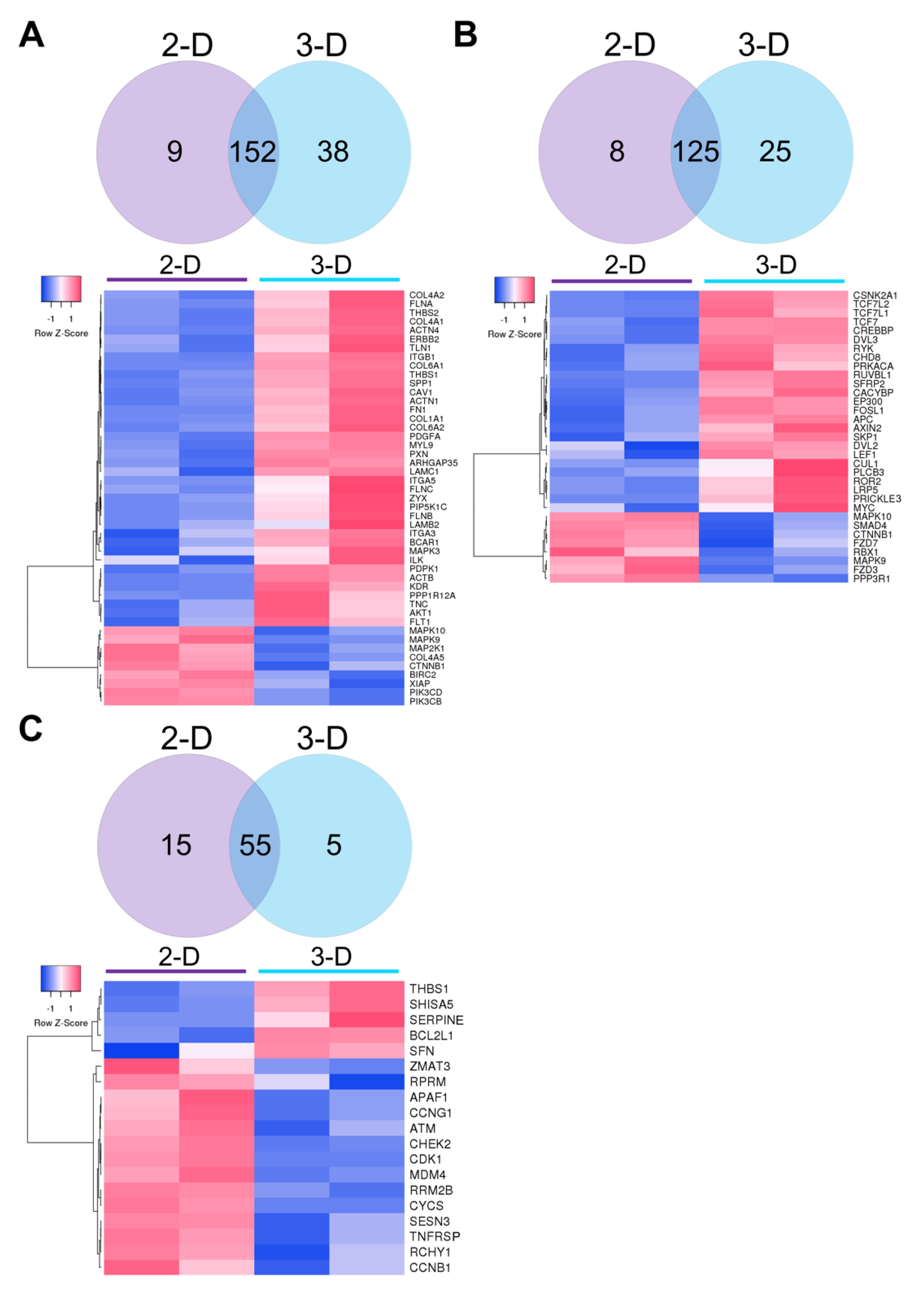
3.2. Transcriptomic Analysis of Naïve ESCs Grown in 2-D and 3-D Culture Conditions
3.3. Functional Analysis of DEGs Using Gene Ontology (GO) and Enrichment Analysis
3.4. KEGG Pathway Analysis of Naïve ESCs Grown in 2-D and 3-D Culture Conditions
3.5. Validation of RNA-seq Results with qRT–PCR
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Evans, M.J.; Kaufman, M.H. Establishment in culture of pluripotential cells from mouse embryos. Nature 1981, 292, 154–156. [Google Scholar] [CrossRef]
- Thomson, J.A.; Itskovitz-Eldor, J.; Shapiro, S.S.; Waknitz, M.A.; Swiergiel, J.J.; Marshall, V.S.; Jones, J.M. Embryonic Stem Cell Lines Derived from Human Blastocysts. Science 1998, 282, 1145–1147. [Google Scholar] [CrossRef] [Green Version]
- Garcia-Leon, J.A.; Vitorica, J.; Gutierrez, A. Use of human pluripotent stem cell-derived cells for neurodegenerative disease modeling and drug screening platform. Future Med. Chem. 2019, 11, 1305–1322. [Google Scholar] [CrossRef]
- Mora, C.; Serzanti, M.; Consiglio, A.; Memo, M.; Dell’Era, P. Clinical potentials of human pluripotent stem cells. Cell Biol. Toxicol. 2017, 33, 351–360. [Google Scholar] [CrossRef] [Green Version]
- Tabar, V.; Studer, L. Pluripotent stem cells in regenerative medicine: Challenges and recent progress. Nat. Rev. Genet. 2014, 15, 82–92. [Google Scholar] [CrossRef] [Green Version]
- McKee, C.; Chaudhry, G.R. Advances and challenges in stem cell culture. Colloids Surf. B Biointerfaces 2017, 159, 62–77. [Google Scholar] [CrossRef] [PubMed]
- Vazin, T.; Freed, W.J. Human embryonic stem cells: Derivation, culture, and differentiation: A review. Restor. Neurol. Neurosci. 2010, 28, 589–603. [Google Scholar] [CrossRef]
- Tesar, P.J.; Chenoweth, J.G.; Brook, F.A.; Davies, T.J.; Evans, E.P.; Mack, D.L.; Gardner, R.L.; McKay, R.D. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature 2007, 448, 196–199. [Google Scholar] [CrossRef]
- Weinberger, L.; Ayyash, M.; Novershtern, N.; Hanna, J.H. Dynamic stem cell states: Naive to primed pluripotency in rodents and humans. Nat. Rev. Mol. Cell Biol. 2016, 17, 155–169. [Google Scholar] [CrossRef]
- Nichols, J.; Smith, A. Naive and Primed Pluripotent States. Cell Stem Cell 2009, 4, 487–492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van der Jeught, M.; Taelman, J.; Duggal, G.; Ghimire, S.; Lierman, S.; Chuva de Sousa Lopes, S.M.; Deforce, D.; Deroo, T.; De Sutter, P.; Heindryckx, B. Application Of Small Molecules Favoring Naïve Pluripotency during Human Embryonic Stem Cell Derivation. Cell. Reprogr. 2015, 17, 170–180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, J.; Yamauchi, T.; Izpisua Belmonte, J.C. An overview of mammalian pluripotency. Development 2016, 143, 1644–1648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, J.; Mai, D.H.; Xie, L. Resetting Human Naïve Pluripotency. Genet. Epigenet. 2016, 8, 37–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zimmerlin, L.; Park, T.S.; Zambidis, E.T. Capturing Human Naïve Pluripotency in the Embryo and in the Dish. Stem Cells Dev. 2017, 26, 1141–1161. [Google Scholar] [CrossRef]
