Revisiting Jatropha curcas Monomeric Esterase: A Dienelactone Hydrolase Compatible with the Electrostatic Catapult Model
Abstract
1. Introduction
2. Materials and Methods
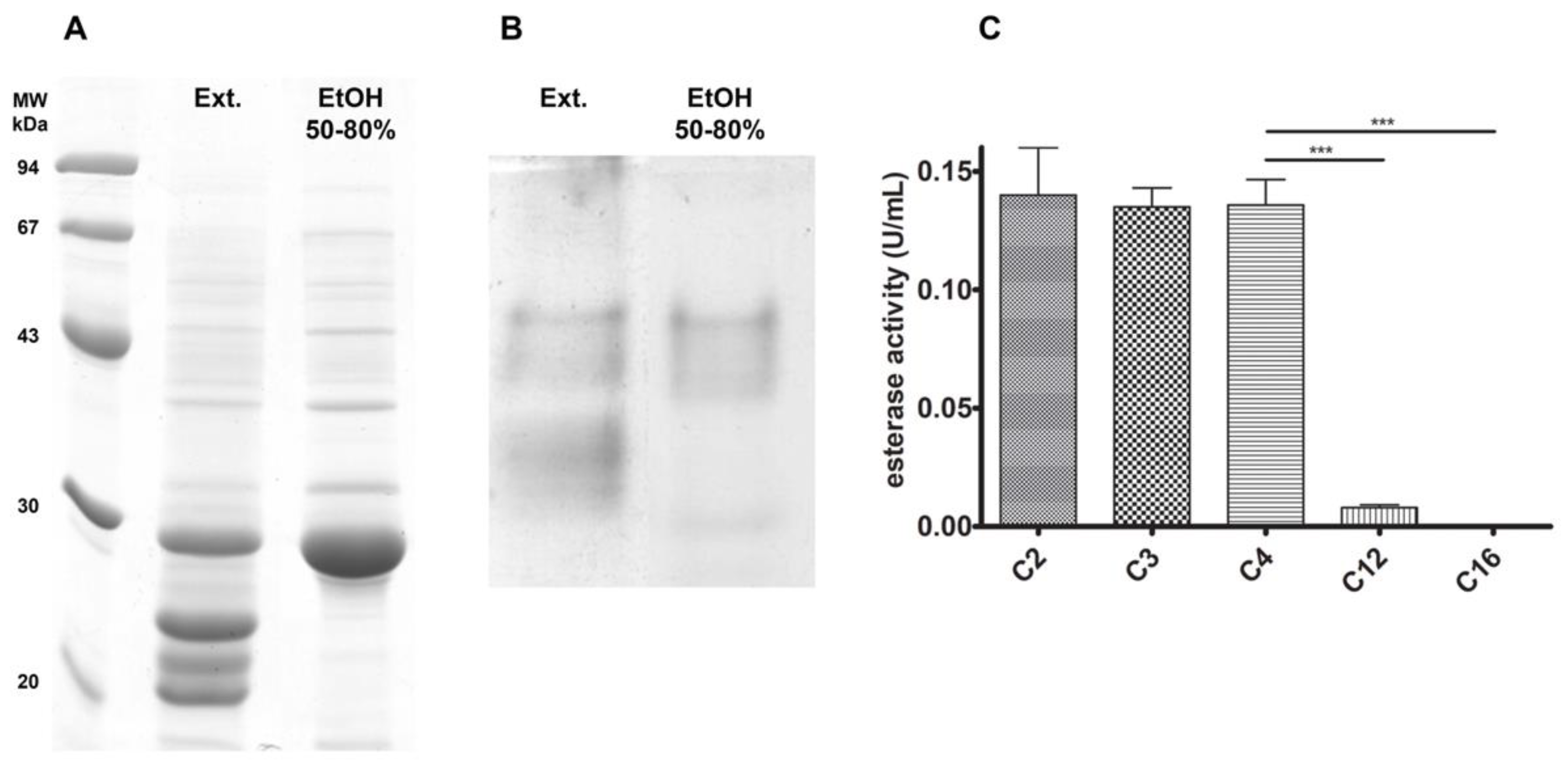
2.1. Crude Enzyme Extract Preparation and Ethanol Precipitation
2.2. Anion Exchange Chromatography
2.3. Enzyme Activity Assays
2.3.1. Esterase
2.3.2. Peptidase
2.4. Polyacrylamide Gel Electrophoresis (PAGE)
2.4.1. Unidimensional SDS-PAGE
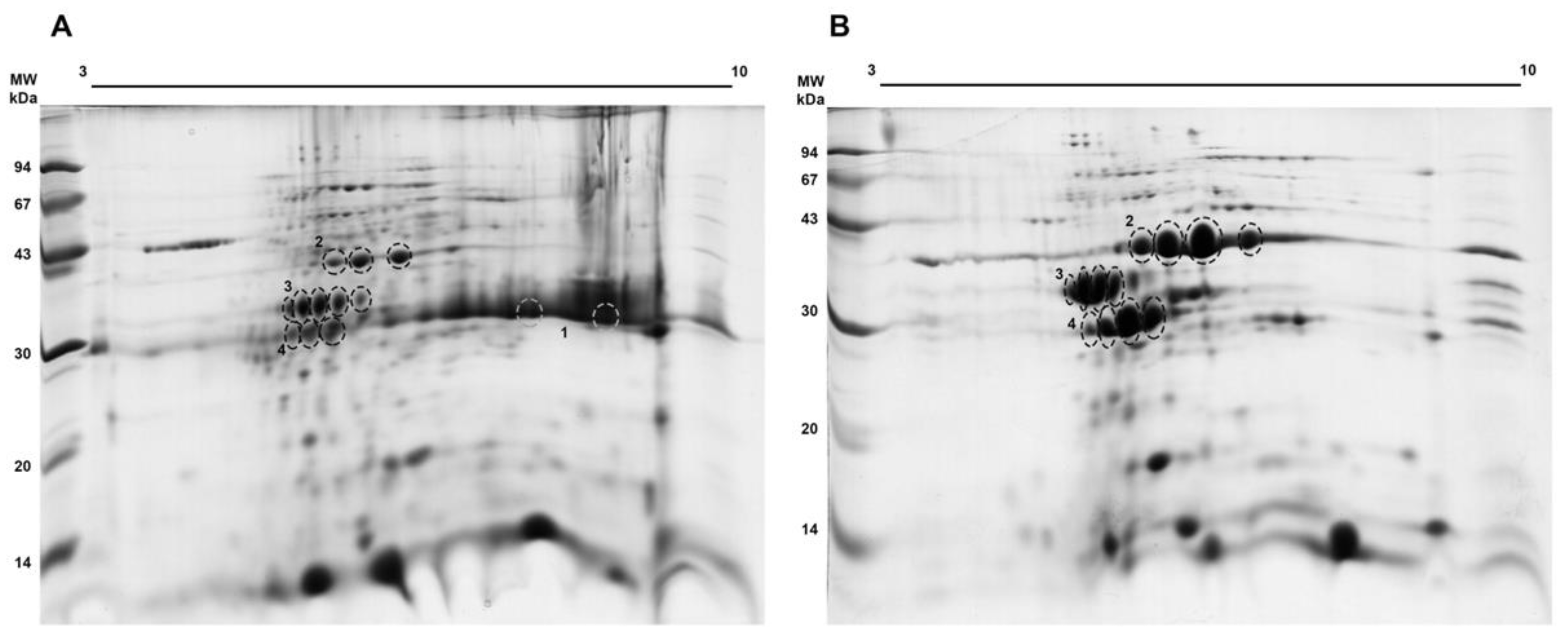
2.4.2. 2D SDS-PAGE
2.4.3. Esterase Zymography
2.5. Protein Digestion, Peptide Extraction, and Mass Spectrometry Analysis
2.6. Hydrolysis of Racemic 1,2-O-Isopropylidene Glycerol (IPG) Ester and Diethyl Phenylmalonate
2.7. Statistical Analysis
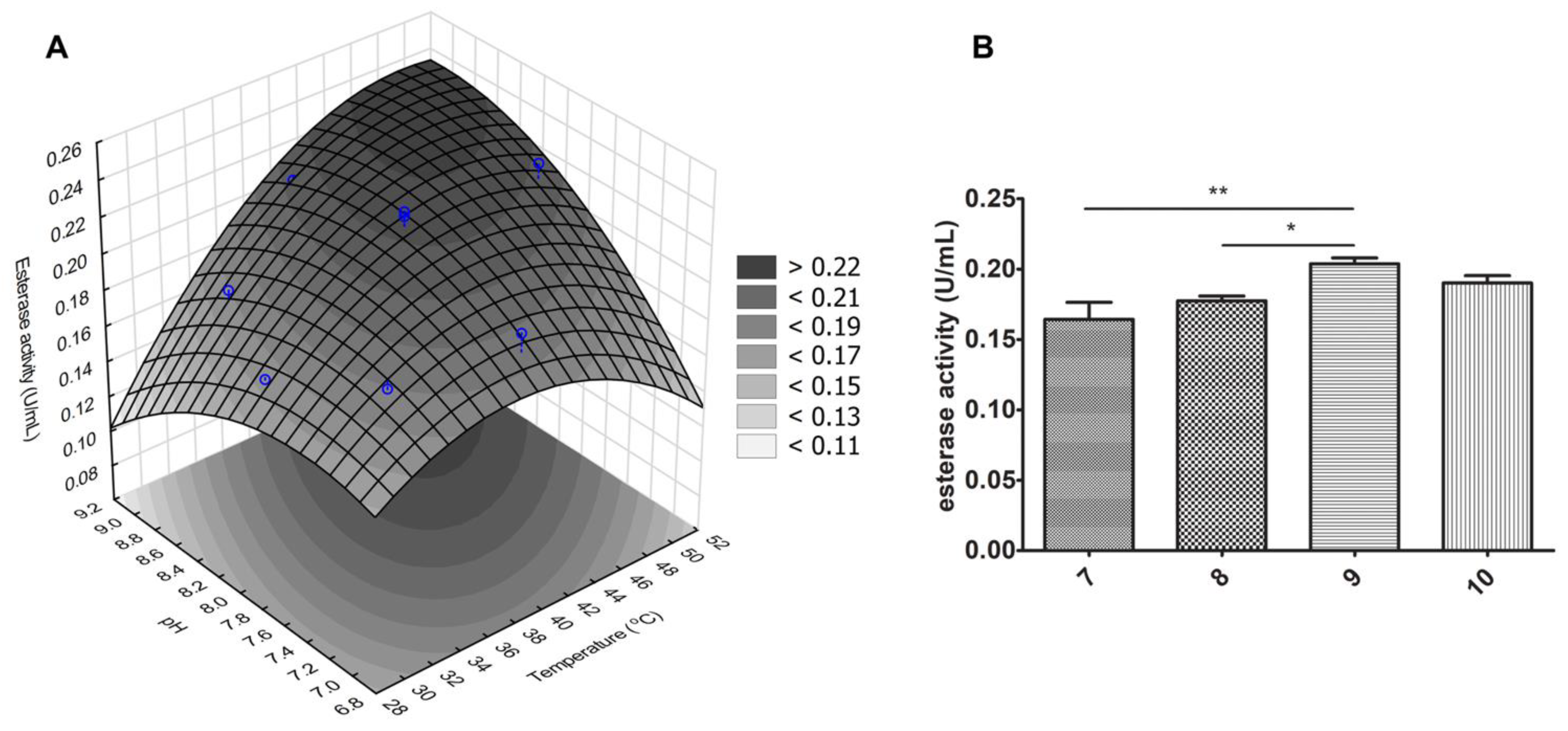
2.8. Central Composite Rotational Design (CCRD)
2.9. Comparative Modeling and Structural Analyses of J. curcas Esterase
3. Results
3.1. Initial Processing and Chain-Length Specificity
3.2. J. curcas L. Esterase B Has an Acidic Isoelectric Point and Belongs to the Dienelactone Hydrolase (DLH) Family
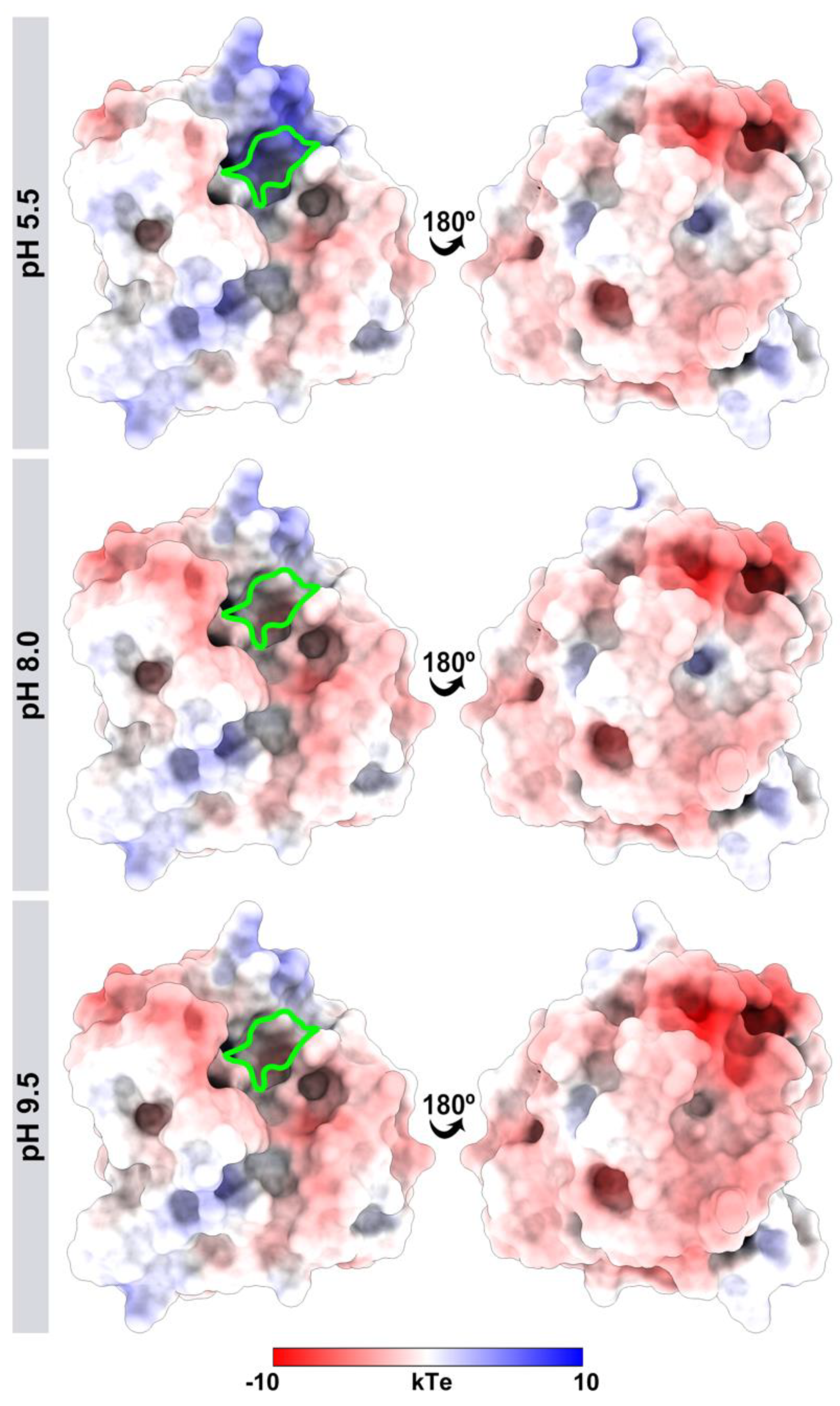
3.3. J. curcas L. Esterase B Activity Increases in Basic pH, Corroborating “the Electrostatic Catapult” Model
3.4. Different pH Values Alter the Electrostatic Potential of the J. curcas Esterase B Catalytic Site
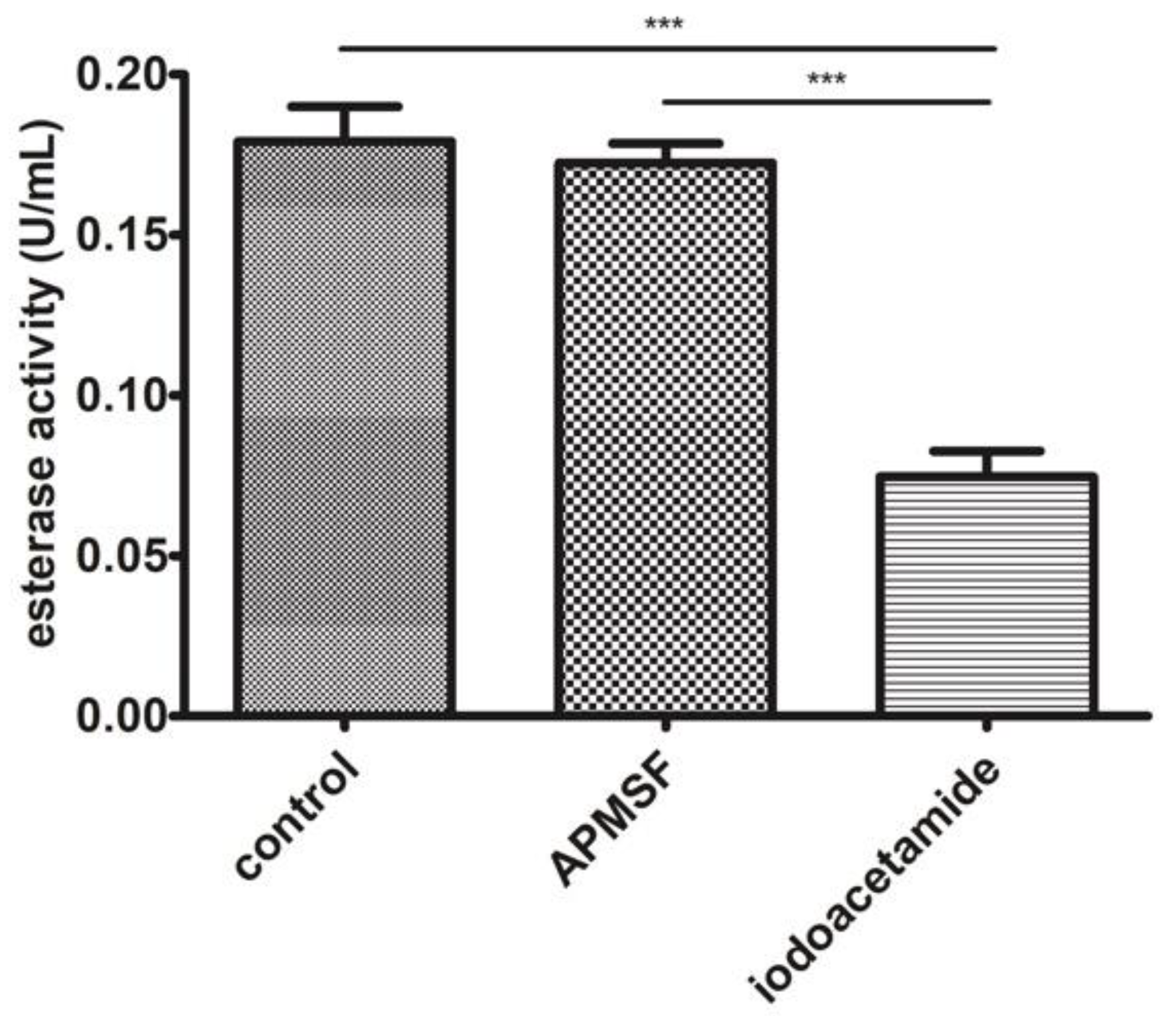
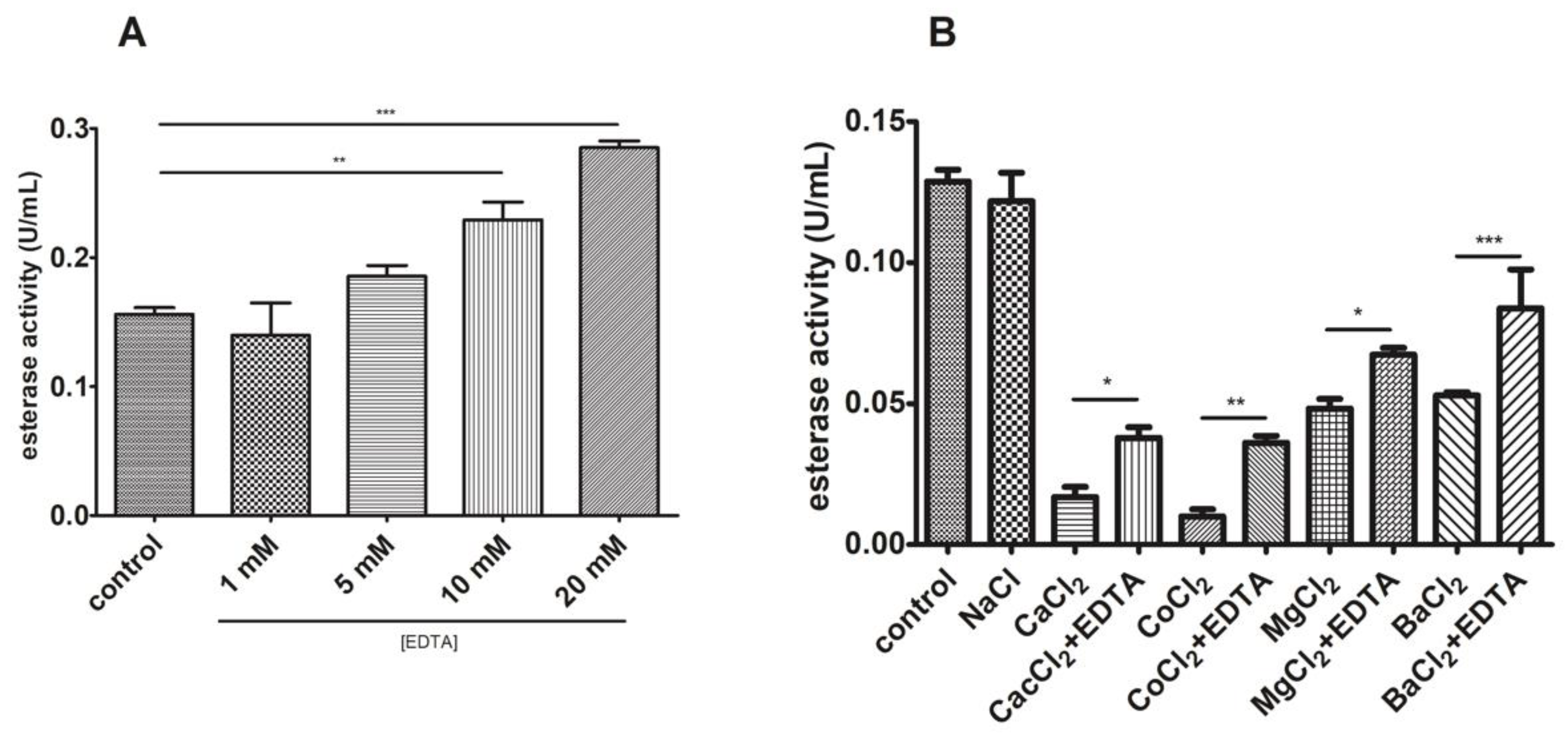
3.5. J. curcas L. Esterase B Has No Proteolytic Activity and Divalent Ions Inhibit the Enzyme
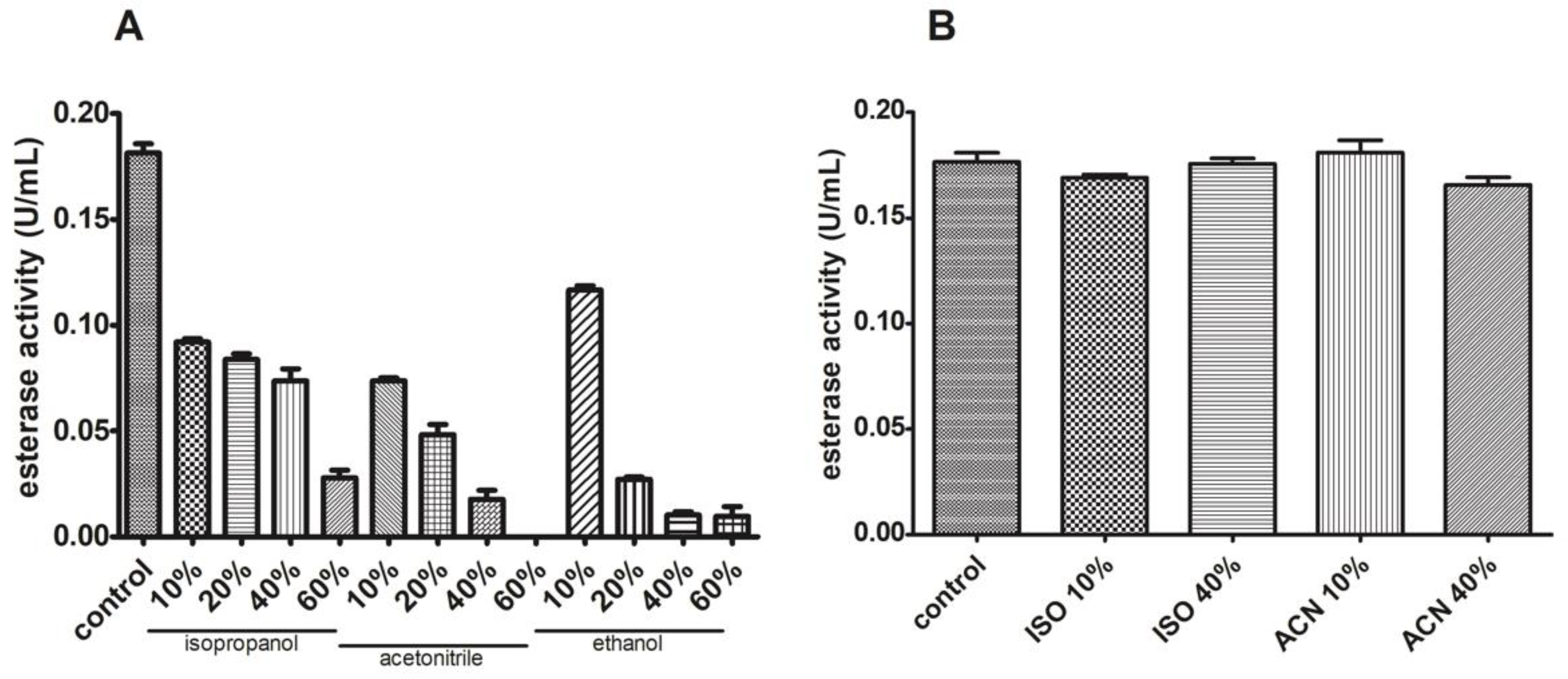
3.6. J. curcas L. Esterase B Has Low Activity in the Presence of Different Solvents
3.7. J. curcas L Esterase B Shows High Hydrolysis Rates but No Enantiospecificity/Selectivity towards Tested Chiral and Prochiral Substrates
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Maghuly, F.; Laimer, M. Jatropha curcas, a Biofuel Crop: Functional Genomics for Understanding Metabolic Pathways and Genetic Improvement. Biotechnol. J. 2013, 8, 1172–1182. [Google Scholar] [CrossRef]
