Lipid Profile of Activated Macrophages and Contribution of Group V Phospholipase A2
Abstract
1. Introduction
2. Materials and Methods
2.1. Macrophage Cultures
2.2. Mass Spectrometry of Lipids
2.3. qPCR
2.4. Statistical Analysis
3. Results
3.1. Phospholipid Metabolism in Bone Marrow-Derived Macrophages Activated by IL-4 or LPS+IFNγ
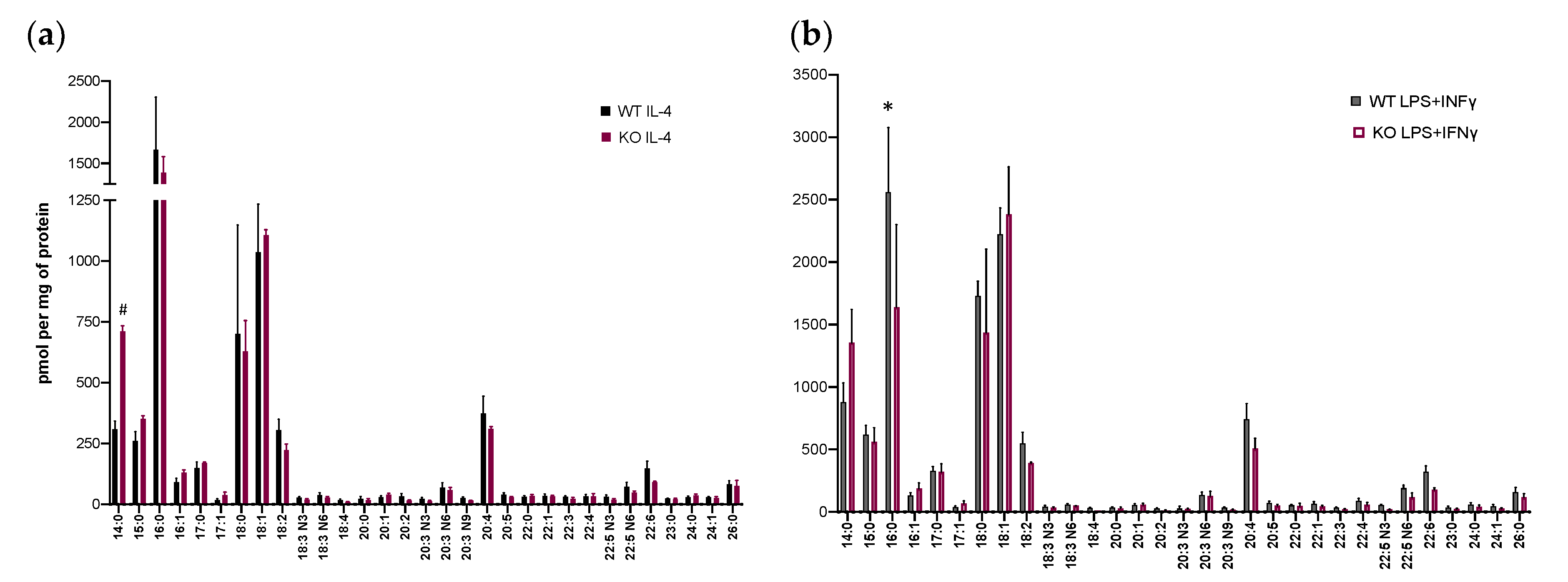
3.2. Lysophospholipid and Fatty Acid Generation in Activated BM-Macs
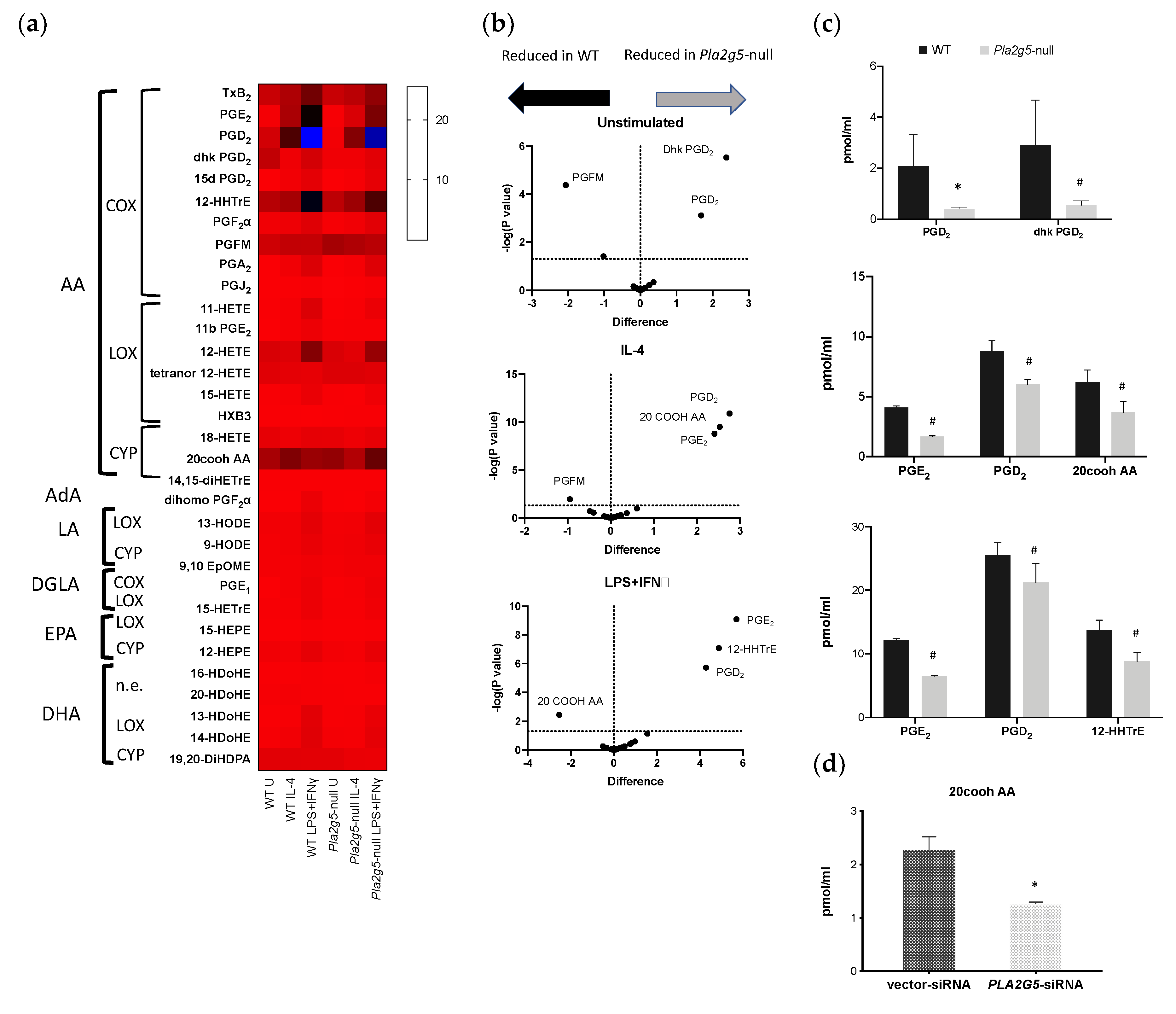
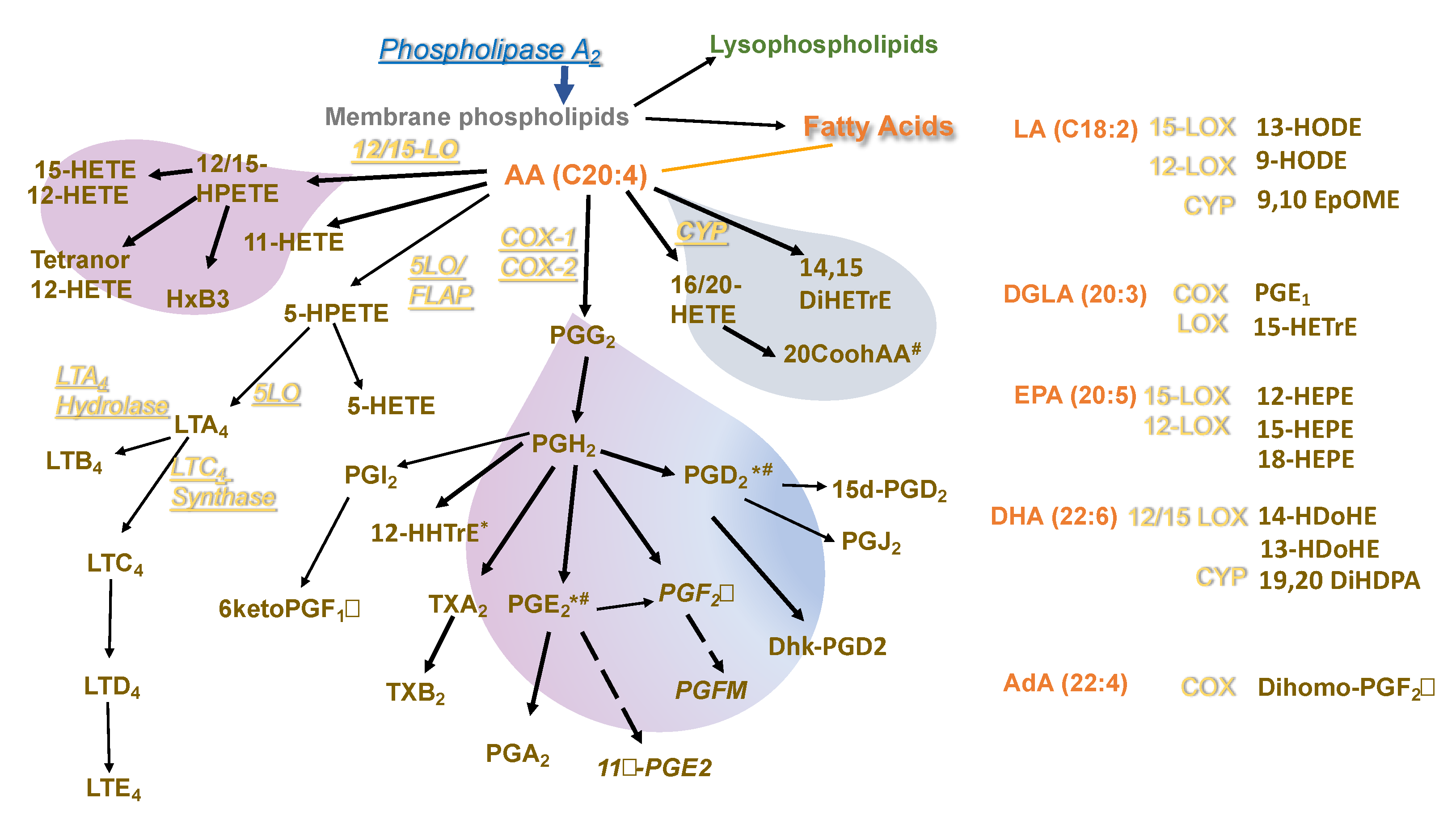
3.3. Differential Eicosanoid Generation in Bone Marrow-Derived Macrophages Activated by IL-4 or LPS+IFNγ
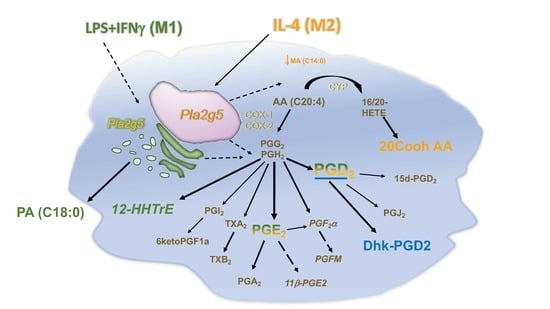
3.4. Pla2g5 Contributes to Selective Eicosanoid Generation in Unstimulated And Activated Macrophages
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Gordon, S.; Pluddemann, A. The mononuclear phagocytic system. Generation of diversity. Front Immunol. 2019, 10, 1893. [Google Scholar] [CrossRef]
- Locati, M.; Curtale, G.; Mantovani, A. Diversity, mechanisms, and significance of macrophage plasticity. Annu. Rev. Pathol. 2020, 15, 123–147. [Google Scholar] [CrossRef] [PubMed]
- Ying, W.; Fu, W.; Lee, Y.S.; Olefsky, J.M. The role of macrophages in obesity-associated islet inflammation and beta-cell abnormalities. Nat. Rev. Endocrinol. 2020, 16, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Gosselin, D.; Link, V.M.; Romanoski, C.E.; Fonseca, G.J.; Eichenfield, D.Z.; Spann, N.J.; Stender, J.D.; Chun, H.B.; Garner, H.; Geissmann, F.; et al. Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell 2014, 159, 1327–1340. [Google Scholar] [CrossRef]
- Ginhoux, F.; Schultze, J.L.; Murray, P.J.; Ochando, J.; Biswas, S.K. New insights into the multidimensional concept of macrophage ontogeny, activation and function. Nat. Immunol. 2016, 17, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Martinez, F.O.; Gordon, S. The M1 and M2 paradigm of macrophage activation: Time for reassessment. F1000Prime Rep. 2014, 6. [Google Scholar] [CrossRef]