- Chan, Y.S.; Göke, J.; Ng, J.H.; Lu, X.; Gonzales, K.A.; Tan, C.P.; Tng, W.Q.; Hong, Z.Z.; Lim, Y.S.; Ng, H.H. Induction of a human pluripotent state with distinct regulatory circuitry that resembles preimplantation epiblast. Cell Stem Cell 2013, 13, 663–675. [Google Scholar] [CrossRef] [Green Version]
- Duggal, G.; Warrier, S.; Ghimire, S.; Broekaert, D.; Van der Jeught, M.; Lierman, S.; Deroo, T.; Peelman, L.; Van Soom, A.; Cornelissen, R.; et al. Alternative Routes to Induce Naïve Pluripotency in Human Embryonic Stem Cells. Stem Cells 2015, 33, 2686–2698. [Google Scholar] [CrossRef]
- Qin, H.; Hejna, M.; Liu, Y.; Percharde, M.; Wossidlo, M.; Blouin, L.; Durruthy-Durruthy, J.; Wong, P.; Qi, Z.; Yu, J.; et al. YAP Induces Human Naive Pluripotency. Cell Rep. 2016, 14, 2301–2312. [Google Scholar] [CrossRef] [Green Version]
- Theunissen, T.W.; Friedli, M.; He, Y.; Planet, E.; O’Neil, R.C.; Markoulaki, S.; Pontis, J.; Wang, H.; Iouranova, A.; Imbeault, M.; et al. Molecular Criteria for Defining the Naive Human Pluripotent State. Cell Stem Cell 2016, 19, 502–515. [Google Scholar] [CrossRef] [Green Version]
- Theunissen, T.W.; Powell, B.E.; Wang, H.; Mitalipova, M.; Faddah, D.A.; Reddy, J.; Fan, Z.P.; Maetzel, D.; Ganz, K.; Shi, L.; et al. Systematic identification of culture conditions for induction and maintenance of naive human pluripotency. Cell Stem Cell 2014, 15, 471–487. [Google Scholar] [CrossRef] [Green Version]
- Gafni, O.; Weinberger, L.; Mansour, A.A.; Manor, Y.S.; Chomsky, E.; Ben-Yosef, D.; Kalma, Y.; Viukov, S.; Maza, I.; Zviran, A.; et al. Derivation of novel human ground state naive pluripotent stem cells. Nature 2013, 504, 282–286. [Google Scholar] [CrossRef]
- Guo, G.; von Meyenn, F.; Santos, F.; Chen, Y.; Reik, W.; Bertone, P.; Smith, A.; Nichols, J. Naive Pluripotent Stem Cells Derived Directly from Isolated Cells of the Human Inner Cell Mass. Stem Cell Rep. 2016, 6, 437–446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ware, C.B.; Nelson, A.M.; Mecham, B.; Hesson, J.; Zhou, W.; Jonlin, E.C.; Jimenez-Caliani, A.J.; Deng, X.; Cavanaugh, C.; Cook, S.; et al. Derivation of naive human embryonic stem cells. Proc. Natl. Acad. Sci. USA 2014, 111, 4484–4489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Antoni, D.; Burckel, H.; Josset, E.; Noel, G. Three-dimensional cell culture: A breakthrough in vivo. Int. J. Mol. Sci. 2015, 16, 5517–5527. [Google Scholar] [CrossRef]
- Kraehenbuehl, T.P.; Langer, R.; Ferreira, L.S. Three-dimensional biomaterials for the study of human pluripotent stem cells. Nat. Methods 2011, 8, 731–736. [Google Scholar] [CrossRef]
- Walma, D.A.C.; Yamada, K.M. The extracellular matrix in development. Development 2020, 147, dev175596. [Google Scholar] [CrossRef]
- Haycock, J.W. 3D cell culture: A review of current approaches and techniques. Methods Mol. Biol. 2011, 695, 1–15. [Google Scholar] [CrossRef]
- Kent, L. Culture and maintenance of human embryonic stem cells. J. Vis. Exp. 2009, 34, e1427. [Google Scholar] [CrossRef]