- Ewunie, G.A.; Morken, J.; Lekang, O.I.; Yigezu, Z.D. Factors Affecting the Potential of Jatropha Curcas for Sustainable Biodiesel Production: A Critical Review. Renew. Sustain. Energy Rev. 2021, 137, 110500. [Google Scholar] [CrossRef]
- Abobatta, W.F. Jatropha curcas, a Novel Crop for Developing the Marginal Lands. In Biofuels and Biodiesel, 1st ed.; Basu, C., Ed.; Springer Nature: New York, NY, USA; Humana Press: New York, NY, USA, 2021; pp. 79–100. [Google Scholar]
- Halmemies-Beauchet-Filleau, A.; Rinne, M.; Lamminen, M.; Mapato, C.; Ampapon, T.; Wanapat, M.; Vanhatalo, A. Review: Alternative and Novel Feeds for Ruminants: Nutritive Value, Product Quality and Environmental Aspects. Animal 2018, 12, s295–s309. [Google Scholar] [CrossRef] [PubMed]
- Gomes, T.G.; Hadi, S.I.I.A.; Costa Alves, G.S.; Mendonça, S.; de Siqueira, F.G.; Miller, R.N.G. Current Strategies for the Detoxification of Jatropha curcas Seed Cake: A Review. J. Agric. Food Chem. 2018, 66, 2510–2522. [Google Scholar] [CrossRef] [PubMed]
- de Sousa, J.S.; Cavalcanti-Oliveira, E.d.; Aranda, D.A.G.; Freire, D.M.G. Application of Lipase from the Physic Nut (Jatropha curcas L.) to a New Hybrid (Enzyme/Chemical) Hydroesterification Process for Biodiesel Production. J. Mol. Catal. B Enzym. 2010, 65, 133–137. [Google Scholar] [CrossRef]
- León-Villanueva, A.; Huerta-Ocampo, J.A.; Barrera-Pacheco, A.; Medina-Godoy, S.; Barba de la Rosa, A.P. Proteomic Analysis of Non-Toxic Jatropha Curcas Byproduct Cake: Fractionation and Identification of the Major Components. Ind. Crop. Prod. 2018, 111, 694–704. [Google Scholar] [CrossRef]
- Yano, H.; Kuroda, M. Disulfide Proteome Yields a Detailed Understanding of Redox Regulations: A Model Study of Thioredoxin-Linked Reactions in Seed Germination. Proteomics 2006, 6, 294–300. [Google Scholar] [CrossRef]
- Urs, A.P.; Rudresha, G.V.; Manjuprasanna, V.N.; Suvilesh, K.N.; Gowda, M.D.M.; Yariswamy, M.; Hiremath, V.; Ramakrishnan, C.; Savitha, M.N.; Jayachandra, K.; et al. Plant Latex Thrombin-like Cysteine Proteases Alleviates Bleeding by Bypassing Factor VIII in Murine Model. J. Cell. Biochem. 2019, 120, 12843–12858. [Google Scholar] [CrossRef]