- Martinez, F.O.; Helming, L.; Milde, R.; Varin, A.; Melgert, B.N.; Draijer, C.; Thomas, B.; Fabbri, M.; Crawshaw, A.; Ho, L.P.; et al. Genetic programs expressed in resting and IL-4 alternatively activated mouse and human macrophages: Similarities and differences. Blood 2013, 121, e57–e69. [Google Scholar] [CrossRef]
- Gieseck, R.L., III; Wilson, M.S.; Wynn, T.A. Type 2 immunity in tissue repair and fibrosis. Nat. Rev. Immunol. 2018, 18, 62–76. [Google Scholar] [CrossRef]
- Lackey, D.E.; Olefsky, J.M. Regulation of metabolism by the innate immune system. Nat. Rev. Endocrinol. 2016, 12, 15–28. [Google Scholar] [CrossRef]
- Wang, F.; Zhang, S.; Jeon, R.; Vuckovic, I.; Jiang, X.; Lerman, A.; Folmes, C.D.; Dzeja, P.D.; Herrmann, J. Interferon gamma induces reversible metabolic reprogramming of M1 macrophages to sustain cell viability and pro-inflammatory activity. EBioMedicine 2018, 30, 303–316. [Google Scholar] [CrossRef]
- Vats, D.; Mukundan, L.; Odegaard, J.I.; Zhang, L.; Smith, K.L.; Morel, C.R.; Wagner, R.A.; Greaves, D.R.; Murray, P.J.; Chawla, A. Oxidative metabolism and PGC-1beta attenuate macrophage-mediated inflammation. Cell Metab. 2006, 4, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Ecker, J.; Liebisch, G.; Grandl, M.; Schmitz, G. Lower SCD expression in dendritic cells compared to macrophages leads to membrane lipids with less mono-unsaturated fatty acids. Immunobiology 2010, 215, 748–755. [Google Scholar] [CrossRef] [PubMed]
- Jha, A.K.; Huang, S.C.; Sergushichev, A.; Lampropoulou, V.; Ivanova, Y.; Loginicheva, E.; Chmielewski, K.; Stewart, K.M.; Ashall, J.; Everts, B.; et al. Network integration of parallel metabolic and transcriptional data reveals metabolic modules that regulate macrophage polarization. Immunity 2015, 42, 419–430. [Google Scholar] [CrossRef] [PubMed]
- Ecker, J.; Liebisch, G.; Englmaier, M.; Grandl, M.; Robenek, H.; Schmitz, G. Induction of fatty acid synthesis is a key requirement for phagocytic differentiation of human monocytes. Proc. Natl. Acad. Sci. USA 2010, 107, 7817–7822. [Google Scholar] [CrossRef]
