Hyaluronan Functions in Wound Repair That Are Captured to Fuel Breast Cancer Progression
Abstract
:1. Background
2. The Hyaluronome
2.1. Hyaluronan
2.2. Hyaluronan Synthases
2.3. Hyaluronidases
2.4. Hyaluronan Receptors, CD44 and RHAMM
3. Functions of Hyaluronan: Size Matters in Cutaneous Repair and Breast Cancer
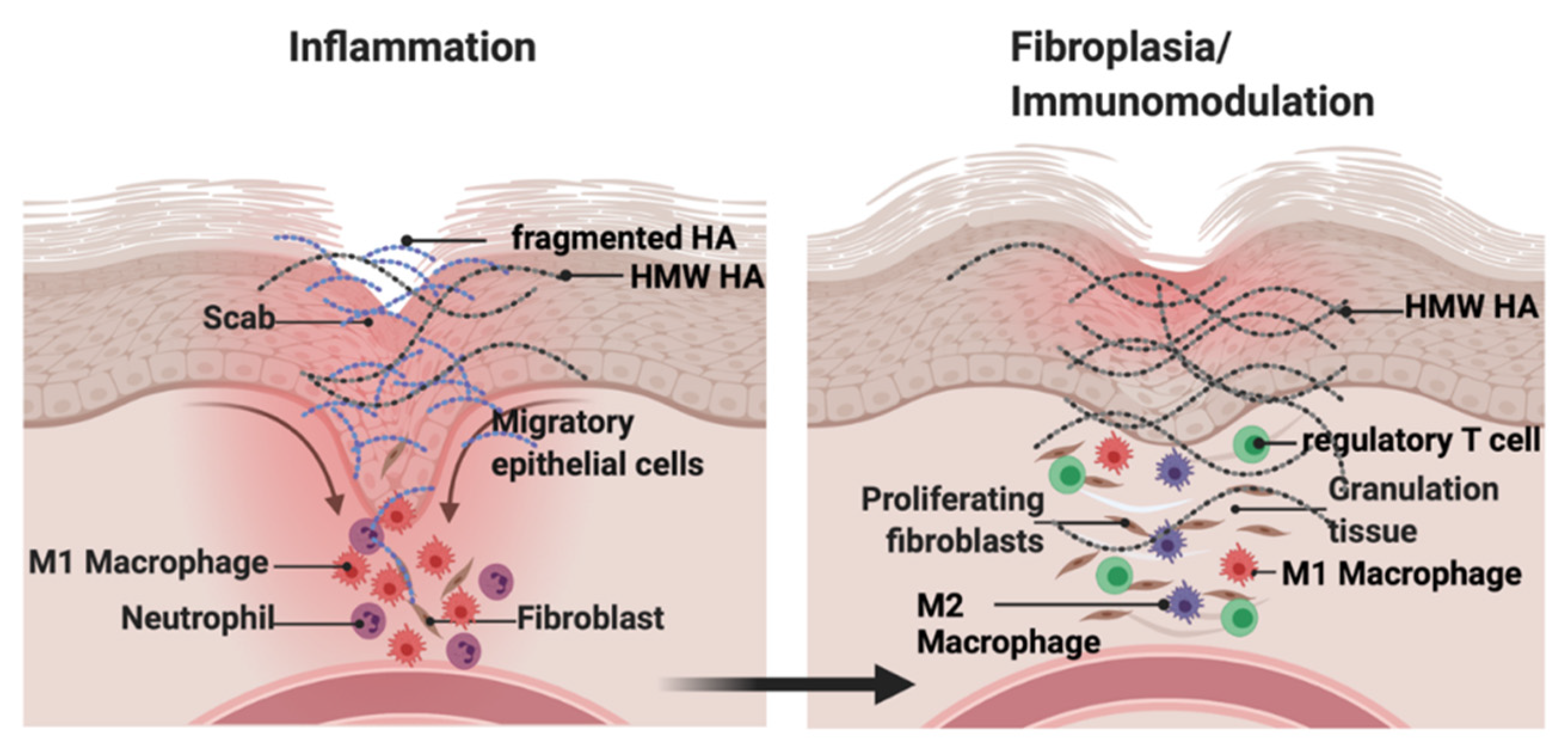
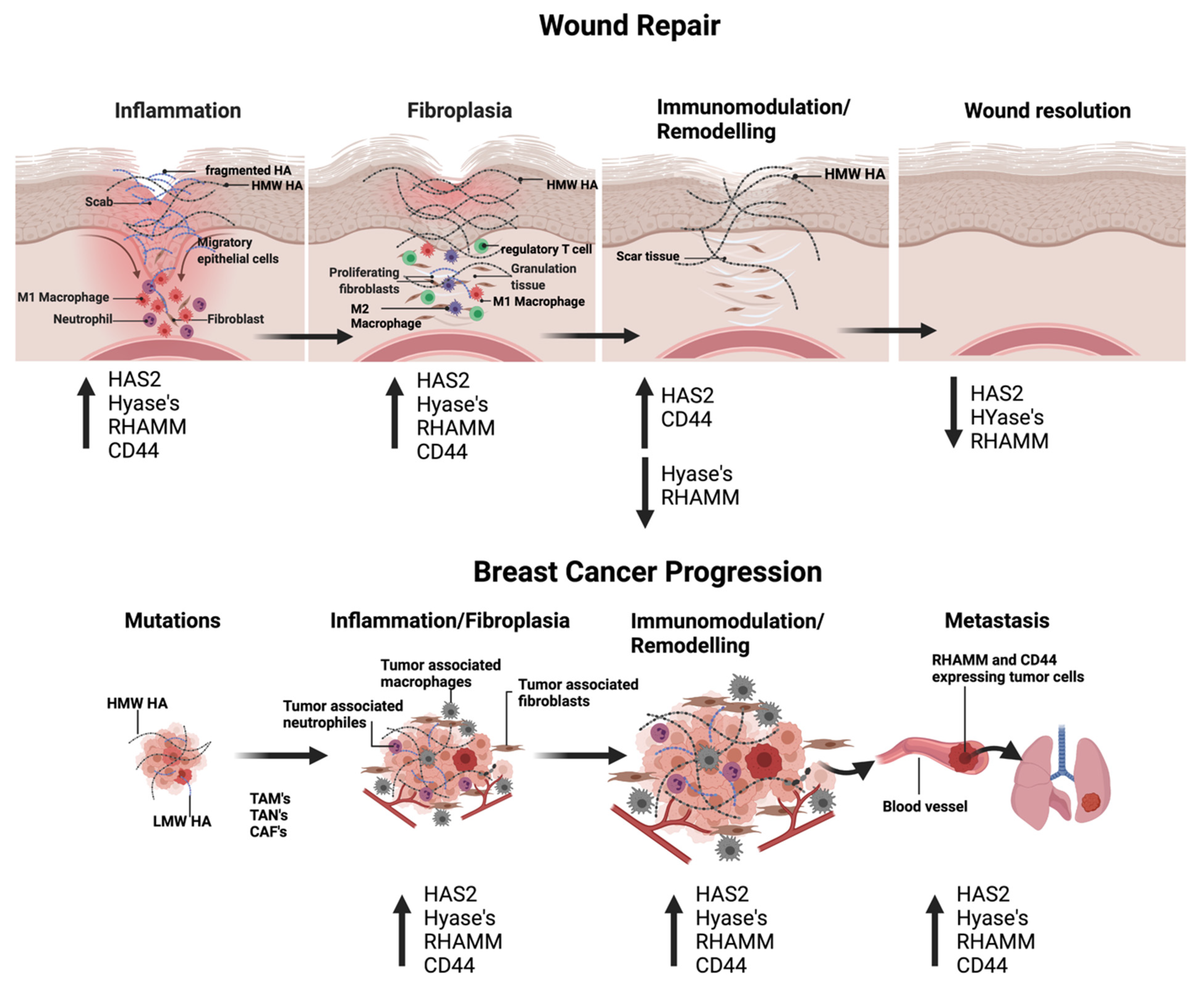
3.1. HA and Cutaneous Injury
3.2. HA and Breast Cancer
4. Roles of HA Receptors in De–Coding HA Polymer Size
4.1. Cutaneous Wound Repair
4.2. Breast Cancer Progression
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Dvorak, H.F. Tumors: Wounds That Do Not Heal–A Historical Perspective with a Focus on the Fundamental Roles of Increased Vascular Permeability and Clotting. Semin. Thromb. Hemost. 2019, 45, 576–592. [Google Scholar] [CrossRef]
- Bissell, M.J.; Hines, W.C. Why don’t we get more cancer? A proposed role of the microenvironment in restraining cancer progression. Nat. Med. 2011, 17, 320–329. [Google Scholar] [CrossRef] [Green Version]
- Allen, M.D.; Jones, L.J. The role of inflammation in progression of breast cancer: Friend or foe? Int. J. Oncol. 2015, 47, 797–805. [Google Scholar] [CrossRef]
- Nakamura, N. A hypothesis: Radiation carcinogenesis may result from tissue injuries and subsequent recovery processes which can act as tumor promoters and lead to an earlier onset of cancer. Br. J. Radiol. 2020, 93, 20190843. [Google Scholar] [CrossRef]
- Liu, M.; Tolg, C.; Turley, E. Dissecting the Dual Nature of Hyaluronan in the Tumor Microenvironment. Front. Immunol. 2019, 10, 947. [Google Scholar] [CrossRef]
- Abatangelo, G.; Vindigni, V.; Avruscio, G.; Pandis, L.; Brun, P. Hyaluronic Acid: Redefining Its Role. Cells 2020, 9, 1743. [Google Scholar] [CrossRef] [PubMed]
- Muto, J.; Sayama, K.; Gallo, R.L.; Kimata, K. Emerging evidence for the essential role of hyaluronan in cutaneous biology. J. Derm. Sci. 2019, 94, 190–195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tolg, C.; McCarthy, J.B.; Yazdani, A.; Turley, E.A. Hyaluronan and RHAMM in wound repair and the “cancerization” of stromal tissues. Biomed. Res. Int. 2014, 2014, 103923. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turley, E.A.; Wood, D.K.; McCarthy, J.B. Carcinoma Cell Hyaluronan as a “Portable” Cancerized Prometastatic Microenvironment. Cancer Res. 2016, 76, 2507–2512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kavasi, R.M.; Berdiaki, A.; Spyridaki, I.; Corsini, E.; Tsatsakis, A.; Tzanakakis, G.; Nikitovic, D. HA metabolism in skin homeostasis and inflammatory disease. Food Chem. Toxicol. 2017, 101, 128–138. [Google Scholar] [CrossRef] [PubMed]
