Characterization of Phosphorylation Status and Kinase Activity of Src Family Kinases Expressed in Cell-Based and Cell-Free Protein Expression Systems
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Plasmid Construction
2.3. Transformation of Plasmids and Protein Expression
2.4. Phos-Tag SDS-PAGE
2.5. Cell Culture and Transfection
2.6. Cell-Free Protein Expression
2.7. In Vitro Kinase Assay
3. Results
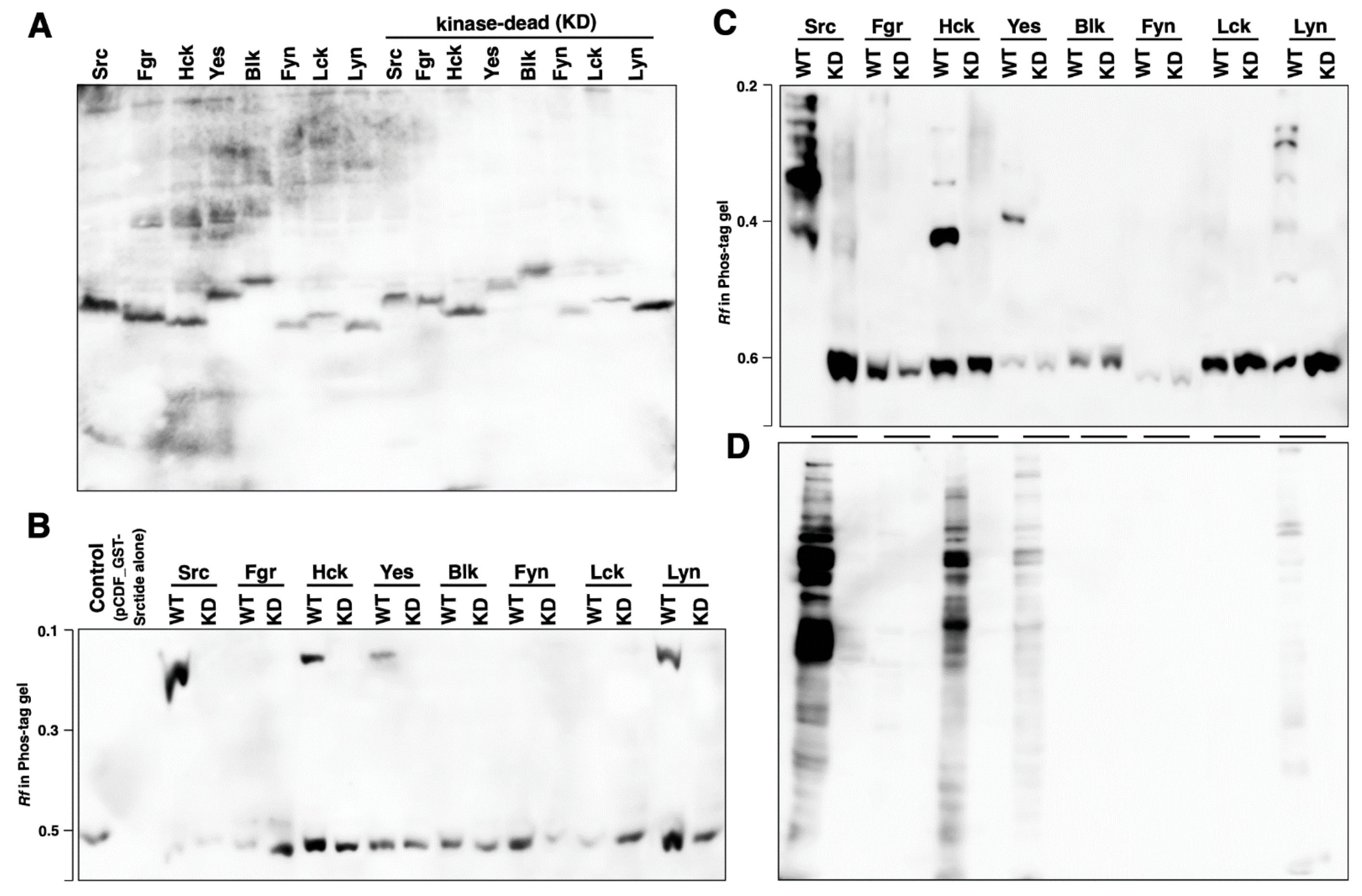
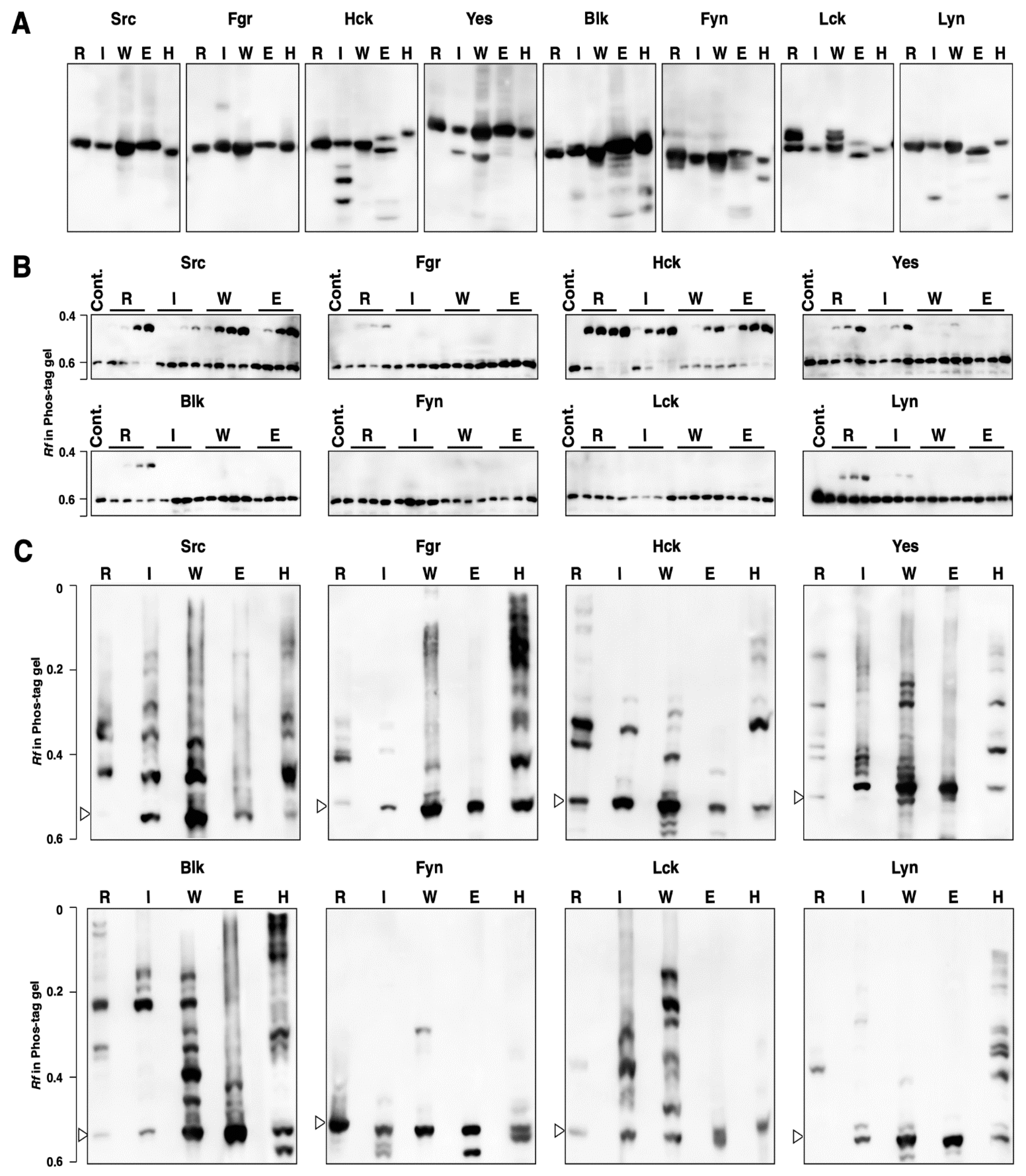
3.1. Kinase Activity and Phosphorylation Status of Human SFKs Expressed in 293 Cells
3.2. Kinase Activity and Phosphorylation Status of Human SFKs Expressed in E. coli
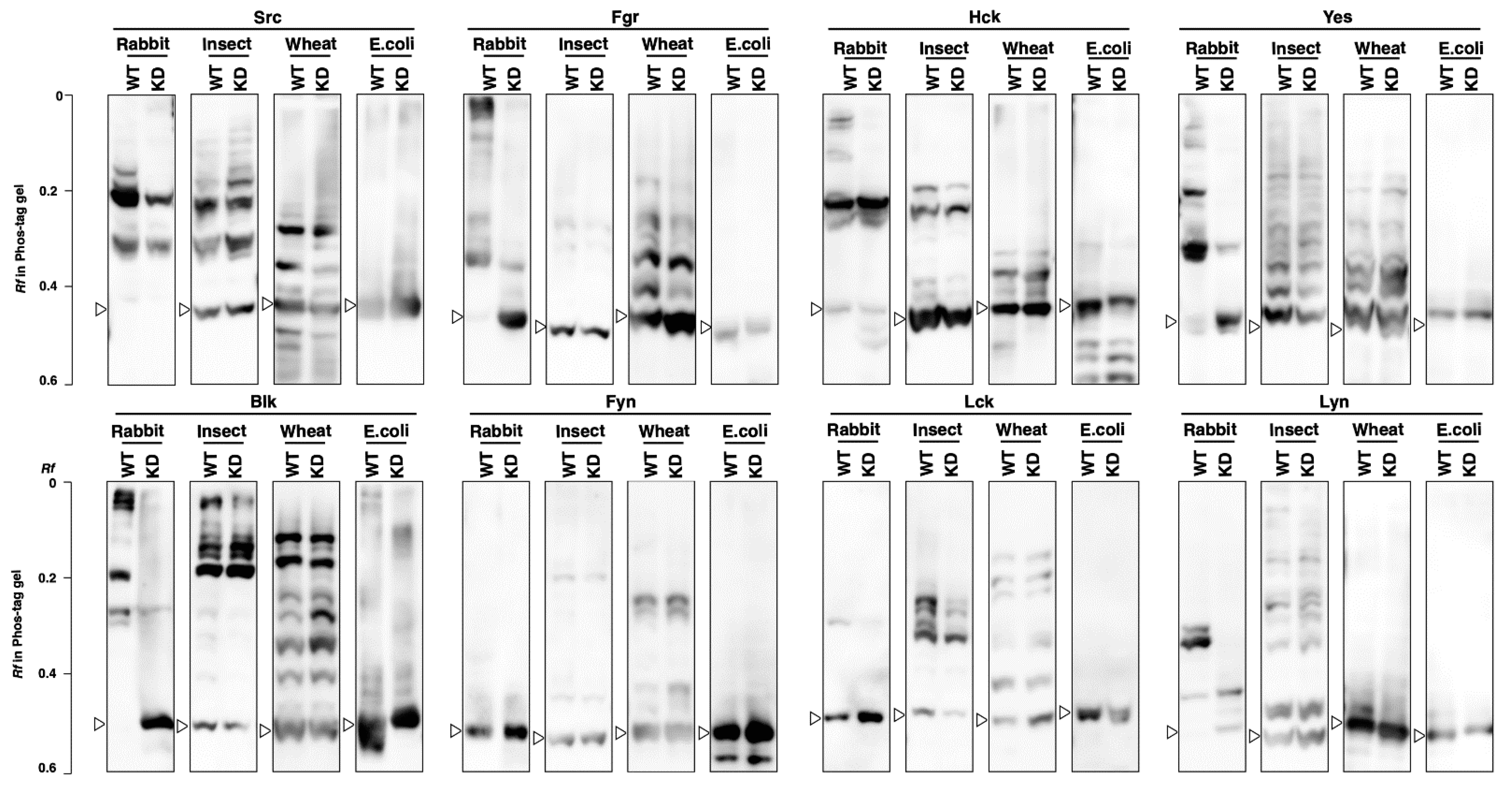
3.3. Kinase Activity and Phosphorylation Status of SFKs Produced by Cell-Free Protein Expression Systems
3.4. Phosphorylation Status of Kinase-Dead Mutants Expressed in Cell-Free Systems
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rosano, G.L.; Ceccarelli, E.A. Recombinant protein expression in Escherichia coli: Advances and challenges. Front. Microbiol. 2014, 17, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Jarvis, D.L. Baculovirus-insect cell expression systems. Methods Enzymol. 2009, 463, 191–222. [Google Scholar] [CrossRef] [PubMed]
- Cereghino, J.L.; Cregg, J.M. Heterologous protein expression in the methylotrophic yeast Pichia pastoris. FEMS Microbiol. Rev. 2000, 24, 45–66. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, N.K.; Shrivastava, A. Recent developments in bioprocessing of recombinant proteins: Expression hosts and process development. Front. Bioeng. Biotechnol. 2019, 20, 420. [Google Scholar] [CrossRef] [Green Version]
