On the Mechanism of Bioinspired Formation of Inorganic Oxides: Structural Evidence of the Electrostatic Nature of the Interaction between a Mononuclear Inorganic Precursor and Lysozyme
Abstract
:1. Introduction
2. Materials and Methods
2.1. Crystallization, Data Collection, and Structure Solution
2.2. DFT Calculations
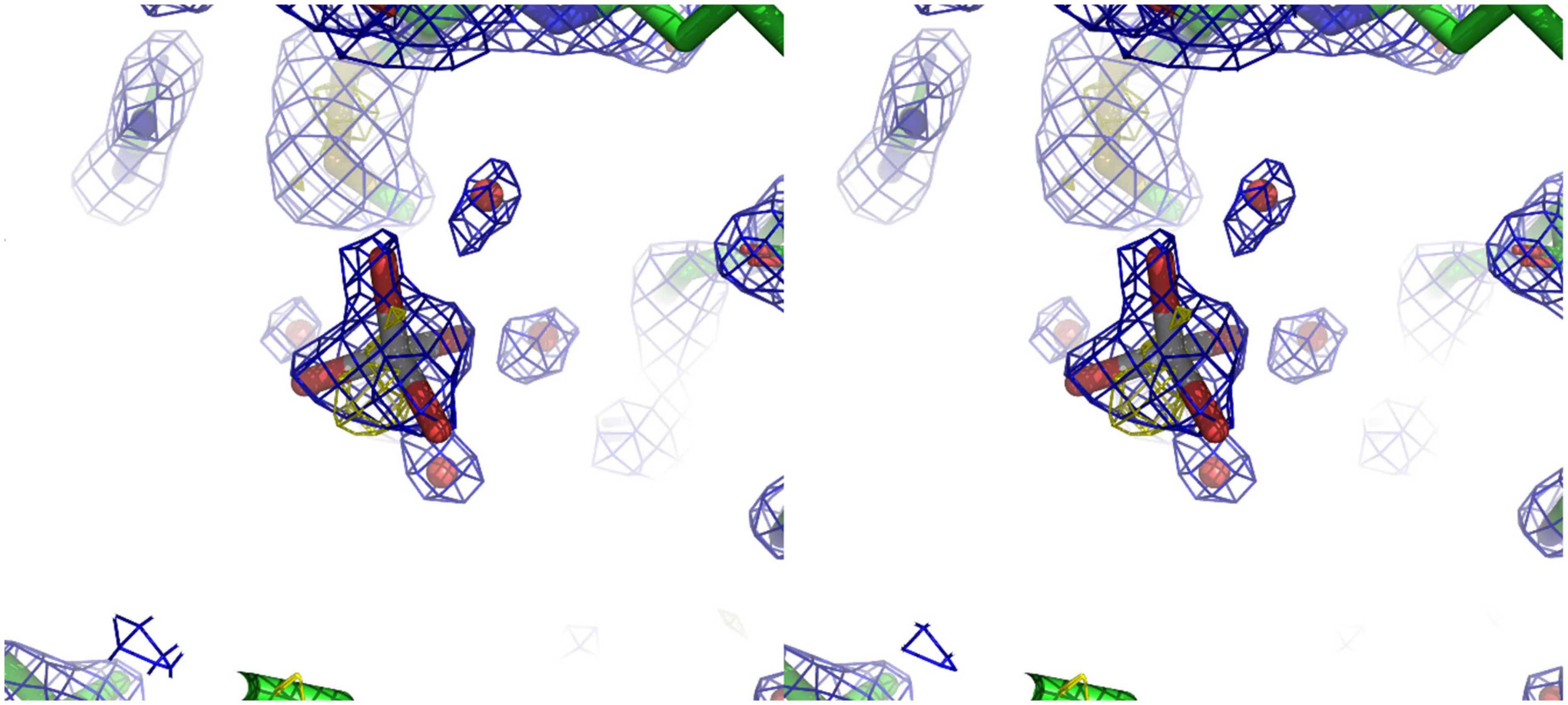
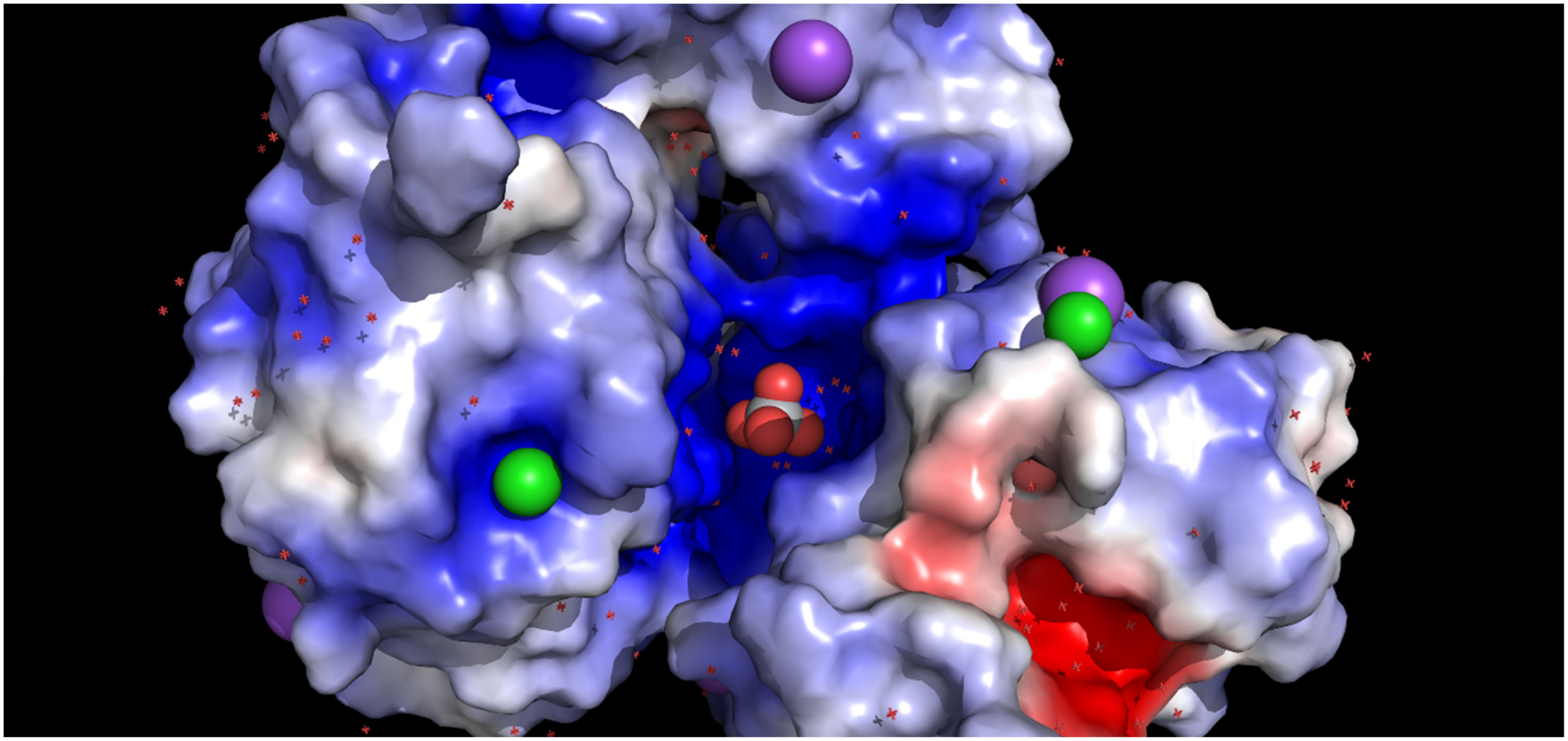
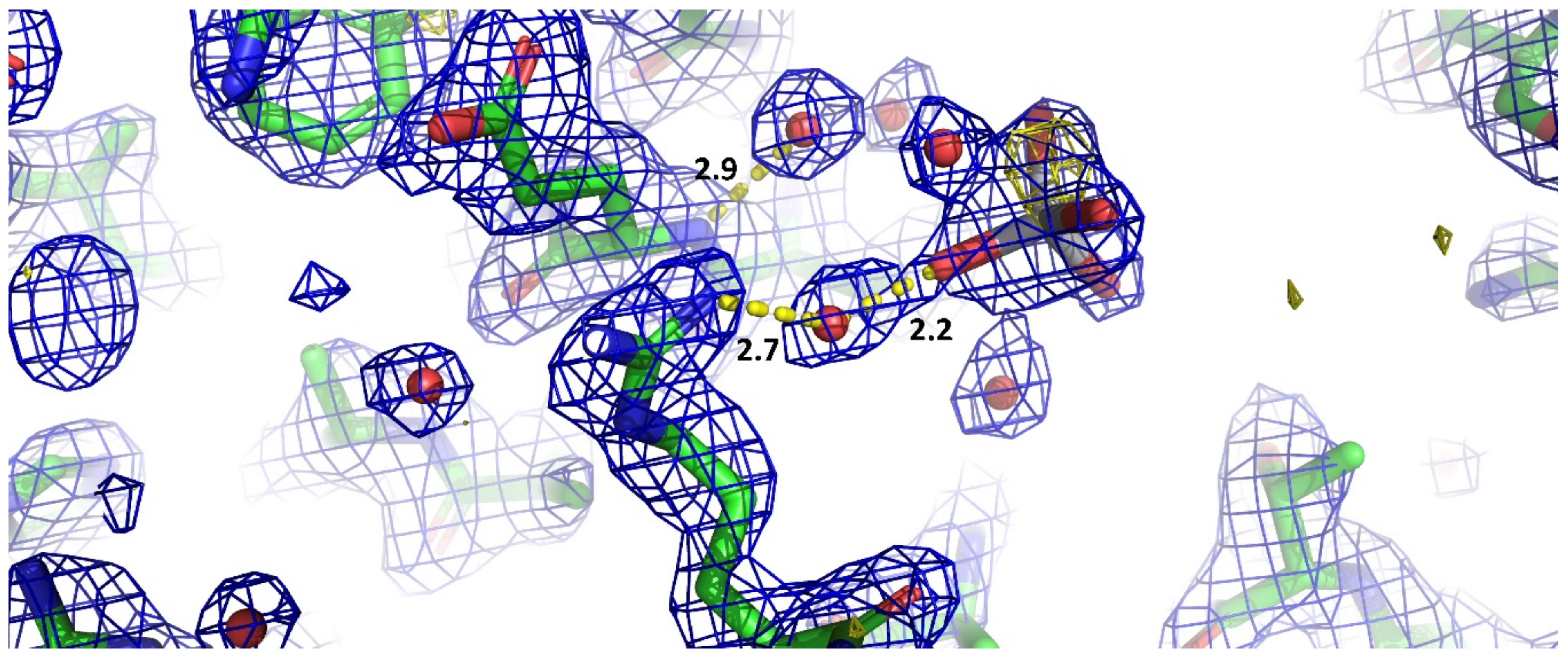
3. Results and Discussion
- (i)
- the presence of an anomalous signal is slightly higher than those attributed to sulfurs and chlorides;
- (ii)
- the shape of the 2Fo-Fc density is clearly not spherical as one would expect for chloride ion but rather tetrahedral as shown in Figure 1, and this becomes even more apparent when slightly lowering its contour value;
- (iii)
- the Ti-O distances refine well at about 1.9 Å, which is in agreement with the theoretical value expected for such bond;
- (iv)
- the Fo-Fc density in that position is absent at ±3.0 σ contour;
- (v)
- the B-factor values for the refined atoms of the titanium moiety have values coherent with those of the other labile or loosely bound atoms in the structure;
- (vi)
- the comparison with several other lysozyme structures with the same space group shows that no density is present in the position that we attributed to the mononuclear titanium species.