- Litwiniuk, M.; Krejner, A.; Speyrer, M.S.; Gauto, A.R.; Grzela, T. Hyaluronic Acid in Inflammation and Tissue Regeneration. Wounds 2016, 28, 78–88. [Google Scholar]
- Zhao, Y.; Zheng, X.; Zheng, Y.; Chen, Y.; Fei, W.; Wang, F.; Zheng, C. Extracellular Matrix: Emerging Roles and Potential Therapeutic Targets for Breast Cancer. Front. Oncol. 2021, 11, 650453. [Google Scholar] [CrossRef]
- Velesiotis, C.; Vasileiou, S.; Vynios, D.H. A guide to hyaluronan and related enzymes in breast cancer: Biological significance and diagnostic value. FEBS J. 2019, 286, 3057–3074. [Google Scholar] [CrossRef] [PubMed]
- Schwertfeger, K.L.; Cowman, M.K.; Telmer, P.G.; Turley, E.A.; McCarthy, J.B. Hyaluronan, Inflammation, and Breast Cancer Progression. Front. Immunol. 2015, 6, 236. [Google Scholar] [CrossRef] [PubMed]
- Tavianatou, A.G.; Caon, I.; Franchi, M.; Piperigkou, Z.; Galesso, D.; Karamanos, N.K. Hyaluronan: Molecular size–dependent signaling and biological functions in inflammation and cancer. FEBS J. 2019, 286, 2883–2908. [Google Scholar] [CrossRef] [PubMed]
- Balazs, E.A. Hyaluronan as an ophthalmic viscoelastic device. Curr. Pharm. Biotechnol. 2008, 9, 236–238. [Google Scholar] [CrossRef] [PubMed]
- Balazs, E.A.; Högberg, B.; Laurent, T.C. The Biological Activity of Hyaluron Sulfuric Acid. Acta Physiol. Scand. 1951, 23, 168–178. [Google Scholar] [CrossRef] [PubMed]
- Snetkov, P.; Zakharova, K.; Morozkina, S.; Olekhnovich, R.; Uspenskaya, M. Hyaluronic Acid: The Influence of Molecular Weight on Structural, Physical, Physico–Chemical, and Degradable Properties of Biopolymer. Polymers 2020, 12, 1800. [Google Scholar] [CrossRef]
- Valachova, K.; Soltes, L. Hyaluronan as a Prominent Biomolecule with Numerous Applications in Medicine. Int. J. Mol. Sci. 2021, 22, 7077. [Google Scholar] [CrossRef]
- Ruiz Martinez, M.A.; Peralta Galisteo, S.; Castan, H.; Morales Hernandez, M.E. Role of proteoglycans on skin ageing: A review. Int. J. Cosmet. Sci. 2020, 42, 529–535. [Google Scholar] [CrossRef]
- Turley, E.A. Hyaluronic acid stimulates protein kinase activity in intact cells and in an isolated protein complex. J. Biol. Chem. 1989, 264, 8951–8955. [Google Scholar] [CrossRef]