- Chiba, C.H.; Knirsch, M.C.; Azzoni, A.R.; Moreira, A.R.; Stephano, M.A. Cell-free protein synthesis: Advances on production process for biopharmaceuticals and immunobiological products. BioTechniques 2021, 70, 126–133. [Google Scholar] [CrossRef] [PubMed]
- Carlson, E.; Gan, R.; Hodgman, C.E.; Jewett, M.C. Cell-free protein synthesis: Applications come of age. Biotechnol. Adv. 2012, 30, 1185–1194. [Google Scholar] [CrossRef] [Green Version]
- Chong, S. Overview of cell-free protein synthesis historic landmarks, commercial systems, and expanding applications. Curr. Protoc. Mol. Biol. 2014, 108, 16.30. 1–16.30. 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, M. Cell-free protein synthesis: Applications in proteomics and biotechnology. New Biotechnol. 2008, 25, 126–132. [Google Scholar] [CrossRef]
- Takahashi, H.; Uematsu, A.; Yamanaka, S.; Imamura, M.; Nakajima, T.; Doi, K.; Yasuoka, S.; Takahashi, C.; Takeda, H.; Sawasaki, T. Establishment of a wheat cell-free synthesized protein array containing 250 human and mouse e3 ubiquitin ligases to identify novel interaction between e3 ligases and substrate proteins. PLoS ONE 2016, 11, e0156718. [Google Scholar] [CrossRef] [Green Version]
- Garenne, D.; Noireaux, V. Cell-free Transcription–Translation: Engineering biology from the nanometer to the millimeter scale. Curr. Opin. Biotechnol. 2019, 58, 19–27. [Google Scholar] [CrossRef]
- Pelham, H.R.B.; Jackson, R.J. An efficient mRNA-dependent translation system from reticulocyte lysates. Eur. J. Biochem. 1976, 67, 247–256. [Google Scholar] [CrossRef] [PubMed]