- Lukic, A.; Larssen, P.; Fauland, A.; Samuelsson, B.; Wheelock, C.E.; Gabrielsson, S.; Radmark, O. GM-CSF- and M-CSF-primed macrophages present similar resolving but distinct inflammatory lipid mediator signatures. FASEB J. 2017, 31, 4370–4381. [Google Scholar] [CrossRef]
- Stein, M.; Keshav, S.; Harris, N.; Gordon, S. Interleukin 4 potently enhances murine macrophage mannose receptor activity: A marker of alternative immunologic macrophage activation. J. Exp. Med. 1992, 176, 287–292. [Google Scholar] [CrossRef]
- Kanaoka, Y.; Austen, K.F. Roles of cysteinyl leukotrienes and their receptors in immune cell-related functions. Adv. Immunol. 2019, 142, 65–84. [Google Scholar] [CrossRef]
- Samuchiwal, S.K.; Balestrieri, B.; Raff, H.; Boyce, J.A. Endogenous prostaglandin E2 amplifies IL-33 production by macrophages through an E prostanoid (EP)2/EP4-cAMP-EPAC-dependent pathway. J. Biol. Chem. 2017, 292, 8195–8206. [Google Scholar] [CrossRef]
- Samuchiwal, S.K.; Boyce, J.A. Role of lipid mediators and control of lymphocyte responses in type 2 immunopathology. J. Allergy Clin. Immunol. 2018, 141, 1182–1190. [Google Scholar] [CrossRef]
- Yamaguchi, M.; Zacharia, J.; Laidlaw, T.M.; Balestrieri, B. PLA2G5 regulates transglutaminase activity of human IL-4-activated M2 macrophages through PGE2 generation. J. Leukoc. Biol. 2016, 100, 131–141. [Google Scholar] [CrossRef]
- Bystrom, J.; Wray, J.A.; Sugden, M.C.; Holness, M.J.; Swales, K.E.; Warner, T.D.; Edin, M.L.; Zeldin, D.C.; Gilroy, D.W.; Bishop-Bailey, D. Endogenous epoxygenases are modulators of monocyte/macrophage activity. PLoS ONE 2011, 6, e26591. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Dillon, J.S.; Hu, S.; Harmon, S.D.; Yao, J.; Anjaiah, S.; Falck, J.R.; Spector, A.A. 20-carboxy-arachidonic acid is a dual activator of peroxisome proliferator-activated receptors alpha and gamma. Prostaglandins Lipid Mediat. 2007, 82, 175–184. [Google Scholar] [CrossRef] [PubMed]