- Nie, Y.; Walsh, P.; Clarke, D.L.; Rowley, J.A.; Fellner, T. Scalable passaging of adherent human pluripotent stem cells. PLoS ONE 2014, 9, e88012. [Google Scholar] [CrossRef] [Green Version]
- Gauthaman, K.; Venugopal, J.R.; Yee, F.C.; Peh, G.S.; Ramakrishna, S.; Bongso, A. Nanofibrous substrates support colony formation and maintain stemness of human embryonic stem cells. J. Cell. Mol. Med. 2009, 13, 3475–3484. [Google Scholar] [CrossRef] [Green Version]
- Gerecht, S.; Burdick, J.A.; Ferreira, L.S.; Townsend, S.A.; Langer, R.; Vunjak-Novakovic, G. Hyaluronic acid hydrogel for controlled self-renewal and differentiation of human embryonic stem cells. Proc. Natl. Acad. Sci. USA 2007, 104, 11298–11303. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Charles, L.F.; Zarembinski, T.I.; Johnson, K.I.; Atzet, S.K.; Wesselschmidt, R.L.; Wight, M.E.; Kuhn, L.T. Modified hyaluronan hydrogels support the maintenance of mouse embryonic stem cells and human induced pluripotent stem cells. Macromol. Biosci. 2012, 12, 1034–1042. [Google Scholar] [CrossRef]
- Serra, M.; Correia, C.; Malpique, R.; Brito, C.; Jensen, J.; Bjorquist, P.; Carrondo, M.J.; Alves, P.M. Microencapsulation technology: A powerful tool for integrating expansion and cryopreservation of human embryonic stem cells. PLoS ONE 2011, 6, e23212. [Google Scholar] [CrossRef]
- Xu, K.; Narayanan, K.; Lee, F.; Bae, K.H.; Gao, S.; Kurisawa, M. Enzyme-mediated hyaluronic acid-tyramine hydrogels for the propagation of human embryonic stem cells in 3D. Acta Biomater. 2015, 24, 159–171. [Google Scholar] [CrossRef]
- Xu, Y.; Chen, C.; Hellwarth, P.B.; Bao, X. Biomaterials for stem cell engineering and biomanufacturing. Bioact. Mater. 2019, 4, 366–379. [Google Scholar] [CrossRef]
- Kharkar, P.M.; Kiick, K.L.; Kloxin, A.M. Designing degradable hydrogels for orthogonal control of cell microenvironments. Chem. Soc. Rev. 2013, 42, 7335–7372. [Google Scholar] [CrossRef] [Green Version]
- McKee, C.; Perez-Cruet, M.; Chavez, F.; Chaudhry, G.R. Simplified three-dimensional culture system for long-term expansion of embryonic stem cells. World J. Stem Cells 2015, 7, 1064–1077. [Google Scholar] [CrossRef]
- McKee, C.; Brown, C.; Chaudhry, G.R. Self-Assembling Scaffolds Supported Long-Term Growth of Human Primed Embryonic Stem Cells and Upregulated Core and Naïve Pluripotent Markers. Cells 2019, 8, 1650. [Google Scholar] [CrossRef] [Green Version]
- Van Meerloo, J.; Kaspers, G.J.; Cloos, J. Cell sensitivity assays: The MTT assay. Methods Mol. Biol. 2011, 731, 237–245. [Google Scholar] [CrossRef]
- Ma, F.; Fuqua, B.K.; Hasin, Y.; Yukhtman, C.; Vulpe, C.D.; Lusis, A.J.; Pellegrini, M. A comparison between whole transcript and 3′ RNA sequencing methods using Kapa and Lexogen library preparation methods. BMC Genom. 2019, 20, 9. [Google Scholar] [CrossRef]
- Afgan, E.; Baker, D.; Batut, B.; van den Beek, M.; Bouvier, D.; Čech, M.; Chilton, J.; Clements, D.; Coraor, N.; Grüning, B.A.; et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018, 46, W537–W544. [Google Scholar] [CrossRef] [Green Version]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 26 December 2020).