- Aruffo, A.; Stamenkovic, I.; Melnick, M.; Underhill, C.B.; Seed, B. CD44 is the principal cell surface receptor for hyaluronate. Cell 1990, 61, 1303–1313. [Google Scholar] [CrossRef]
- Hardwick, C.; Hoare, K.; Owens, R.; Hohn, H.P.; Hook, M.; Moore, D.; Cripps, V.; Austen, L.; Nance, D.M.; Turley, E.A. Molecular cloning of a novel hyaluronan receptor that mediates tumor cell motility. J. Cell Biol. 1992, 117, 1343–1350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turley, E.A.; Noble, P.W.; Bourguignon, L.Y. Signaling properties of hyaluronan receptors. J. Biol. Chem. 2002, 277, 4589–4592. [Google Scholar] [CrossRef] [Green Version]
- Kaul, A.; Short, W.D.; Keswani, S.G.; Wang, X. Immunologic Roles of Hyaluronan in Dermal Wound Healing. Biomolecules 2021, 11, 1234. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Geng, C.; Hou, S.; Fan, H.; Gong, Y. Damage–Associated Molecular Patterns and Their Signaling Pathways in Primary Blast Lung Injury: New Research Progress and Future Directions. Int. J. Mol. Sci. 2020, 21, 6303. [Google Scholar] [CrossRef] [PubMed]
- Roedig, H.; Damiescu, R.; Zeng–Brouwers, J.; Kutija, I.; Trebicka, J.; Wygrecka, M.; Schaefer, L. Danger matrix molecules orchestrate CD14/CD44 signaling in cancer development. Semin. Cancer Biol. 2020, 62, 31–47. [Google Scholar] [CrossRef] [PubMed]
- Garantziotis, S.; Savani, R.C. Hyaluronan biology: A complex balancing act of structure, function, location and context. Matrix Biol. 2019, 78, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Savani, R.C. Modulators of inflammation in Bronchopulmonary Dysplasia. Semin. Perinatol. 2018, 42, 459–470. [Google Scholar] [CrossRef]
- Zheng, H.; Siddharth, S.; Parida, S.; Wu, X.; Sharma, D. Tumor Microenvironment: Key Players in Triple Negative Breast Cancer Immunomodulation. Cancers 2021, 13, 3357. [Google Scholar] [CrossRef]
- Camenisch, T.D.; Spicer, A.P.; Brehm–Gibson, T.; Biesterfeldt, J.; Augustine, M.L.; Calabro, A., Jr.; Kubalak, S.; Klewer, S.E.; McDonald, J.A. Disruption of hyaluronan synthase–2 abrogates normal cardiac morphogenesis and hyaluronan–mediated transformation of epithelium to mesenchyme. J. Clin. Investig. 2000, 106, 349–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bai, K.J.; Spicer, A.P.; Mascarenhas, M.M.; Yu, L.; Ochoa, C.D.; Garg, H.G.; Quinn, D.A. The role of hyaluronan synthase 3 in ventilator–induced lung injury. Am. J. Respir. Crit. Care Med. 2005, 172, 92–98. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, N.; Miyoshi, S.; Mikami, T.; Koyama, H.; Kitazawa, M.; Takeoka, M.; Sano, K.; Amano, J.; Isogai, Z.; Niida, S.; et al. Hyaluronan deficiency in tumor stroma impairs macrophage trafficking and tumor neovascularization. Cancer Res. 2010, 70, 7073–7083. [Google Scholar] [CrossRef] [Green Version]
- Mack, J.A.; Feldman, R.J.; Itano, N.; Kimata, K.; Lauer, M.; Hascall, V.C.; Maytin, E.V. Enhanced inflammation and accelerated wound closure following tetraphorbol ester application or full–thickness wounding in mice lacking hyaluronan synthases Has1 and Has3. J. Investig. Dermatol. 2012, 132, 198–207. [Google Scholar] [CrossRef] [Green Version]
- Tolg, C.; Yuan, H.; Flynn, S.M.; Basu, K.; Ma, J.; Tse, K.C.K.; Kowalska, B.; Vulkanesku, D.; Cowman, M.K.; McCarthy, J.B.; et al. Hyaluronan modulates growth factor induced mammary gland branching in a size dependent manner. Matrix Biol. 2017, 63, 117–132. [Google Scholar] [CrossRef]
- Heldin, P.; Basu, K.; Olofsson, B.; Porsch, H.; Kozlova, I.; Kahata, K. Deregulation of hyaluronan synthesis, degradation and binding promotes breast cancer. J. Biochem. 2013, 154, 395–408. [Google Scholar] [CrossRef] [Green Version]
- Sayo, T.; Sugiyama, Y.; Takahashi, Y.; Ozawa, N.; Sakai, S.; Ishikawa, O.; Tamura, M.; Inoue, S. Hyaluronan synthase 3 regulates hyaluronan synthesis in cultured human keratinocytes. J. Investig. Dermatol. 2002, 118, 43–48. [Google Scholar] [CrossRef] [Green Version]
- Sugiyama, Y.; Shimada, A.; Sayo, T.; Sakai, S.; Inoue, S. Putative hyaluronan synthase mRNA are expressed in mouse skin and TGF–beta upregulates their expression in cultured human skin cells. J. Investig. Dermatol. 1998, 110, 116–121. [Google Scholar] [CrossRef] [Green Version]
- Tammi, R.; Pasonen–Seppanen, S.; Kolehmainen, E.; Tammi, M. Hyaluronan synthase induction and hyaluronan accumulation in mouse epidermis following skin injury. J. Investig. Dermatol. 2005, 124, 898–905. [Google Scholar] [CrossRef] [PubMed]
- Yamada, Y.; Itano, N.; Narimatsu, H.; Kudo, T.; Morozumi, K.; Hirohashi, S.; Ochiai, A.; Ueda, M.; Kimata, K. Elevated transcript level of hyaluronan synthase1 gene correlates with poor prognosis of human colon cancer. Clin. Exp. Metastasis 2004, 21, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Heldin, P.; Lin, C.Y.; Kolliopoulos, C.; Chen, Y.H.; Skandalis, S.S. Regulation of hyaluronan biosynthesis and clinical impact of excessive hyaluronan production. Matrix Biol. 2019, 78, 100–117. [Google Scholar] [CrossRef]