- Garcia, V.; Gilani, A.; Shkolnik, B.; Pandey, V.; Zhang, F.F.; Dakarapu, R.; Gandham, S.K.; Reddy, N.R.; Graves, J.P.; Gruzdev, A.; et al. 20-HETE signals through g-protein-coupled receptor GPR75 (Gq) to affect vascular function and trigger hypertension. Circ. Res. 2017, 120, 1776–1788. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.C.; Everts, B.; Ivanova, Y.; O’Sullivan, D.; Nascimento, M.; Smith, A.M.; Beatty, W.; Love-Gregory, L.; Lam, W.Y.; O’Neill, C.M.; et al. Cell-intrinsic lysosomal lipolysis is essential for alternative activation of macrophages. Nat. Immunol. 2014, 15, 846–855. [Google Scholar] [CrossRef] [PubMed]
- Samuchiwal, S.K.; Balestrieri, B. Harmful and protective roles of group V phospholipase A2: Current perspectives and future directions. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2019, 1864, 819–826. [Google Scholar] [CrossRef]
- Murakami, M.; Sato, H.; Taketomi, Y. Updating phospholipase A2 biology. Biomolecules 2020, 10, 1457. [Google Scholar] [CrossRef]
- Balestrieri, B.; Hsu, V.W.; Gilbert, H.; Leslie, C.C.; Han, W.K.; Bonventre, J.V.; Arm, J.P. Group V secretory phospholipase A2 translocates to the phagosome after zymosan stimulation of mouse peritoneal macrophages and regulates phagocytosis. J. Biol. Chem. 2006, 281, 6691–6698. [Google Scholar] [CrossRef]
- Ruiperez, V.; Astudillo, A.M.; Balboa, M.A.; Balsinde, J. Coordinate regulation of TLR-mediated arachidonic acid mobilization in macrophages by group IVA and group V phospholipase A2s. J. Immunol. 2009, 182, 3877–3883. [Google Scholar] [CrossRef]
- Balestrieri, B.; Maekawa, A.; Xing, W.; Gelb, M.H.; Katz, H.R.; Arm, J.P. Group V secretory phospholipase A2 modulates phagosome maturation and regulates the innate immune response against Candida albicans. J. Immunol. 2009, 182, 4891–4898. [Google Scholar] [CrossRef]
- Lapointe, S.; Brkovic, A.; Cloutier, I.; Tanguay, J.F.; Arm, J.P.; Sirois, M.G. Group V secreted phospholipase A2 contributes to LPS-induced leukocyte recruitment. J. Cell. Physiol. 2010, 224, 127–134. [Google Scholar] [CrossRef]