- Krueger, F. Trim Galore!: A Wrapper Tool around Cutadapt and FastQC to Consistently Apply Quality and Adapter Trimming to FastQ Files. 2015. Available online: http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ (accessed on 26 December 2020).
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [Green Version]
- Frankish, A.; Diekhans, M.; Ferreira, A.M.; Johnson, R.; Jungreis, I.; Loveland, J.; Mudge, J.M.; Sisu, C.; Wright, J.; Armstrong, J.; et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019, 47, D766–D773. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Mi, H.; Muruganujan, A.; Ebert, D.; Huang, X.; Thomas, P.D. PANTHER version 14: More genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2018, 47, D419–D426. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’ayan, A. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef] [Green Version]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef] [Green Version]
- Babicki, S.; Arndt, D.; Marcu, A.; Liang, Y.; Grant, J.R.; Maciejewski, A.; Wishart, D.S. Heatmapper: Web-enabled heat mapping for all. Nucleic Acids Res. 2016, 44, W147–W153. [Google Scholar] [CrossRef]
- Rivero, R.E.; Capella, V.; Cecilia Liaudat, A.; Bosch, P.; Barbero, C.A.; Rodríguez, N.; Rivarola, C.R. Mechanical and physicochemical behavior of a 3D hydrogel scaffold during cell growth and proliferation. RSC Adv. 2020, 10, 5827–5837. [Google Scholar] [CrossRef]
- Bratt-Leal, A.M.; Carpenedo, R.L.; McDevitt, T.C. Engineering the embryoid body microenvironment to direct embryonic stem cell differentiation. Biotechnol. Prog. 2009, 25, 43–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abbasalizadeh, S.; Larijani, M.R.; Samadian, A.; Baharvand, H. Bioprocess development for mass production of size-controlled human pluripotent stem cell aggregates in stirred suspension bioreactor. Tissue Eng. Part C Methods 2012, 18, 831–851. [Google Scholar] [CrossRef]
- Lee, Y.-L.; Fong, S.-W.; Chen, A.C.H.; Li, T.; Yue, C.; Lee, C.-L.; Ng, E.H.Y.; Yeung, W.S.B.; Lee, K.-F. Establishment of a novel human embryonic stem cell-derived trophoblastic spheroid implantation model. Hum. Reprod. 2015, 30, 2614–2626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, H.; Li, Q.; Lei, Y. Three-dimensional tissues using human pluripotent stem cell spheroids as biofabrication building blocks. Biofabrication 2017, 9, 025007. [Google Scholar] [CrossRef] [Green Version]
- Mohr, J.C.; Zhang, J.; Azarin, S.M.; Soerens, A.G.; de Pablo, J.J.; Thomson, J.A.; Lyons, G.E.; Palecek, S.P.; Kamp, T.J. The microwell control of embryoid body size in order to regulate cardiac differentiation of human embryonic stem cells. Biomaterials 2010, 31, 1885–1893. [Google Scholar] [CrossRef] [Green Version]
- Caliari, S.R.; Burdick, J.A. A practical guide to hydrogels for cell culture. Nat. Methods 2016, 13, 405–414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsou, Y.H.; Khoneisser, J.; Huang, P.C.; Xu, X. Hydrogel as a bioactive material to regulate stem cell fate. Bioact. Mater. 2016, 1, 39–55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Leung, M.; Hopper, R.; Ellenbogen, R.; Zhang, M. Feeder-free self-renewal of human embryonic stem cells in 3D porous natural polymer scaffolds. Biomaterials 2010, 31, 404–412. [Google Scholar] [CrossRef] [PubMed]