- Krieg, P.A.; Melton, D.A. Functional messenger RNAs are produced by SP6 in vitro transcription of cloned cDNAs. Nucleic Acids Res. 1984, 12, 7057–7070. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ezure, T.; Suzuki, T.; Higashide, S.; Shintani, E.; Endo, K.; Kobayashi, S.; Shikata, M.; Ito, M.; Tanimizu, K.; Nishimura, O. Cell-free protein synthesis system prepared from insect cells by freeze-thawing. Biotechnol. Prog. 2006, 22, 1570–1577. [Google Scholar] [CrossRef] [PubMed]
- Zubay, G. In vitro synthesis of protein in microbial systems. Annu. Rev. Genet. 1973, 7, 267–287. [Google Scholar] [CrossRef] [PubMed]
- Zubay, G. The isolation and properties of CAP, the catabolite gene activator. Methods Enzymol. 1980, 65, 856–877. [Google Scholar] [CrossRef]
- Lesley, S.A.; Brow, M.A.; Burgess, R.R. Use of in vitro protein synthesis from polymerase chain reaction-generated templates to study interaction of Escherichia coli transcription factors with core RNA polymerase and for epitope mapping of monoclonal antibodies. J. Biol. Chem. 1991, 266, 2632–2638. [Google Scholar] [CrossRef]
- Thomas, S.M.; Brugge, J.S. Cellular functions regulated by Src family kinases. Annu. Rev. Cell Dev. Biol. 1997, 13, 513–609. [Google Scholar] [CrossRef] [Green Version]
- Kinoshita, E.; Kinoshita-Kikuta, E.; Takiyama, K.; Koike, T. Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Mol. Cell. Proteom. 2006, 5, 749–757. [Google Scholar] [CrossRef] [Green Version]
- Kinoshita-Kikuta, E.; Aoki, Y.; Kinoshita, E.; Koike, T. Label-free kinase profiling using phosphate affinity polyacrylamide gel electrophoresis. Mol. Cell. Proteom. 2007, 6, 356–366. [Google Scholar] [CrossRef] [Green Version]
- Kinoshita, E.; Kinoshita-Kikuta, E.; Koike, T. Separation and detection of large phosphoproteins using phos-tag SDS-PAGE. Nat. Protoc. 2009, 4, 1513–1521. [Google Scholar] [CrossRef]