- Staubmann, R.; Ncube, I.; Gübitz, G.M.; Steiner, W.; Read, J.S. Esterase and Lipase Activity in Jatropha curcas L. Seeds. J. Biotechnol. 1999, 75, 117–126. [Google Scholar] [CrossRef]
- Subramani, T.; Chandrashekharaiah, K.S.; Swamy, N.R.; Murthy, K.R.S. Purification and Characterization of Carboxylesterase from the Seeds of Jatropha Curcas. Protein J. 2012, 31, 120–128. [Google Scholar] [CrossRef]
- Nanssou Kouteu, P.A.; Baréa, B.; Barouh, N.; Blin, J.; Villeneuve, P. Lipase Activity of Tropical Oilseed Plants for Ethyl Biodiesel Synthesis and Their Typo- and Regioselectivity. J. Agric. Food Chem. 2016, 64, 8838–8847. [Google Scholar] [CrossRef]
- Chen, Y.; Black, D.S.; Reilly, P.J. Carboxylic Ester Hydrolases: Classification and Database Derived from Their Primary, Secondary, and Tertiary Structures. Protein Sci. 2016, 25, 1942–1953. [Google Scholar] [CrossRef] [PubMed]
- Casas-Godoy, L.; Gasteazoro, F.; Duquesne, S.; Bordes, F.; Marty, A.; Sandoval, G. Lipases: An Overview. In Lipases and Phospholipases, 2nd ed.; Sandoval, G., Ed.; Springer Nature: New York, NY, USA; Humana Press: New York, NY, USA, 2018; pp. 3–38. [Google Scholar]
- Bracco, P.; van Midden, N.; Arango, E.; Torrelo, G.; Ferrario, V.; Gardossi, L.; Hanefeld, U. Bacillus Subtilis Lipase A—Lipase or Esterase? Catalysts 2020, 10, 308. [Google Scholar] [CrossRef]
- Carvalho, A.; Fonseca, T.; Mattos, M.; Oliveira, M.; Lemos, T.; Molinari, F.; Romano, D.; Serra, I. Recent Advances in Lipase-Mediated Preparation of Pharmaceuticals and Their Intermediates. Int. J. Mol. Sci. 2015, 16, 29682–29716. [Google Scholar] [CrossRef] [PubMed]
- Neves Petersen, M.T.; Fojan, P.; Petersen, S.B. How Do Lipases and Esterases Work: The Electrostatic Contribution. J. Biotechnol. 2001, 85, 115–147. [Google Scholar] [CrossRef]