- Heldin, P.; Basu, K.; Kozlova, I.; Porsch, H. HAS2 and CD44 in breast tumorigenesis. Adv. Cancer Res. 2014, 123, 211–229. [Google Scholar]
- Sindelar, M.; Jilkova, J.; Kubala, L.; Velebny, V.; Turkova, K. Hyaluronidases and hyaluronate lyases: From humans to bacteriophages. Colloids Surf. B Biointerfaces 2021, 208, 112095. [Google Scholar] [CrossRef]
- Triggs–Raine, B.; Natowicz, M.R. Biology of hyaluronan: Insights from genetic disorders of hyaluronan metabolism. World J. Biol. Chem. 2015, 6, 110–120. [Google Scholar] [CrossRef] [PubMed]
- Pibuel, M.A.; Poodts, D.; Diaz, M.; Hajos, S.E.; Lompardia, S.L. The scrambled story between hyaluronan and glioblastoma. J. Biol. Chem. 2021, 296, 100549. [Google Scholar] [CrossRef] [PubMed]
- Piperigkou, Z.; Kyriakopoulou, K.; Koutsakis, C.; Mastronikolis, S.; Karamanos, N.K. Key Matrix Remodeling Enzymes: Functions and Targeting in Cancer. Cancers 2021, 13, 1441. [Google Scholar] [CrossRef]
- Irie, F.; Tobisawa, Y.; Murao, A.; Yamamoto, H.; Ohyama, C.; Yamaguchi, Y. The cell surface hyaluronidase TMEM2 regulates cell adhesion and migration via degradation of hyaluronan at focal adhesion sites. J. Biol. Chem. 2021, 296, 100481. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Yan, W.; Han, P.; Tian, D. The emerging role of KIAA1199 in cancer development and therapy. Biomed. Pharmacother. 2021, 138, 111507. [Google Scholar] [CrossRef] [PubMed]
- Bono, P.; Rubin, K.; Higgins, J.M.; Hynes, R.O. Layilin, a novel integral membrane protein, is a hyaluronan receptor. Mol. Biol. Cell 2001, 12, 891–900. [Google Scholar] [CrossRef] [Green Version]
- Soroosh, A.; Albeiroti, S.; West, G.A.; Willard, B.; Fiocchi, C.; de la Motte, C.A. Crohn’s Disease Fibroblasts Overproduce the Novel Protein KIAA1199 to Create Proinflammatory Hyaluronan Fragments. Cell. Mol. Gastroenterol. Hepatol. 2016, 2, 358–368.e4. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, H.; Nagaoka, A.; Kusaka–Kikushima, A.; Tobiishi, M.; Kawabata, K.; Sayo, T.; Sakai, S.; Sugiyama, Y.; Enomoto, H.; Okada, Y.; et al. KIAA1199, a deafness gene of unknown function, is a new hyaluronan binding protein involved in hyaluronan depolymerization. Proc. Natl. Acad. Sci. USA 2013, 110, 5612–5617. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, H.; Okada, Y. Role of HYBID (Hyaluronan Binding Protein Involved in Hyaluronan Depolymerization), Alias KIAA1199/CEMIP, in Hyaluronan Degradation in Normal and Photoaged Skin. Int. J. Mol. Sci. 2019, 20, 5804. [Google Scholar] [CrossRef] [Green Version]