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Koch, C.C. Nanostructured Materials Processing, Properties and Potential Applications; Noyes Publications: Norwich, NY, USA, 2002; pp. 3–40. [Google Scholar]
- Rao, C.N.R.; Raveau, B. Transition Metal Oxides: Structure, Properties, and Synthesis of Ceramic Oxides, 2nd ed.; Wiley-VCH: New York, NY, USA, 1998; p. xi, 373p. [Google Scholar]
- Mann, S. Biomineralization Principles and Concepts in Bioinorganic Materials Chemistry; Oxford University Press: Oxford, NY, USA, 2001; p. 198S. [Google Scholar]
- Nalwa, H.S. Encyclopedia of Nanoscience and Nanotechnology. v. 1-10; American Scientific Publishers: Stevenson Ranch, CA, USA, 2004; Volume 1, pp. 293–308. [Google Scholar]
- Sumerel, J.L.; Yang, W.J.; Kisailus, D.; Weaver, J.C.; Choi, J.H.; Morse, D.E. Biocatalytically templated synthesis of titanium dioxide. Chem. Mater. 2003, 15, 4804–4809. [Google Scholar] [CrossRef]
- Sewell, S.L.; Wright, D.W. Biomimetic synthesis of titanium dioxide utilizing the R5 peptide derived from Cylindrotheca fusiformis. Chem. Mater. 2006, 18, 3108–3113. [Google Scholar] [CrossRef]
- Luckarift, H.R.; Dickerson, M.B.; Sandhage, K.H.; Spain, J.C. Rapid, room-temperature synthesis of antibacterial bionanocomposites of lysozyme with amorphous silica or titania. Small 2006, 2, 640–643. [Google Scholar] [CrossRef] [PubMed]
- Tong, Z.W.; Jiang, Y.J.; Yang, D.; Shi, J.F.; Zhang, S.H.; Liu, C.; Jiang, Z.Y. Biomimetic and bioinspired synthesis of titania and titania-based materials. RSC Adv. 2014, 4, 12388–12403. [Google Scholar] [CrossRef]
- Wang, C.; Jiao, K.; Yan, J.; Wan, M.; Wan, Q.; Breschi, L.; Chen, J.; Tay, F.R.; Niu, L. Biological and synthetic template-directed syntheses of mineralized hybrid and inorganic materials. Prog. Mater. Sci. 2020, 100712. [Google Scholar] [CrossRef]
- Kim, J.K.; Jang, J.R.; Choi, N.; Hong, D.; Nam, C.H.; Yoo, P.J.; Park, J.H.; Choe, W.S. Lysozyme-mediated biomineralization of titanium-tungsten oxide hybrid nanoparticles with high photocatalytic activity. Chem. Commun. 2014, 50, 12392–12395. [Google Scholar] [CrossRef]
- Nadeem, M.; Tungmunnithum, D.; Hano, C.; Abbasi, B.H.; Hashmi, S.S.; Ahmad, W.; Zahir, A. The current trends in the green syntheses of titanium oxide nanoparticles and their applications. Green Chem. Lett. Rev. 2018, 11, 492–502. [Google Scholar] [CrossRef] [Green Version]
- Salman, M.S.; Park, A.R.; Cha, M.J.; Choi, Y.; Jang, S.K.; Tan, L.H.; Yoo, P.J.; Choe, W.S. Lysozyme-Templated Meso-Macroporous Hollow TiO2 for Lithium Ion Battery Anode. Acs Appl. Nano Mater. 2018, 1, 698–710. [Google Scholar] [CrossRef]
- Choi, N.; Tan, L.H.; Jang, J.R.; Um, Y.M.; Yoo, P.J.; Choe, W.S. The interplay of peptide sequence and local structure in TiO2 biomineralization. J. Inorg. Biochem. 2012, 115, 20–27. [Google Scholar] [CrossRef]
- Kim, J.K.; Jang, J.R.; Salman, M.S.; Tan, L.H.; Nam, C.H.; Yoo, P.J.; Choe, W.S. Harnessing designer biotemplates for biomineralization of TiO2 with tunable photocatalytic activity. Ceram. Int. 2019, 45, 6467–6476. [Google Scholar] [CrossRef]
- Kuroda, A.; Ogino, K. Development and application of amorphous titanium dioxide. Fragr. J. 1994, 22, 17. [Google Scholar]
- Kröger, N.; Deutzmann, R.; Bergsdorf, C.; Sumper, M. Species-specific polyamines from diatoms control silica morphology. Proc. Natl. Acad. Sci. USA 2000, 97, 14133–14138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kröger, N.; Deutzmann, R.; Sumper, M. Polycationic Peptides from Diatom Biosilica That Direct Silica Nanosphere Formation. Science 1999, 286, 1129–1132. [Google Scholar] [PubMed] [Green Version]