- Kinoshita, E.; Kinoshita-Kikuta, E. Improved phos-tag SDS-PAGE under neutral ph conditions for advanced protein phosphorylation profiling. Proteomics 2011, 11, 319–323. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, E.; Kinoshita-Kikuta, E.; Koike, T. Phos-tag SDS-PAGE systems for phosphorylation profiling of proteins with a wide range of molecular masses under neutral pH conditions. Proteomics 2012, 12, 192–202. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita-Kikuta, E.; Utsumi, T.; Miyazaki, A.; Tokumoto, C.; Doi, K.; Harada, H.; Kinoshita, E.; Koike, T. Protein-N-myristoylation-dependent phosphorylation of serine 13 of tyrosine kinase lyn by casein kinase 1γ at the golgi during intracellular protein traffic. Sci. Rep. 2020, 10, 16273. [Google Scholar] [CrossRef]
- Kinoshita-Kikuta, E.; Yoshimoto, M.; Yano, M.; Kinoshita, E.; Koike, T. An assay of human tyrosine protein kinase activities by using an Escherichia coli protein-expression system. BioTechniques 2021, 70, 209–217. [Google Scholar] [CrossRef]
- Johnson, T.M.; Perich, J.W.; Bjorge, J.D.; Fujita, D.J.; Cheng, H.C. Common and differential recognition of structural features in synthetic peptides by the catalytic domain and the Src-homology 2 (SH2) domain of pp60c-src. J. Pept. Res. 1997, 50, 365–371. [Google Scholar] [CrossRef]
- Kinoshita-Kikuta, E.; Kinoshita, E.; Matsuda, A.; Koike, T. Tips on improving the efficiency of electrotransfer of target proteins from Phos-tag SDS-PAGE gel. Proteomics 2014, 14, 2437–2442. [Google Scholar] [CrossRef]