- Johnson, P.; Arif, A.A.; Lee–Sayer, S.S.M.; Dong, Y. Hyaluronan and Its Interactions with Immune Cells in the Healthy and Inflamed Lung. Front. Immunol. 2018, 9, 2787. [Google Scholar] [CrossRef] [PubMed]
- Johnson, B.Z.; Stevenson, A.W.; Prele, C.M.; Fear, M.W.; Wood, F.M. The Role of IL–6 in Skin Fibrosis and Cutaneous Wound Healing. Biomedicines 2020, 8, 101. [Google Scholar] [CrossRef] [PubMed]
- Abaurrea, A.; Araujo, A.M.; Caffarel, M.M. The Role of the IL–6 Cytokine Family in Epithelial–Mesenchymal Plasticity in Cancer Progression. Int. J. Mol. Sci. 2021, 22, 8334. [Google Scholar] [CrossRef] [PubMed]
- Toole, B.P. The CD147–HYALURONAN Axis in Cancer. Anat. Rec. 2020, 303, 1573–1583. [Google Scholar] [CrossRef]
- Jackson, D.G. Hyaluronan in the lymphatics: The key role of the hyaluronan receptor LYVE–1 in leucocyte trafficking. Matrix Biol. 2019, 78, 219–235. [Google Scholar] [CrossRef]
- Garantziotis, S.; Matalon, S. Sugarcoating Lung Injury: A Novel Role for High–Molecular–Weight Hyaluronan in Pneumonia. Am. J. Respir. Crit. Care Med. 2019, 200, 1197–1198. [Google Scholar] [CrossRef]
- Weigel, P.H. Planning, evaluating and vetting receptor signaling studies to assess hyaluronan size–dependence and specificity. Glycobiology 2017, 27, 796–799. [Google Scholar] [CrossRef]
- Weigel, P.H.; Baggenstoss, B.A. What is special about 200 kDa hyaluronan that activates hyaluronan receptor signaling? Glycobiology 2017, 27, 868–877. [Google Scholar] [CrossRef] [Green Version]
- Ziebell, M.R.; Prestwich, G.D. Interactions of peptide mimics of hyaluronic acid with the receptor for hyaluronan mediated motility (RHAMM). J. Comput. Aided. Mol. Des. 2004, 18, 597–614. [Google Scholar] [CrossRef]
- Yang, B.; Yang, B.L.; Savani, R.C.; Turley, E.A. Identification of a common hyaluronan binding motif in the hyaluronan binding proteins RHAMM, CD44 and link protein. EMBO J. 1994, 13, 286–296. [Google Scholar] [CrossRef]
- Johnson, L.A.; Jackson, D.G. Hyaluronan and Its Receptors: Key Mediators of Immune Cell Entry and Trafficking in the Lymphatic. System Cells 2021, 10, 2061. [Google Scholar]