- Knecht, M.R.; Wright, D.W. Functional analysis of the biomimetic silica precipitating activity of the R5 peptide from Cylindrotheca fusiformis. Chem. Commun. 2003, 24, 3038–3039. [Google Scholar] [CrossRef]
- Naik, R.R.; Whitlock, P.W.; Rodriguez, F.; Brott, L.L.; Glawe, D.D.; Clarson, S.J.; Stone, M.O. Controlled formation of biosilica structures in vitro. Chem. Commun. 2003, 238–239. [Google Scholar] [CrossRef]
- Patwardhan, S.V.; Mukherjee, N.; Steintz-Kannan, M.; Clarson, S.J. Bioinspired synthesis of new silica structures. Chem. Commun. 2003, 1122–1123. [Google Scholar] [CrossRef]
- Lechner, C.C.; Becker, C.F. A sequence-function analysis of the silica precipitating silaffin R5 peptide. J. Pept. Sci. 2014, 20, 152–158. [Google Scholar] [CrossRef]
- Ndao, M.; Goobes, G.; Emani, P.S.; Drobny, G.P. A REDOR ssNMR Investigation of the Role of an N-Terminus Lysine in R5 Silica Recognition. Langmuir 2018, 34, 8678–8684. [Google Scholar] [CrossRef]
- Sprenger, K.G.; Prakash, A.; Drobny, G.; Pfaendtner, J. Investigating the Role of Phosphorylation in the Binding of Silaffin Peptide R5 to Silica with Molecular Dynamics Simulations. Langmuir 2018, 34, 1199–1207. [Google Scholar] [CrossRef]
- Bruckner, S.I.; Donets, S.; Dianat, A.; Bobeth, M.; Gutierrez, R.; Cuniberti, G.; Brunner, E. Probing Silica-Biomolecule Interactions by Solid-State NMR and Molecular Dynamics Simulations. Langmuir 2016, 32, 11698–11705. [Google Scholar] [CrossRef]
- Jantschke, A.; Koers, E.; Mance, D.; Weingarth, M.; Brunner, E.; Baldus, M. Insight into the Supramolecular Architecture of Intact Diatom Biosilica from DNP-Supported Solid-State NMR Spectroscopy. Angew. Chem. Int. Ed. 2015, 54, 15069–15073. [Google Scholar] [CrossRef] [PubMed]
- Geiger, Y.; Gottlieb, H.E.; Akbey, U.; Oschkinat, H.; Goobes, G. Studying the Conformation of a Silaffin-Derived Pentalysine Peptide Embedded in Bioinspired Silica using Solution and Dynamic Nuclear Polarization Magic-Angle Spinning NMR. J. Am. Chem. Soc. 2016, 138, 5561–5567. [Google Scholar] [CrossRef] [PubMed]
- Ravera, E.; Michaelis, V.K.; Ong, T.C.; Keeler, E.G.; Martelli, T.; Fragai, M.; Griffin, R.G.; Luchinat, C. Biosilica-entrapped enzymes can be studied by DNP-enhanced high-field NMR. Chem. Phys. Chem. 2015, 16, 2751–2754. [Google Scholar] [PubMed]
- Buckle, E.L.; Lum, J.S.; Roehrich, A.M.; Stote, R.E.; Vandermoon, B.; Dracinsky, M.; Filocamo, S.F.; Drobny, G.P. Serine-Lysine Peptides as Mediators for the Production of Titanium Dioxide: Investigating the Effects of Primary and Secondary Structures Using Solid-State NMR Spectroscopy and DFT Calculations. J. Phys. Chem. B. 2018, 122, 4708–4718. [Google Scholar] [CrossRef]
- Ravera, E.; Cerofolini, L.; Martelli, T.; Louka, A.; Fragai, M.; Luchinat, C. 1H detected solid state NMR of proteins entrapped in bioinspired silica: A new tool for biomaterials. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [Green Version]
- Patwardhan, S.V.; Clarson, S.J. Silicification and biosilicification-Part 6. Poly-L-histidine mediated synthesis of silica at neutral pH. J. Inorg. Organomet. Polymers 2003, 13, 49–53. [Google Scholar] [CrossRef]
- Kabsch, W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 125–132. [Google Scholar] [CrossRef] [Green Version]
- Vagin, A.A.; Teplyakov, A. An approach to multi-copy search in molecular replacement. Acta Crystallogr. D Biol. Crystallogr. 2000, 56, 1622–1624. [Google Scholar] [CrossRef]