- Lei, Y.; Schaffer, D.V. A fully defined and scalable 3D culture system for human pluripotent stem cell expansion and differentiation. Proc. Natl. Acad. Sci. USA 2013, 110, e5039–e5048. [Google Scholar] [CrossRef] [Green Version]
- Wei, J.; Han, J.; Zhao, Y.; Cui, Y.; Wang, B.; Xiao, Z.; Chen, B.; Dai, J. The importance of three-dimensional scaffold structure on stemness maintenance of mouse embryonic stem cells. Biomaterials 2014, 35, 7724–7733. [Google Scholar] [CrossRef]
- Jang, M.; Lee, S.T.; Kim, J.W.; Yang, J.H.; Yoon, J.K.; Park, J.C.; Ryoo, H.M.; van der Vlies, A.J.; Ahn, J.Y.; Hubbell, J.A.; et al. A feeder-free, defined three-dimensional polyethylene glycol-based extracellular matrix niche for culture of human embryonic stem cells. Biomaterials 2013, 34, 3571–3580. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Ma, R.; Gu, Q.; Liang, L.; Wang, L.; Zhang, Y.; Wang, X.; Liu, X.; Li, Z.; Fang, J.; et al. A fully defined static suspension culture system for large-scale human embryonic stem cell production. Cell Death Dis. 2018, 9, 892. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Reinberg, D. Transcription regulation by histone methylation: Interplay between different covalent modifications of the core histone tails. Genes Dev. 2001, 15, 2343–2360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plath, K.; Lowry, W.E. Progress in understanding reprogramming to the induced pluripotent state. Nat. Rev. Genet. 2011, 12, 253–265. [Google Scholar] [CrossRef] [Green Version]
- Battle, S.L.; Doni Jayavelu, N.; Azad, R.N.; Hesson, J.; Ahmed, F.N.; Overbey, E.G.; Zoller, J.A.; Mathieu, J.; Ruohola-Baker, H.; Ware, C.B.; et al. Enhancer Chromatin and 3D Genome Architecture Changes from Naive to Primed Human Embryonic Stem Cell States. Stem Cell Rep. 2019, 12, 1129–1144. [Google Scholar] [CrossRef] [Green Version]
- Hayashi, Y.; Furue, M.K. Biological Effects of Culture Substrates on Human Pluripotent Stem Cells. Stem Cells Int. 2016, 2016, 5380560. [Google Scholar] [CrossRef] [Green Version]
- Werner, M.; Kurniawan, N.A.; Bouten, C.V.C. Cellular Geometry Sensing at Different Length Scales and its Implications for Scaffold Design. Materials 2020, 13, 963. [Google Scholar] [CrossRef] [Green Version]
- Harkness, L.; Chen, X.; Gillard, M.; Gray, P.P.; Davies, A.M. Media composition modulates human embryonic stem cell morphology and may influence preferential lineage differentiation potential. PLoS ONE 2019, 14, e0213678. [Google Scholar] [CrossRef] [Green Version]
- Vitillo, L.; Kimber, S.J. Integrin and FAK Regulation of Human Pluripotent Stem Cells. Curr. Stem Cell Rep. 2017, 3, 358–365. [Google Scholar] [CrossRef] [Green Version]
- Mair, B.; Tomic, J.; Masud, S.N.; Tonge, P.; Weiss, A.; Usaj, M.; Tong, A.H.Y.; Kwan, J.J.; Brown, K.R.; Titus, E.; et al. Essential Gene Profiles for Human Pluripotent Stem Cells Identify Uncharacterized Genes and Substrate Dependencies. Cell Rep. 2019, 27, 599–615.e512. [Google Scholar] [CrossRef] [Green Version]
- Wrighton, P.J.; Klim, J.R.; Hernandez, B.A.; Koonce, C.H.; Kamp, T.J.; Kiessling, L.L. Signals from the surface modulate differentiation of human pluripotent stem cells through glycosaminoglycans and integrins. Proc. Natl. Acad. Sci. USA 2014, 111, 18126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.G.; Li, Z.; Wang, X.F. Where PI3K/Akt meets Smads: The crosstalk determines human embryonic stem cell fate. Cell Stem Cell 2012, 10, 231–232. [Google Scholar] [CrossRef] [Green Version]