- Tammi, M.I.; Oikari, S.; Pasonen–Seppanen, S.; Rilla, K.; Auvinen, P.; Tammi, R.H. Activated hyaluronan metabolism in the tumor matrix—Causes and consequences. Matrix Biol. 2019, 78, 147–164. [Google Scholar] [CrossRef]
- Altman, R.; Bedi, A.; Manjoo, A.; Niazi, F.; Shaw, P.; Mease, P. Anti–Inflammatory Effects of Intra–Articular Hyaluronic Acid: A Systematic Review. Cartilage 2019, 10, 4352. [Google Scholar] [CrossRef] [PubMed]
- Heldin, P.; Kolliopoulos, C.; Lin, C.Y.; Heldin, C.H. Involvement of hyaluronan and CD44 in cancer and viral infections. Cell Signal. 2020, 65, 109427. [Google Scholar] [CrossRef] [PubMed]
- Marozzi, M.; Parnigoni, A.; Negri, A.; Viola, M.; Vigetti, D.; Passi, A.; Karousou, E.; Rizzi, F. Inflammation, Extracellular Matrix Remodeling, and Proteostasis in Tumor Microenvironment. Int. J. Mol. Sci. 2021, 22, 8102. [Google Scholar] [CrossRef]
- Anderegg, U.; Simon, J.C.; Averbeck, M. More than just a filler—The role of hyaluronan for skin homeostasis. Exp. Dermatol. 2014, 23, 295–303. [Google Scholar] [CrossRef]
- Kleiser, S.; Nystrom, A. Interplay between Cell–Surface Receptors and Extracellular Matrix in Skin. Biomolecules 2020, 10, 1170. [Google Scholar] [CrossRef] [PubMed]
- Banerji, S.; Wright, A.J.; Noble, M.; Mahoney, D.J.; Campbell, I.D.; Day, A.J.; Jackson, D.G. Structures of the CD44–hyaluronan complex provide insight into a fundamental carbohydrate–protein interaction. Nat. Struct. Mol. Biol. 2007, 14, 234–239. [Google Scholar] [CrossRef]
- Senbanjo, L.T.; AlJohani, H.; AlQranei, M.; Majumdar, S.; Ma, T.; Chellaiah, M.A. Identification of sequence–specific interactions of the CD44–intracellular domain with RUNX2 in the transcription of matrix metalloprotease–9 in human prostate cancer cells. Cancer Drug. Resist. 2020, 3, 586–602. [Google Scholar] [CrossRef]
- Al–Othman, N.; Alhendi, A.; Ihbaisha, M.; Barahmeh, M.; Alqaraleh, M.; Al–Momany, B.Z. Role of CD44 in breast cancer. Breast Dis. 2020, 39, 1–13. [Google Scholar] [CrossRef]
- Ooki, T.; Murata–Kamiya, N.; Takahashi–Kanemitsu, A.; Wu, W.; Hatakeyama, M. High–Molecular–Weight Hyaluronan Is a Hippo Pathway Ligand Directing Cell Density–Dependent Growth Inhibition via PAR1b. Dev. Cell 2019, 49, 590–604.e9. [Google Scholar] [CrossRef]
- Jou, I.M.; Wu, T.T.; Hsu, C.C.; Yang, C.C.; Huang, J.S.; Tu, Y.K.; Lee, J.S.; Su, F.C.; Kuo, Y.L. High molecular weight form of hyaluronic acid reduces neuroinflammatory response in injured sciatic nerve via the intracellular domain of CD44. J. Biomed. Mater. Res. B Appl. Biomater. 2021, 109, 673–680. [Google Scholar] [CrossRef]