- Adams, P.D.; Afonine, P.V.; Bunkoczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 213–221. [Google Scholar] [CrossRef] [Green Version]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef] [Green Version]
- Langer, G.; Cohen, S.X.; Lamzin, V.S.; Perrakis, A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat. Protoc. 2008, 3, 1171–1179. [Google Scholar] [CrossRef] [PubMed]
- Chen, V.B.; Arendall, W.B., III; Headd, J.J.; Keedy, D.A.; Immormino, R.M.; Kapral, G.J.; Murray, L.W.; Richardson, J.S.; Richardson, D.C. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr 2010, 66, 12–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Becke, A.D. A New Mixing of Hartree-Fock and Local Density-Functional Theories. J. Chem. Phys. 1993, 98, 1372–1377. [Google Scholar] [CrossRef]
- Lee, C.; Yang, W.; Parr, R.G. Development of the Colle-Salvetti correlation-energy formula into a functional of electron density. Phys. Rev. B 1988, 37, 785–789. [Google Scholar] [CrossRef] [Green Version]
- Stephens, P.J.; Devlin, F.J.; Chabalowski, C.F.; Frisch, M.J. Ab-Initio Calculation of Vibrational Absorption and Circular-Dichroism Spectra Using Density-Functional Force-Fields. J. Phys. Chem. 1994, 98, 11623–11627. [Google Scholar] [CrossRef]
- Vosko, S.H.; Wilk, L.; Nusair, M. Accurate spin-dependent electron liquid correlation energies for local spin density calculations: A critical analysis. Can. J. Phys. 1980, 58, 1200–1211. [Google Scholar] [CrossRef] [Green Version]
- Schafer, A.; Huber, C.; Ahlrichs, R. Fully Optimized Contracted Gaussian-Basis Sets of Triple Zeta Valence Quality for Atoms Li to Kr. J. Chem. Phys. 1994, 100, 5829–5835. [Google Scholar] [CrossRef]
- Weigend, F.; Ahlrichs, R. Balanced basis sets of split valence, triple zeta valence and quadruple zeta valence quality for H to Rn: Design and assessment of accuracy. Phys. Chem. Chem. Phys. 2005, 7, 3297–3305. [Google Scholar] [CrossRef]
- Grimme, S.; Antony, J.; Ehrlich, S.; Krieg, H. A consistent and accurate ab initio parametrization of density functional dispersion correction (DFT-D) for the 94 elements H-Pu. J. Chem. Phys. 2010, 132. [Google Scholar] [CrossRef] [Green Version]
- Grimme, S.; Ehrlich, S.; Goerigk, L. Effect of the Damping Function in Dispersion Corrected Density Functional Theory. J. Comput. Chem. 2011, 32, 1456–1465. [Google Scholar] [CrossRef]
- Kendall, R.A.; Fruchtl, H.A. The impact of the resolution of the identity approximate integral method on modern ab initio algorithm development. Theor. Chem. Acc. 1997, 97, 158–163. [Google Scholar] [CrossRef]
- Vahtras, O.; Almlof, J.; Feyereisen, M.W. Integral Approximations for Lcao-Scf Calculations. Chem. Phys. Lett. 1993, 213, 514–518. [Google Scholar] [CrossRef]
- Barone, V.; Cossi, M. Quantum calculation of molecular energies and energy gradients in solution by a conductor solvent model. J. Phys. Chem. A 1998, 102, 1995–2001. [Google Scholar] [CrossRef]
- Neese, F. The ORCA program system. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2012, 2, 73–78. [Google Scholar] [CrossRef]
- Neese, F. Software update: The ORCA program system, version 4.0. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2018, 8. [Google Scholar] [CrossRef]
- Martelli, T.; Ravera, E.; Louka, A.; Cerofolini, L.; Hafner, M.; Fragai, M.; Becker, C.F.W.; Luchinat, C. Atomic level quality assessment of biosilica encapsulated and autoencapsulated enzymes. Chem. Eur. J. 2016, 22, 425–432. [Google Scholar] [CrossRef]