- Watanabe, S.; Umehara, H.; Murayama, K.; Okabe, M.; Kimura, T.; Nakano, T. Activation of Akt signaling is sufficient to maintain pluripotency in mouse and primate embryonic stem cells. Oncogene 2006, 25, 2697–2707. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martino, F.; Perestrelo, A.R.; Vinarský, V.; Pagliari, S.; Forte, G. Cellular Mechanotransduction: From Tension to Function. Front. Physiol. 2018, 9, 824. [Google Scholar] [CrossRef] [PubMed]
- Hoon, J.L.; Tan, M.H.; Koh, C.-G. The Regulation of Cellular Responses to Mechanical Cues by Rho GTPases. Cells 2016, 5, 17. [Google Scholar] [CrossRef] [Green Version]
- Hsiao, C.; Lampe, M.; Nillasithanukroh, S.; Han, W.; Lian, X.; Palecek, S.P. Human pluripotent stem cell culture density modulates YAP signaling. Biotechnol. J. 2016, 11, 662–675. [Google Scholar] [CrossRef] [Green Version]
- Ohgushi, M.; Minaguchi, M.; Sasai, Y. Rho-Signaling-Directed YAP/TAZ Activity Underlies the Long-Term Survival and Expansion of Human Embryonic Stem Cells. Cell Stem Cell 2015, 17, 448–461. [Google Scholar] [CrossRef] [Green Version]
- Cha, B.; Geng, X.; Mahamud, M.R.; Fu, J.; Mukherjee, A.; Kim, Y.; Jho, E.H.; Kim, T.H.; Kahn, M.L.; Xia, L.; et al. Mechanotransduction activates canonical Wnt/β-catenin signaling to promote lymphatic vascular patterning and the development of lymphatic and lymphovenous valves. Genes Dev. 2016, 30, 1454–1469. [Google Scholar] [CrossRef] [Green Version]
- Galli, C.; Piemontese, M.; Lumetti, S.; Ravanetti, F.; Macaluso, G.M.; Passeri, G. Actin cytoskeleton controls activation of Wnt/β-catenin signaling in mesenchymal cells on implant surfaces with different topographies. Acta Biomater. 2012, 8, 2963–2968. [Google Scholar] [CrossRef]
- Xu, H.G.; Zheng, Q.; Song, J.X.; Li, J.; Wang, H.; Liu, P.; Wang, J.; Wang, C.D.; Zhang, X.L. Intermittent cyclic mechanical tension promotes endplate cartilage degeneration via canonical Wnt signaling pathway and E-cadherin/β-catenin complex cross-talk. Osteoarthr. Cartil. 2016, 24, 158–168. [Google Scholar] [CrossRef] [Green Version]
- Huang, T.-S.; Li, L.; Moalim-Nour, L.; Jia, D.; Bai, J.; Yao, Z.; Bennett, S.A.L.; Figeys, D.; Wang, L. A Regulatory Network Involving β-Catenin, e-Cadherin, PI3k/Akt, and Slug Balances Self-Renewal and Differentiation of Human Pluripotent Stem Cells In Response to Wnt Signaling. Stem Cells 2015, 33, 1419–1433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Z.; Robitaille, A.M.; Berndt, J.D.; Davidson, K.C.; Fischer, K.A.; Mathieu, J.; Potter, J.C.; Ruohola-Baker, H.; Moon, R.T. Wnt/β-catenin signaling promotes self-renewal and inhibits the primed state transition in naïve human embryonic stem cells. Proc. Natl. Acad. Sci. USA 2016, 113, E6382–E6390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sperber, H.; Mathieu, J.; Wang, Y.; Ferreccio, A.; Hesson, J.; Xu, Z.; Fischer, K.A.; Devi, A.; Detraux, D.; Gu, H.; et al. The metabolome regulates the epigenetic landscape during naive-to-primed human embryonic stem cell transition. Nat. Cell Biol. 2015, 17, 1523–1535. [Google Scholar] [CrossRef]
- Theka, I.; Sottile, F.; Cammisa, M.; Bonnin, S.; Sanchez-Delgado, M.; Di Vicino, U.; Neguembor, M.V.; Arumugam, K.; Aulicino, F.; Monk, D.; et al. Wnt/β-catenin signaling pathway safeguards epigenetic stability and homeostasis of mouse embryonic stem cells. Sci. Rep. 2019, 9, 948. [Google Scholar] [CrossRef] [PubMed]