- Hata, A.; Sabe, H.; Kurosaki, T.; Takata, M.; Hanafusa, H. Functional analysis of Csk in signal transduction through the B-Cell antigen receptor. Mol. Cell. Biol. 1994, 14, 7306–7313. [Google Scholar] [CrossRef] [Green Version]
- Palacious, E.H.; Weiss, A. Function of the Src-family kinases, Lck and Fyn, in T-cell development and activation. Oncogene 2004, 23, 7990–8000. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seeliger, M.A.; Young, M.; Henderson, M.N.; Pellicena, P.; King, D.S.; Falick, A.M.; Kuriyan, J. High yield bacterial expression of active c-Abl and c-Src tyrosine kinases. Protein Sci. 2005, 14, 3135–3139. [Google Scholar] [CrossRef] [Green Version]
- Díaz Galicia, M.E.; Aldehaiman, A.; Hong, S.B.; Arold, S.T.; Grünberg, R. Methods for the recombinant expression of active tyrosine kinase domains: Guidelines and pitfalls. Methods Enzymol. 2019, 62, 1131–1152. [Google Scholar] [CrossRef] [Green Version]
- Hancock, J.F. Reticulocyte lysate assay for in vitro translation and posttranslational modification of ras proteins. Methods Enzymol. 1995, 255, 60–65. [Google Scholar] [CrossRef]
- Suzuki, T.; Ito, M.; Ezure, T.; Shikata, M.; Ando, E.; Utsumi, T.; Tsunasawa, S.; Nishimura, O. Protein prenylation in an insect cell-free protein synthesis system and identification of products by mass spectrometry. Proteomics 2007, 7, 1942–1950. [Google Scholar] [CrossRef]
- Gibbs, P.E.M.; Zouzias, D.C.; Freedberg, I.M. Differential post-translational modification of human type-I deratins synthesized in a rabbit reticulocyte cell-free system. Biochim. Biophys. Acta 1985, 824, 247–255. [Google Scholar] [CrossRef]