- Bruno, F.; Francischello, R.; Bellomo, G.; Gigli, L.; Flori, A.; Menichetti, L.; Tenori, L.; Luchinat, C.; Ravera, E. Multivariate Curve Resolution for 2D Solid-State NMR spectra. Anal. Chem. 2020, 92, 4451–4458. [Google Scholar] [CrossRef]
- Fragai, M.; Luchinat, C.; Martelli, T.; Ravera, E.; Sagi, I.; Solomonov, I.; Udi, Y. SSNMR of biosilica-entrapped enzymes permits an easy assessment of preservation of native conformation in atomic detail. Chem. Commun. 2014, 50, 421–423. [Google Scholar] [CrossRef]
- Bernado, P.; Blackledge, M. Anisotropic small amplitude peptide plane dynamics in proteins from residual dipolar couplings. J. Am. Chem. Soc. 2004, 126, 4907–4920. [Google Scholar] [CrossRef]
- Schwalbe, H.; Grimshaw, S.B.; Spencer, A.; Buck, M.; Boyd, J.; Dobson, C.M.; Redfield, C.; Smith, L.J. A refined solution structure of hen lysozyme determined using residual dipolar coupling data. Protein Sci. 2001, 10, 677–688. [Google Scholar] [CrossRef] [Green Version]
- Saijo, S.; Yamada, Y.; Sato, T.; Tanaka, N.; Matsui, T.; Sazaki, G.; Nakajima, K.; Matsuura, Y. Structural consequences of hen egg-white lysozyme orthorhombic crystal growth in a high magnetic field: Validation of X-ray diffraction intensity, conformational energy searching and quantitative analysis of B factors and mosaicity. Acta Crystallogr. D 2005, 61, 207–217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schiro, A.; Carlon, A.; Parigi, G.; Murshudov, G.; Calderone, V.; Ravera, E.; Luchinat, C. On the complementarity of X-ray and NMR data. J. Struct. Biol. X 2020, 4, 100019. [Google Scholar] [CrossRef] [PubMed]
- Kroger, N.; Dickerson, M.B.; Ahmad, G.; Cai, Y.; Haluska, M.S.; Sandhage, K.H.; Poulsen, N.; Sheppard, V.C. Bioenabled synthesis of rutile (TiO2) at ambient temperature and neutral pH. Angew. Chem. Int. Ed. 2006, 45, 7239–7243. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.X.; Yu, L.; Middelberg, A.P.J. Design of low-charge peptide sequences for high-yield formation of titania nanoparticles. RSC Adv. 2012, 2, 1292–1295. [Google Scholar] [CrossRef]
- Dickerson, M.B.; Jones, S.E.; Cai, Y.; Ahmad, G.; Naik, R.R.; Kroger, N.; Sandhage, K.H. Identification and design of peptides for the rapid, high-yield formation of nanoparticulate TiO2 from aqueous solutions at room temperature. Chem. Mater. 2008, 20, 1578–1584. [Google Scholar] [CrossRef]
- Yan, Y.; Hao, B.; Wang, X.B.; Chen, G. Bio-inspired synthesis of titania with polyamine induced morphology and phase transformation at room-temperature: Insight into the role of the protonated amino group. Dalton Trans. 2013, 42, 12179–12184. [Google Scholar] [CrossRef]
- Schmidt, J.; Vogelsberger, W. Aqueous Long-Term Solubility of Titania Nanoparticles and Titanium(IV) Hydrolysis in a Sodium Chloride System Studied by Adsorptive Stripping Voltammetry. J. Solution Chem. 2009, 38, 1267–1282. [Google Scholar] [CrossRef]
- Hernandez-Gordillo, A.; Hernandez-Arana, A.; Campero-Celis, A.; Vera-Robles, L.I. TiBALDH as a precursor for biomimetic TiO2 synthesis: Stability aspects in aqueous media. RSC Adv. 2019, 9, 34559–34566. [Google Scholar] [CrossRef] [Green Version]
- Kakihana, M.; Tomita, K.; Petrykin, V.; Tada, M.; Sasaki, S.; Nakamura, Y. Chelating of titanium by lactic acid in the water-soluble diammonium tris(2-hydroxypropionato)titanate(IV). Inorg. Chem. 2004, 43, 4546–4548. [Google Scholar] [CrossRef]
- Forgacs, A.; Moldovan, K.; Herman, P.; Baranyai, E.; Fabian, I.; Lente, G.; Kalmar, J. Kinetic Model for Hydrolytic Nucleation and Growth of TiO2 Nanoparticles. J. Phys. Chem. C 2018, 122, 19161–19170. [Google Scholar] [CrossRef]