- Yi, F.; Pereira, L.; Hoffman, J.A.; Shy, B.R.; Yuen, C.M.; Liu, D.R.; Merrill, B.J. Opposing effects of Tcf3 and Tcf1 control Wnt stimulation of embryonic stem cell self-renewal. Nat. Cell Biol. 2011, 13, 762–770. [Google Scholar] [CrossRef] [PubMed]
- Sierra, R.A.; Hoverter, N.P.; Ramirez, R.N.; Vuong, L.M.; Mortazavi, A.; Merrill, B.J.; Waterman, M.L.; Donovan, P.J. TCF7L1 suppresses primitive streak gene expression to support human embryonic stem cell pluripotency. Development 2018, 145, dev161075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cevallos, R.R.; Rodríguez-Martínez, G.; Gazarian, K. Wnt/β-Catenin/TCF Pathway Is a Phase-Dependent Promoter of Colony Formation and Mesendodermal Differentiation During Human Somatic Cell Reprogramming. Stem Cells 2018, 36, 683–695. [Google Scholar] [CrossRef] [Green Version]
- Kelly, K.F.; Ng, D.Y.; Jayakumaran, G.; Wood, G.A.; Koide, H.; Doble, B.W. β-catenin enhances Oct-4 activity and reinforces pluripotency through a TCF-independent mechanism. Cell Stem Cell 2011, 8, 214–227. [Google Scholar] [CrossRef] [Green Version]
- Fang, F.; Xu, Y.; Chew, K.K.; Chen, X.; Ng, H.H.; Matsudaira, P. Coactivators p300 and CBP maintain the identity of mouse embryonic stem cells by mediating long-range chromatin structure. Stem Cells 2014, 32, 1805–1816. [Google Scholar] [CrossRef]
- Chen, X.; Xu, H.; Yuan, P.; Fang, F.; Huss, M.; Vega, V.B.; Wong, E.; Orlov, Y.L.; Zhang, W.; Jiang, J.; et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 2008, 133, 1106–1117. [Google Scholar] [CrossRef] [Green Version]
- Del Valle, I.; Rudloff, S.; Carles, A.; Li, Y.; Liszewska, E.; Vogt, R.; Kemler, R. E-cadherin is required for the proper activation of the Lifr/Gp130 signaling pathway in mouse embryonic stem cells. Development 2013, 140, 1684–1692. [Google Scholar] [CrossRef] [Green Version]
- Zimmerlin, L.; Park, T.S.; Huo, J.S.; Verma, K.; Pather, S.R.; Talbot, C.C.; Agarwal, J.; Steppan, D.; Zhang, Y.W.; Considine, M.; et al. Tankyrase inhibition promotes a stable human naïve pluripotent state with improved functionality. Development 2016, 143, 4368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Altshuler, A.; Verbuk, M.; Bhattacharya, S.; Abramovich, I.; Haklai, R.; Hanna, J.H.; Kloog, Y.; Gottlieb, E.; Shalom-Feuerstein, R. RAS Regulates the Transition from Naive to Primed Pluripotent Stem Cells. Stem Cell Rep. 2018, 10, 1088–1101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, C.; Xiao, G.; Hu, J. Regulation of Wnt/β-catenin signaling by posttranslational modifications. Cell Biosci. 2014, 4, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, X.; Wu, S.; Li, B.; Xu, Y.; Liu, J. Functions of p53 in pluripotent stem cells. Protein Cell 2020, 11, 71–78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, H.; Takahashi, K.; Ichisaka, T.; Aoi, T.; Kanagawa, O.; Nakagawa, M.; Okita, K.; Yamanaka, S. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature 2009, 460, 1132–1135. [Google Scholar] [CrossRef] [PubMed]
- Jain, A.K.; Allton, K.; Iacovino, M.; Mahen, E.; Milczarek, R.J.; Zwaka, T.P.; Kyba, M.; Barton, M.C. p53 Regulates Cell Cycle and MicroRNAs to Promote Differentiation of Human Embryonic Stem Cells. PLoS Biol. 2012, 10, e1001268. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.-N.; Chung, S.-K.; Xu, Z.; Xu, Y. Oct4 maintains the pluripotency of human embryonic stem cells by inactivating p53 through Sirt1-mediated deacetylation. Stem Cells 2014, 32, 157–165. [Google Scholar] [CrossRef] [Green Version]