- Munoz, N.M.; Meliton, A.Y.; Meliton, L.N.; Dudek, S.M.; Leff, A.R. Secretory group V phospholipase A2 regulates acute lung injury and neutrophilic inflammation caused by LPS in mice. Am. J. Physiol. Lung Cell Mol. Physiol. 2009, 296, L879–L887. [Google Scholar] [CrossRef] [PubMed]
- Ohta, S.; Imamura, M.; Xing, W.; Boyce, J.A.; Balestrieri, B. Group V secretory phospholipase A2 is involved in macrophage activation and is sufficient for macrophage effector functions in allergic pulmonary inflammation. J. Immunol. 2013, 190, 5927–5938. [Google Scholar] [CrossRef] [PubMed]
- Rubio, J.M.; Rodriguez, J.P.; Gil-de-Gomez, L.; Guijas, C.; Balboa, M.A.; Balsinde, J. Group V secreted phospholipase A2 is upregulated by IL-4 in human macrophages and mediates phagocytosis via hydrolysis of ethanolamine phospholipids. J. Immunol. 2015, 194, 3327–3339. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, M.; Samuchiwal, S.K.; Quehenberger, O.; Boyce, J.A.; Balestrieri, B. Macrophages regulate lung ILC2 activation via Pla2g5-dependent mechanisms. Mucosal. Immunol. 2017. [Google Scholar] [CrossRef]
- Satake, Y.; Diaz, B.L.; Balestrieri, B.; Lam, B.K.; Kanaoka, Y.; Grusby, M.J.; Arm, J.P. Role of group V phospholipase A2 in zymosan-induced eicosanoid generation and vascular permeability revealed by targeted gene disruption. J. Biol. Chem. 2004, 279, 16488–16494. [Google Scholar] [CrossRef]
- Xue, J.; Schmidt, S.V.; Sander, J.; Draffehn, A.; Krebs, W.; Quester, I.; de Nardo, D.; Gohel, T.D.; Emde, M.; Schmidleithner, L.; et al. Transcriptome-based network analysis reveals a spectrum model of human macrophage activation. Immunity 2014, 40, 274–288. [Google Scholar] [CrossRef]
- Quehenberger, O.; Armando, A.M.; Brown, A.H.; Milne, S.B.; Myers, D.S.; Merrill, A.H.; Bandyopadhyay, S.; Jones, K.N.; Kelly, S.; Shaner, R.L.; et al. Lipidomics reveals a remarkable diversity of lipids in human plasma. J. Lipid Res. 2010, 51, 3299–3305. [Google Scholar] [CrossRef]