- Neves-Petersen, M.T.; Petersen, E.I.; Fojan, P.; Noronha, M.; Madsen, R.G.; Petersen, S.B. Engineering the PH-Optimum of a Triglyceride Lipase: From Predictions Based on Electrostatic Computations to Experimental Results. J. Biotechnol. 2001, 87, 225–254. [Google Scholar] [CrossRef]
- Moura-da-Silva, A.M.; Baldo, C. Jararhagin, a Hemorrhagic Snake Venom Metalloproteinase from Bothrops Jararaca. Toxicon 2012, 60, 280–289. [Google Scholar] [CrossRef] [PubMed]
- LAEMMLI, U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Rabilloud, T.; Charmont, S. Proteome Research: Two-Dimensional Gel Electrophoresis and Identification Methods; Rabilloud, T., Ed.; Springer: Berlin/Heidelberg, Germany, 2000; ISBN 978-3-540-65792-7. [Google Scholar]
- Shevchenko, A.; Wilm, M.; Vorm, O.; Mann, M. Mass Spectrometric Sequencing of Proteins from Silver-Stained Polyacrylamide Gels. Anal. Chem. 1996, 68, 850–858. [Google Scholar] [CrossRef] [PubMed]
- Gharahdaghi, F.; Weinberg, C.R.; Meagher, D.A.; Imai, B.S.; Mische, S.M. Mass Spectrometric Identification of Proteins from Silver-Stained Polyacrylamide Gel: A Method for the Removal of Silver Ions to Enhance Sensitivity. Electrophoresis 1999, 20, 601–605. [Google Scholar] [CrossRef]
- Machado, A.C.O.; da Silva, A.A.T.; Borges, C.P.; Simas, A.B.C.; Freire, D.M.G. Kinetic Resolution of (R,S)-1,2-Isopropylidene Glycerol (Solketal) Ester Derivatives by Lipases. J. Mol. Catal. B Enzym. 2011, 69, 42–46. [Google Scholar] [CrossRef][Green Version]
- Manoel, E.A.; Ribeiro, M.F.P.; dos Santos, J.C.S.; Coelho, M.A.Z.; Simas, A.B.C.; Fernandez-Lafuente, R.; Freire, D.M.G. Accurel MP 1000 as a Support for the Immobilization of Lipase from Burkholderia cepacia: Application to the Kinetic Resolution of myo-Inositol Derivatives. Process Biochem. 2015, 50, 1557–1564. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology Modelling of Protein Structures and Complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Feig, M. Local Protein Structure Refinement via Molecular Dynamics Simulations with LocPREFMD. J. Chem. Inf. Model. 2016, 56, 1304–1312. [Google Scholar] [CrossRef]