- Vitillo, L.; Baxter, M.; Iskender, B.; Whiting, P.; Kimber, S.J. Integrin-Associated Focal Adhesion Kinase Protects Human Embryonic Stem Cells from Apoptosis, Detachment, and Differentiation. Stem Cell Rep. 2016, 7, 167–176. [Google Scholar] [CrossRef] [Green Version]
- Wawryk-Gawda, E.; Chylińska-Wrzos, P.; Lis-Sochocka, M.; Chłapek, K.; Bulak, K.; Jędrych, M.; Jodłowska-Jędrych, B. P53 protein in proliferation, repair and apoptosis of cells. Protoplasma 2014, 251, 525–533. [Google Scholar] [CrossRef] [Green Version]





| Gene | Primer Sequence | ||
|---|---|---|---|
| Forward (5′-3′) | Reverse (5′-3′) | Product Length | |
| ACTN4 | TGGCTGCTGAATGAGATCCG | GGCTTTGATGTCCGATAGTGT | 156 |
| ATM | CCTACCAAATCCCTCCACC | CCTTGAGCATCCCTTGTGTT | 236 |
| CCNG1 | AATGAAGGTACAGCCCAAGCA | GCTTTGACTTTCCAACACACC | 195 |
| CDK1 | TGCTGGGGTCAGCTCGTTAC | TGGGATGCTAGGCTTCCTGG | 232 |
| COL6A1 | TCAAGAGCCTGCAGTGGATG | TGGACACTTCTTGTCTATGC | 377 |
| CREBBP | AGTAACGGCACAGCCTCTCA | CCTGTCGATACAGTGCTTCTAG | 115 |
| DNMT3L | CTGCTCCATCTGCTGCTCC | ATCCACACACTCGAAGCAGT | 85 |
| DPPA3 | AGACCAACAAACAAGGAGCCT | CCCATCCATTAGACACGCAGA | 88 |
| DVL3 | ACAATGCCAAGCTACCATGCTTC | AGCTCCGATGGGTTATCAGCAC | 103 |
| FZD7 | GCAAAGCAGCGCAAATCTGA | AACCTCTGGCTTAACGGTGTGTG | 116 |
| GAPDH | ACAACTTTGGTATCGTGGAAGG | GCCATCACGCCACAGTTTC | 101 |
| HMBS | AGGAGTTCAGTGCCATCATCCT | CACAGCATACATGCATTCCTCA | 104 |
| ITGB1 | GGATTCTCCAGAAGGTGGTTT | TGCCACCAAGTTTCCCATCT | 143 |
| KLF17 | TCAGGAAGGGACTGGTAGAA | GTACCCGCATATGTCGTCTAAG | 206 |
| KLF4 | CGAACCCACACAGGTGAGAA | TACGGTAGTGCCTGGTCAGTTC | 75 |
| MDM4 | CTAAGTCCTTAAGTGATGATACCGATG | AACTTTGAACAATCTGAATACCAATCC | 151 |
| MIXL1 | CCGAGTCCAGGATCCAGGTA | CTCTGACGCCGAGACTTGG | 58 |
| MYL9 | GAGCCCAAGCGCCTTCT | GTCAATGAAGCCATCACGGT | 202 |
| NANOG | AAAGAATCTTCACCTATGCC | GAAGGAAGAGGAGAGACAGT | 110 |
| N-CADHERIN | TGTTTGGCCTGGCGTTCTTT | AGGAGACAGAAACGAAGCCA | 156 |
| NCAM | AGGAGACAGAAACGAAGCCA | GGTGTTGGAAATGCTCTGGT | 161 |
| OCT4 | CCCCTGGTGCCGTGAA | GCAAATTGCTCGAGTTCTTTCTG | 97 |
| PAX6 | CTTTGCTTGGGAAATCCGAG | AGCCAGGTTGCGAAGAACTC | 103 |
| RUVBL1 | TTGCTCAGGAGCTGGGTAGT | CCCATGGGATTCTCTGTCTC | 196 |
| sFRP2 | ACGGCATCGAATACCAGAACA | CTCGTCTAGGTCATCGAGGCA | 176 |
| SMAD4 | CCAGGATCAGTAGGTGGAAT | GTCTAAAGGTTGTGGGTCTG | 243 |
| SOX17 | CGCACGGAATTTGAACAGTA | GGATCAGGGACCTGTCACAC | 182 |
| SOX2 | TTGCTGCCTCTTTAAGACTAGGA | CTGGGGCTCAAACTTCTCTC | 75 |
| SOX7 | ACGCCGAGCTCAGCAAGAT | TCCACGTACGGCCTCTTCTG | 73 |
| TFCP2L1 | TTTGTGGGACCCTGCGAAG | TGCTTAAACGTGTCAATCTGGA | 129 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
McKee, C.; Brown, C.; Bakshi, S.; Walker, K.; Govind, C.K.; Chaudhry, G.R. Transcriptomic Analysis of Naïve Human Embryonic Stem Cells Cultured in Three-Dimensional PEG Scaffolds. Biomolecules 2021, 11, 21. https://doi.org/10.3390/biom11010021
McKee C, Brown C, Bakshi S, Walker K, Govind CK, Chaudhry GR. Transcriptomic Analysis of Naïve Human Embryonic Stem Cells Cultured in Three-Dimensional PEG Scaffolds. Biomolecules. 2021; 11(1):21. https://doi.org/10.3390/biom11010021
Chicago/Turabian StyleMcKee, Christina, Christina Brown, Shreeya Bakshi, Keegan Walker, Chhabi K. Govind, and G. Rasul Chaudhry. 2021. "Transcriptomic Analysis of Naïve Human Embryonic Stem Cells Cultured in Three-Dimensional PEG Scaffolds" Biomolecules 11, no. 1: 21. https://doi.org/10.3390/biom11010021