- Wang, Y.; Armando, A.M.; Quehenberger, O.; Yan, C.; Dennis, E.A. Comprehensive ultra-performance liquid chromatographic separation and mass spectrometric analysis of eicosanoid metabolites in human samples. J. Chromatogr. A 2014, 1359, 60–69. [Google Scholar] [CrossRef]
- Quehenberger, O.; Armando, A.; Dumlao, D.; Stephens, D.L.; Dennis, E.A. Lipidomics analysis of essential fatty acids in macrophages. Prostaglandins Leukot. Essent. Fat. Acids 2008, 79, 123–129. [Google Scholar] [CrossRef]
- Murray, P.J.; Allen, J.E.; Biswas, S.K.; Fisher, E.A.; Gilroy, D.W.; Goerdt, S.; Gordon, S.; Hamilton, J.A.; Ivashkiv, L.B.; Lawrence, T.; et al. Macrophage activation and polarization: Nomenclature and experimental guidelines. Immunity 2014, 41, 14–20. [Google Scholar] [CrossRef]
- Shindou, H.; Hishikawa, D.; Harayama, T.; Eto, M.; Shimizu, T. Generation of membrane diversity by lysophospholipid acyltransferases. J. Biochem. 2013, 154, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Quehenberger, O.; Armando, A.M.; Dennis, E.A. High sensitivity quantitative lipidomics analysis of fatty acids in biological samples by gas chromatography-mass spectrometry. Biochim. Biophys. Acta 2011, 1811, 648–656. [Google Scholar] [CrossRef] [PubMed]
- Dennis, E.A.; Deems, R.A.; Harkewicz, R.; Quehenberger, O.; Brown, H.A.; Milne, S.B.; Myers, D.S.; Glass, C.K.; Hardiman, G.; Reichart, D.; et al. A mouse macrophage lipidome. J. Biol. Chem. 2010, 285, 39976–39985. [Google Scholar] [CrossRef] [PubMed]
- Montenegro-Burke, J.R.; Sutton, J.A.; Rogers, L.M.; Milne, G.L.; McLean, J.A.; Aronoff, D.M. Lipid profiling of polarized human monocyte-derived macrophages. Prostaglandins Other Lipid Mediat. 2016, 127, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Nolin, J.D.; Lai, Y.; Ogden, H.L.; Manicone, A.M.; Murphy, R.C.; An, D.; Frevert, C.W.; Ghomashchi, F.; Naika, G.S.; Gelb, M.H.; et al. Secreted PLA2 group X orchestrates innate and adaptive immune responses to inhaled allergen. JCI Insight 2017, 2. [Google Scholar] [CrossRef] [PubMed]