- Bond, C.S.; Schüttelkopf, A.W. ALINE: A WYSIWYG Protein-Sequence Alignment Editor for Publication-Quality Alignments. Acta Crystallogr. Sect. D Biol. Crystallogr. 2009, 65, 510–512. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Goddard, T.D.; Huang, C.C.; Meng, E.C.; Pettersen, E.F.; Couch, G.S.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Meeting Modern Challenges in Visualization and Analysis. Protein Sci. 2018, 27, 14–25. [Google Scholar] [CrossRef] [PubMed]
- Baker, N.A.; Sept, D.; Joseph, S.; Holst, M.J.; McCammon, J.A. Electrostatics of Nanosystems: Application to Microtubules and the Ribosome. Proc. Natl. Acad. Sci. USA 2001, 98, 10037–10041. [Google Scholar] [CrossRef] [PubMed]
- Dolinsky, T.J.; Nielsen, J.E.; McCammon, J.A.; Baker, N.A. PDB2PQR: An Automated Pipeline for the Setup of Poisson-Boltzmann Electrostatics Calculations. Nucleic Acids Res. 2004, 32, W665–W667. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yang, Q.; Li, C.; Ding, M.; Lv, X.; Tao, C.; Yu, H.; Chen, F.; Xu, Y. Curcin C, a Novel Type I Ribosome-Inactivating Protein from the Post-Germinating Cotyledons of Jatropha curcas. Amino Acids 2017, 49, 1619–1631. [Google Scholar] [CrossRef] [PubMed]
- Pathak, D.; Ollis, D. Refined Structure of Dienelactone Hydrolase at 1.8A°. J. Mol. Biol. 1990, 214, 497–525. [Google Scholar] [CrossRef]
- Porter, J.L.; Carr, P.D.; Collyer, C.A.; Ollis, D.L. Crystallization of Dienelactone Hydrolase in Two Space Groups: Structural Changes Caused by Crystal Packing. Acta Crystallogr. Sect. F Struct. Biol. Commun. 2014, 70, 884–889. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Pillay, B.; Olaniran, A.O. Two Structurally Different Dienelactone Hydrolases (TfdEI and TfdEII) from Cupriavidus Necator JMP134 Plasmid PJP4 Catalyse Cis- and Trans-Dienelactones with Similar Efficiency. PLoS ONE 2014, 9, e101801. [Google Scholar] [CrossRef] [PubMed]