- Suzuki, T.; Ito, M.; Ezure, T.; Shikata, M.; Ando, E.; Utsumi, T.; Tsunasawa, S.; Nishimura, O. N-terminal protein modifications in an insect cell-free protein synthesis system and their identification by mass spectrometry. Proteomics 2006, 6, 4486–4495. [Google Scholar] [CrossRef]
- Safer, B.; Jagus, R. Control of eIF-2 phosphatase activity in rabbit reticulocyte lysate. Proc. Natl. Acad. Sci. USA 1979, 76, 1094–1098. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, T.; Ezure, T.; Ando, E.; Nishimura, O.; Utsumi, T.; Tsunasawa, S. Preparation of ubiquitin-conjugated proteins using an insect cell-free protein synthesis system. J. Biotechnol. 2010, 145, 73–78. [Google Scholar] [CrossRef]
- Shields, D.; Blobel, G. Efficient cleavage and segregation of nascent presecretory proteins in a reticulocyte lysate supplemented with microsomal membranes. J. Biol. Chem. 1978, 253, 3753–3756. [Google Scholar] [CrossRef]
- Tarui, H.; Murata, M.; Tani, I.; Imanishi, S.; Nishikawa, S.; Hara, T. Establishment and characterization of cell-free translation/glycosylation in insect cell (Spodoptera frugiperda 21) extract prepared with high pressure treatment. Appl. Microbiol. Biotechnol. 2001, 55, 446–453. [Google Scholar] [CrossRef] [PubMed]
- Sawasaki, T.; Ogasawara, T.; Morishita, R.; Endo, Y. A Cell-free protein synthesis system for high-throughput proteomics. Proc. Natl. Acad. Sci. USA 2002, 99, 14652–14657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
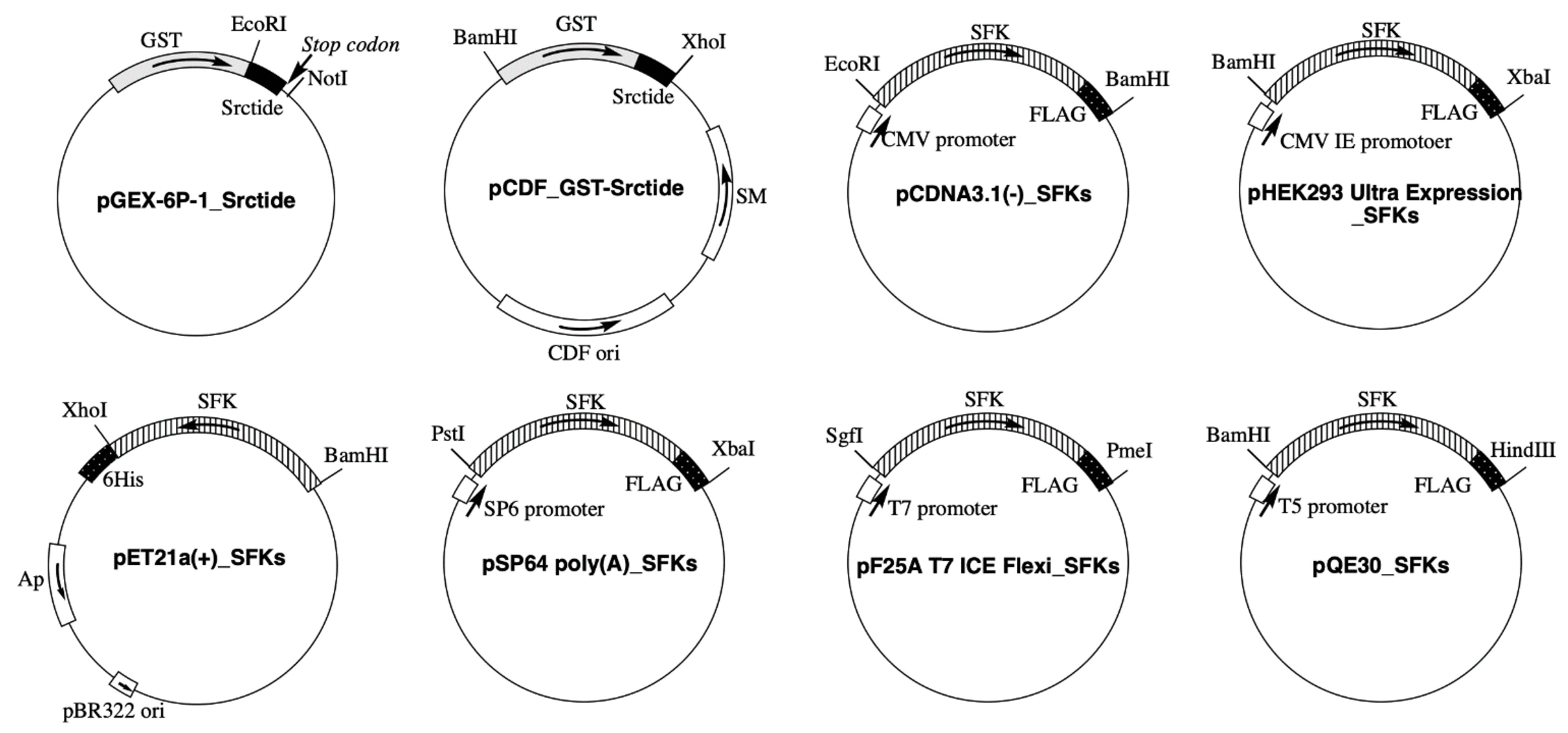

| SFK | UniProt Accession No. | Expression Vector | ||||
|---|---|---|---|---|---|---|
| E. coli BL21(DE3) | 293 Cells | For Cell-Free Protein Expression System | ||||
| Rabbit Reticulocytes, Wheat Germ | Insect Cells | E. coli | ||||
| Src | P12931 | pET21a(+) | pcDNA3.1(−) | pSP64 poly(A) Vector | pF25A ICE T7 Flexi Vector | pQE30 |