- Shindou, H.; Hishikawa, D.; Harayama, T.; Yuki, K.; Shimizu, T. Recent progress on acyl CoA: Lysophospholipid acyltransferase research. J. Lipid Res. 2009, 50, S46–S51. [Google Scholar] [CrossRef]
- Alvarez-Curto, E.; Milligan, G. Metabolism meets immunity: The role of free fatty acid receptors in the immune system. Biochem. Pharm. 2016, 114, 3–13. [Google Scholar] [CrossRef]
- Sato, H.; Taketomi, Y.; Ushida, A.; Isogai, Y.; Kojima, T.; Hirabayashi, T.; Miki, Y.; Yamamoto, K.; Nishito, Y.; Kobayashi, T.; et al. The adipocyte-inducible secreted phospholipases PLA2G5 and PLA2G2E play distinct roles in obesity. Cell Metab. 2014, 20, 119–132. [Google Scholar] [CrossRef]
- Watanabe, K.; Taketomi, Y.; Miki, Y.; Kugiyama, K.; Murakami, M. Group V secreted phospholipase A2 plays a protective role against aortic dissection. J. Biol. Chem. 2020, 295, 10092–10111. [Google Scholar] [CrossRef]
- Norris, P.C.; Reichart, D.; Dumlao, D.S.; Glass, C.K.; Dennis, E.A. Specificity of eicosanoid production depends on the TLR-4-stimulated macrophage phenotype. J. Leukoc. Biol. 2011, 90, 563–574. [Google Scholar] [CrossRef]
- Qi, H.Y.; Shelhamer, J.H. Toll-like receptor 4 signaling regulates cytosolic phospholipase A2 activation and lipid generation in lipopolysaccharide-stimulated macrophages. J. Biol. Chem. 2005, 280, 38969–38975. [Google Scholar] [CrossRef] [PubMed]
- Ueno, N.; Takegoshi, Y.; Kamei, D.; Kudo, I.; Murakami, M. Coupling between cyclooxygenases and terminal prostanoid synthases. Biochem. Biophys. Res. Commun. 2005, 338, 70–76. [Google Scholar] [CrossRef] [PubMed]
- Buczynski, M.W.; Stephens, D.L.; Bowers-Gentry, R.C.; Grkovich, A.; Deems, R.A.; Dennis, E.A. TLR-4 and sustained calcium agonists synergistically produce eicosanoids independent of protein synthesis in RAW264. 7 cells. J. Biol. Chem. 2007, 282, 22834–22847. [Google Scholar] [CrossRef] [PubMed]