- Holmquist, M. Alpha Beta-Hydrolase Fold Enzymes Structures, Functions and Mechanisms. Curr. Protein Pept. Sci. 2000, 1, 209–235. [Google Scholar] [CrossRef]
- Walker, I.; Hennessy, J.E.; Ollis, D.L.; Easton, C.J. Substrate-Induced Conformational Change and Isomerase Activity of Dienelactone Hydrolase and Its Site-Specific Mutants. ChemBioChem 2012, 13, 1645–1651. [Google Scholar] [CrossRef] [PubMed]
- Fahmy, A.S.; Abo-Zeid, A.Z.; Mohamed, T.M.; Ghanem, H.M.; Borai, I.H.; Mohamed, S.A. Characterization of Esterases from Cucurbita pepo Cv. “Eskandrani.”. Bioresour. Technol. 2008, 99, 437–443. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, M.A.; Mohamed, T.M.; Mohamed, S.A.; Fahmy, A.S. Distribution of Lipases in the Gramineae. Partial Purification and Characterization of Esterase from Avena Fatua. Bioresour. Technol. 2000, 73, 227–234. [Google Scholar] [CrossRef]
- Hedstrom, L. Serine Protease Mechanism and Specificity. Chem. Rev. 2002, 102, 4501–4524. [Google Scholar] [CrossRef]
- Seth, S.; Chakravorty, D.; Dubey, V.K.; Patra, S. An Insight into Plant Lipase Research—Challenges Encountered. Protein Expr. Purif. 2014, 95, 13–21. [Google Scholar] [CrossRef]
- Hou, C.; He, K.; Yang, L.; Huo, D.; Yang, M.; Huang, S.; Zhang, L.; Shen, C. Catalytic Characteristics of Plant-Esterase from Wheat Flour. World J. Microbiol. Biotechnol. 2012, 28, 541–548. [Google Scholar] [CrossRef]
- Eron, S.J.; MacPherson, D.J.; Dagbay, K.B.; Hardy, J.A. Multiple Mechanisms of Zinc-Mediated Inhibition for the Apoptotic Caspases-3, -6, -7, and -8. ACS Chem. Biol. 2018, 13, 1279–1290. [Google Scholar] [CrossRef]
- Bowles, I.E.; Pool, E.H.; Lancaster, B.S.; Lawson, E.K.; Savas, C.P.; Kartje, Z.J.; Severinac, L.; Cho, D.H.; Macbeth, M.R.; Johnson, R.J.; et al. Transition Metal Cation Inhibition of Mycobacterium tuberculosis Esterase RV0045C. Protein Sci. 2021, 30, 1554–1565. [Google Scholar] [CrossRef] [PubMed]
- Hertadi, R.; Widhyastuti, H. Effect of Ca2+ Ion to the Activity and Stability of Lipase Isolated from Chromohalobacter japonicus BK-AB18. Procedia Chem. 2015, 16, 306–313. [Google Scholar] [CrossRef]