| Lck | P06239 | |||||
| Hck | P08631 | |||||
| Blk | P51451 | |||||
| Yes | P07947 | pHEK293 Ultra Expression Vector I | ||||
| Fyn | P06241 | |||||
| Lyn | P07948 | |||||
| Fgr | P09769 | |||||
| SFK | 293 Cells | E. coli BL21(DE3) | Cell-Free Protein Expression System | |||
|---|---|---|---|---|---|---|
| Rabbit Reticulocytes | Insect Cells | Wheat Germ | E. coli | |||
| Src | + | + | + | + | + | + |
| Fgr | − | − | + | − | − | − |
| Hck | + | + | + | + | + | + |
| Yes | − | + | + | + | + | − |
| Blk | − | − | + | + | − | − |
| Fyn | − | − | − | − | − | − |
| Lck | − | − | − | − | − | − |
| Lyn | + | + | + | + | − | − |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kinoshita-Kikuta, E.; Kinoshita, E.; Suga, M.; Higashida, M.; Yamane, Y.; Nakamura, T.; Koike, T. Characterization of Phosphorylation Status and Kinase Activity of Src Family Kinases Expressed in Cell-Based and Cell-Free Protein Expression Systems. Biomolecules 2021, 11, 1448. https://doi.org/10.3390/biom11101448
Kinoshita-Kikuta E, Kinoshita E, Suga M, Higashida M, Yamane Y, Nakamura T, Koike T. Characterization of Phosphorylation Status and Kinase Activity of Src Family Kinases Expressed in Cell-Based and Cell-Free Protein Expression Systems. Biomolecules. 2021; 11(10):1448. https://doi.org/10.3390/biom11101448
Chicago/Turabian StyleKinoshita-Kikuta, Emiko, Eiji Kinoshita, Misaki Suga, Mana Higashida, Yuka Yamane, Tomoka Nakamura, and Tohru Koike. 2021. "Characterization of Phosphorylation Status and Kinase Activity of Src Family Kinases Expressed in Cell-Based and Cell-Free Protein Expression Systems" Biomolecules 11, no. 10: 1448. https://doi.org/10.3390/biom11101448