- Fromel, T.; Kohlstedt, K.; Popp, R.; Yin, X.; Awwad, K.; Barbosa-Sicard, E.; Thomas, A.C.; Lieberz, R.; Mayr, M.; Fleming, I. Cytochrome P4502S1: A novel monocyte/macrophage fatty acid epoxygenase in human atherosclerotic plaques. Basic Res. Cardiol. 2013, 108, 319. [Google Scholar] [CrossRef]
- Odegaard, J.I.; Ricardo-Gonzalez, R.R.; Goforth, M.H.; Morel, C.R.; Subramanian, V.; Mukundan, L.; Eagle, A.R.; Vats, D.; Brombacher, F.; Ferrante, A.W.; et al. Macrophage-specific PPARgamma controls alternative activation and improves insulin resistance. Nature 2007, 447, 1116–1120. [Google Scholar] [CrossRef]
- Buczynski, M.W.; Dumlao, D.S.; Dennis, E.A. Thematic review series: Proteomics. An integrated omics analysis of eicosanoid biology. J. Lipid Res. 2009, 50, 1015–1038. [Google Scholar] [CrossRef]
- Boilard, E.; Lai, Y.; Larabee, K.; Balestrieri, B.; Ghomashchi, F.; Fujioka, D.; Gobezie, R.; Coblyn, J.S.; Weinblatt, M.E.; Massarotti, E.M.; et al. A novel anti-inflammatory role for secretory phospholipase A2 in immune complex-mediated arthritis. EMBO Mol. Med. 2010, 2, 172–187. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koganesawa, M.; Yamaguchi, M.; Samuchiwal, S.K.; Balestrieri, B. Lipid Profile of Activated Macrophages and Contribution of Group V Phospholipase A2. Biomolecules 2021, 11, 25. https://doi.org/10.3390/biom11010025
Koganesawa M, Yamaguchi M, Samuchiwal SK, Balestrieri B. Lipid Profile of Activated Macrophages and Contribution of Group V Phospholipase A2. Biomolecules. 2021; 11(1):25. https://doi.org/10.3390/biom11010025
Chicago/Turabian StyleKoganesawa, Masaya, Munehiro Yamaguchi, Sachin K. Samuchiwal, and Barbara Balestrieri. 2021. "Lipid Profile of Activated Macrophages and Contribution of Group V Phospholipase A2" Biomolecules 11, no. 1: 25. https://doi.org/10.3390/biom11010025
APA StyleKoganesawa, M., Yamaguchi, M., Samuchiwal, S. K., & Balestrieri, B. (2021). Lipid Profile of Activated Macrophages and Contribution of Group V Phospholipase A2. Biomolecules, 11(1), 25. https://doi.org/10.3390/biom11010025