- García-Cano, I.; Rocha-Mendoza, D.; Kosmerl, E.; Jiménez-Flores, R. Purification and Characterization of a Phospholipid-Hydrolyzing Phosphoesterase Produced by Pediococcus acidilactici Isolated from Gouda Cheese. J. Dairy Sci. 2020, 103, 3912–3923. [Google Scholar] [CrossRef] [PubMed]





| Factor | Level | ||||
|---|---|---|---|---|---|
| −1.41 | −1 | 0 | +1 | +1.41 | |
| pH | 7.0 | 7.3 | 8.0 | 8.7 | 9.0 |
| Temperature (°C) | 30 | 33 | 40 | 47 | 50 |
| Spot n° | Protein | Organism | Theoretical Mass (Da) | p-Value | Coverage (%) | Peptides | |
|---|---|---|---|---|---|---|---|
| N | Sequence | ||||||
| I/1 | Curcin precursor | Jatropha curcas | 32,514.338 | 2.42 × 10−224 | 24.91 | 5 | QQTLSFTGSYADFLSR EAFGFSYSSHEIPVLR VGGTSYFFNDPESLADAK SSKPADIAKPLVGFIEMVPEAAR SSKPADIAKPLVGFIEM(+15.99)VPEAAR |
| II/2 | Malate dehydrogenase | Ricinus communis | 36,103.72 | 8.3 × 10−149 | 22.61 | 4 | LFGVTTLDVVR DDLFNINAGIVK GYVGEDQLGKALEGSDVVIIPAGVPR LNPLVSNLALYDIANTPGVAADVSHINTR |
| III/3 | Lactoylglutathione lyase | Ricinus communis | 31,547.15 | 2.1 × 10−149 | 11.79 | 4 | FYTEC(+57.02)FGMK ITSFLDPDGWK GPTPEPLC(+57.02)QVMLR GPTPEPLC(+71.04)QVMLR |
| IV/4 | Putative carboxymethylenebutenolidase | Arabidopsis thaliana | 25,893.30 | 2.4 × 10−59 | 12.97 | 2 | ALIPDLYR APIQAHFGELDNFVGFSDVTAAK |
| ANOVA; R2 = 0.82238; (2 Factors with 2 Levels Each) Central Composite, nc = 4, ns = 4, n0 = 2, and Runs = 10) | |||||
|---|---|---|---|---|---|
| Factor | S.S. | Df | M.S. | F | P |
| (1) Temperature (L) | 0.001857 | 1 | 0.001857 | 11.68790 | 0.014165 |
| Temperature (Q) | 0.001155 | 1 | 0.001155 | 7.26803 | 0.035769 |
| (2) pH (L) | 0.000377 | 1 | 0.000377 | 2.37215 | 0.174446 |
| pH (Q) | 0.000558 | 1 | 0.000558 | 3.51329 | 0.110009 |
| 1L by 2L | 0.000730 | 1 | 0.000730 | 4.59470 | 0.075781 |
| Error | 0.000953 | 6 | 0.000159 | ||
| Total SS | 0.005367 | 11 | |||
| Temperature (°C) | pH | Esterase Activity (U/mL) |
|---|---|---|
| 33 (−1) | 7.3 (−1) | 0.186869 |