- Sbirkova-Dimitrova, H.I.; Georgieva, S.; Ganev, V.; Shivachev, B.L. Crystallization and crystal structure of lysozyme in the presence of nanosized Titanium dioxide. Bulg. Chem. Commun. 2018, 50, 7–14. [Google Scholar]
- Buettner, K.M.; Valentine, A.M. Bioinorganic chemistry of titanium. Chem. Rev. 2012, 112, 1863–1881. [Google Scholar] [CrossRef] [PubMed]
- Summers, A.Z.; Iacovella, C.R.; Cane, O.M.; Cummings, P.T.; McCabe, C. A Transferable, Multi-Resolution Coarse-Grained Model for Amorphous Silica Nanoparticles. J. Chem. Theory Comput. 2019, 15, 3260–3271. [Google Scholar] [CrossRef] [PubMed]
- Perez-Sanchez, G.; Chien, S.C.; Gomes, J.R.B.; Cordeiro, M.N.D.S.; Auerbach, S.M.; Monson, P.A.; Jorge, M. Multiscale Model for the Templated Synthesis of Mesoporous Silica: The Essential Role of Silica Oligomers. Chem. Mater. 2016, 28, 2715–2727. [Google Scholar] [CrossRef]
- Centi, A.; Jorge, M. Molecular Simulation Study of the Early Stages of Formation of Bioinspired Mesoporous Silica Materials. Langmuir 2016, 32, 7228–7240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buck, M.; Boyd, J.; Redfield, C.; MacKenzie, D.A.; Jeenes, D.J.; Archer, D.B.; Dobson, C.M. Structural Determinants of Protein Dynamics: Analysis of 15N NMR Relaxation Measurements for Main-Chain and Side-Chain Nuclei of Hen Egg White Lysozyme. Biochemistry 1995, 34, 4041–4055. [Google Scholar] [CrossRef]
- Kamatari, Y.O.; Yamada, H.; Akasaka, K.; Jones, J.A.; Dobson, C.M.; Smith, L.J. Response of Native and Denatured Hen Lysozyme to High Pressure Studied by 15N/1H NMR Spectroscopy. Eur. J. Biochem 2001, 268, 1782–1793. [Google Scholar] [CrossRef]
| Diffraction Source | BRUKER D8 Venture |
|---|---|
| Wavelength (Å) | 1.541 |
| Temperature (K) | 100 |
| Detector | PHOTON II |
| Crystal-detector distance (mm) | 50 |
| Oscillation range (°) | 0.5 |
| Total rotation range (°) | 360 |
| Exposure time/image (s) | 30 |
| Space group | P43212 |
| a, b, c (Å) | 78.1, 78.1, 37.3 |
| Mosaicity (°) | 0.3 |
| Resolution range (Å) | 50.00–1.80 (1.91–1.80) |
| Total reflections | 274,135 (24,437) |
| Unique reflections | 20,052 (2909) |
| Completeness (%) | 97.8 (89) |
| CC1/2 | 99.9 (45.1) |
| I/(σI) | 16.1 (1.7) |
| Rmerge † | 0.12 (0.91) |
| Wilson B factor (Å2) | 29.9 |
| Rcryst/Rfree ‡ (%) | 19.6/25.6 |
| Protein atoms | 1001 |
| Water molecules | 82 |
| Ligand atoms | 13 |
| RMSD bond lengths (Å) | 0.010 |
| RMSD bond angles (º) | 1.950 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gigli, L.; Ravera, E.; Calderone, V.; Luchinat, C. On the Mechanism of Bioinspired Formation of Inorganic Oxides: Structural Evidence of the Electrostatic Nature of the Interaction between a Mononuclear Inorganic Precursor and Lysozyme. Biomolecules 2021, 11, 43. https://doi.org/10.3390/biom11010043
Gigli L, Ravera E, Calderone V, Luchinat C. On the Mechanism of Bioinspired Formation of Inorganic Oxides: Structural Evidence of the Electrostatic Nature of the Interaction between a Mononuclear Inorganic Precursor and Lysozyme. Biomolecules. 2021; 11(1):43. https://doi.org/10.3390/biom11010043
Chicago/Turabian StyleGigli, Lucia, Enrico Ravera, Vito Calderone, and Claudio Luchinat. 2021. "On the Mechanism of Bioinspired Formation of Inorganic Oxides: Structural Evidence of the Electrostatic Nature of the Interaction between a Mononuclear Inorganic Precursor and Lysozyme" Biomolecules 11, no. 1: 43. https://doi.org/10.3390/biom11010043