| 33 (−1) | 8.7 (+1) | 0.183168 |
| 47 (+1) | 7.3 (−1) | 0.183168 |
| 47 (+1) | 8.7 (+1) | 0.233503 |
| 30 (−1.41) | 8 (0) | 0.17268 |
| 50 (+1.41) | 8 (0) | 0.225888 |
| 40 (0) | 7 (−1.41) | 0.204545 |
| 40 (0) | 9 (+1.41) | 0.210396 |
| 40 (0) | 8 (0) | 0.202073 |
| 40 (0) | 8 (0) | 0.23057 |
| 40 (0) | 8 (0) | 0.23057 |
| 40 (0) | 8 (0) | 0.227979 |
| Assessment | PDB ID: 4ZV9 | Model | Model Refined |
|---|---|---|---|
| MolProbity Score | 0.98 | 1.72 | 0.88 |
| Clash Score | 1.37 | 4.62 | 0.00 |
| Ramachandran Favoured | 97.42% | 91.92% | 94.44% |
| Ramachandran Outliers | 0.00% | 1.52% | 0.64% |
| Rotamer Outliers | 0.54% | 0.00% | 0.00% |
| QMEAN | 0.03 | −2.40 | −2.39 |
| Time (h) | ||||
|---|---|---|---|---|
| 2 | 5 | |||
| X (%) | ee (%) | X (%) | ee (%) | |
| 50–80% EtOH fraction | 50 | <0.1 | 70 | <0.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schwarz, M.G.A.; Antunes, D.; Brêda, G.C.; Valente, R.H.; Freire, D.M.G. Revisiting Jatropha curcas Monomeric Esterase: A Dienelactone Hydrolase Compatible with the Electrostatic Catapult Model. Biomolecules 2021, 11, 1486. https://doi.org/10.3390/biom11101486
Schwarz MGA, Antunes D, Brêda GC, Valente RH, Freire DMG. Revisiting Jatropha curcas Monomeric Esterase: A Dienelactone Hydrolase Compatible with the Electrostatic Catapult Model. Biomolecules. 2021; 11(10):1486. https://doi.org/10.3390/biom11101486
Chicago/Turabian StyleSchwarz, Marcos Gustavo Araujo, Deborah Antunes, Gabriela Coelho Brêda, Richard Hemmi Valente, and Denise Maria Guimarães Freire. 2021. "Revisiting Jatropha curcas Monomeric Esterase: A Dienelactone Hydrolase Compatible with the Electrostatic Catapult Model" Biomolecules 11, no. 10: 1486. https://doi.org/10.3390/biom11101486
APA StyleSchwarz, M. G. A., Antunes, D., Brêda, G. C., Valente, R. H., & Freire, D. M. G. (2021). Revisiting Jatropha curcas Monomeric Esterase: A Dienelactone Hydrolase Compatible with the Electrostatic Catapult Model. Biomolecules, 11(10), 1486. https://doi.org/10.3390/biom11101486