- Hauser–Kawaguchi, A.; Luyt, L.G.; Turley, E. Design of peptide mimetics to block pro–inflammatory functions of HA fragments. Matrix Biol. 2019, 78, 346–356. [Google Scholar] [CrossRef]
- Gulati, K.; Jamsandekar, M.; Poluri, K.M. Mechanistic insights into molecular evolution of species–specific differential glycosaminoglycan binding surfaces in growth–related oncogene chemokines. R. Soc. Open Sci. 2017, 4, 171059. [Google Scholar] [CrossRef] [Green Version]
- Boittier, E.D.; Gandhi, N.S.; Ferro, V.; Coombe, D.R. Cross–Species Analysis of Glycosaminoglycan Binding Proteins Reveals Some Animal Models Are ”More Equal“ than Others. Molecules 2019, 24, 924. [Google Scholar] [CrossRef] [Green Version]
- Carvalho, A.M.; Soares da Costa, D.; Paulo, P.M.R.; Reis, R.L.; Pashkuleva, I. Co–localization and crosstalk between CD44 and RHAMM depend on hyaluronan presentation. Acta Biomater. 2021, 119, 114–124. [Google Scholar] [CrossRef] [PubMed]
- Proksch, E.; Brandner, J.M.; Jensen, J.M. The skin: An indispensable barrier. Exp. Dermatol. 2008, 17, 1063–1072. [Google Scholar] [CrossRef] [PubMed]
- Singampalli, K.L.; Balaji, S.; Wang, X.; Parikh, U.M.; Kaul, A.; Gilley, J.; Birla, R.K.; Bollyky, P.L.; Keswani, S.G. The Role of an IL–10/Hyaluronan Axis in Dermal Wound Healing. Front. Cell Dev. Biol. 2020, 8, 636. [Google Scholar] [CrossRef] [PubMed]
- Tolg, C.; Hamilton, S.R.; Zalinska, E.; McCulloch, L.; Amin, R.; Akentieva, N.; Winnik, F.; Savani, R.; Bagli, D.J.; Luyt, L.G.; et al. A RHAMM mimetic peptide blocks hyaluronan signaling and reduces inflammation and fibrogenesis in excisional skin wounds. Am. J. Pathol. 2012, 181, 1250–1270. [Google Scholar] [CrossRef] [Green Version]
- Condorelli, A.G.; El Hachem, M.; Zambruno, G.; Nystrom, A.; Candi, E.; Castiglia, D. Notch–ing up knowledge on molecular mechanisms of skin fibrosis: Focus on the multifaceted Notch signalling pathway. J. Biomed. Sci. 2021, 28, 36. [Google Scholar] [CrossRef]
- Caskey, R.C.; Allukian, M.; Lind, R.C.; Herdrich, B.J.; Xu, J.; Radu, A.; Mitchell, M.E.; Liechty, K.W. Lentiviral–mediated over–expression of hyaluronan synthase–1 (HAS–1) decreases the cellular inflammatory response and results in regenerative wound repair. Cell Tissue Res. 2013, 351, 117–125. [Google Scholar] [CrossRef]
- Lee, B.M.; Park, S.J.; Noh, I.; Kim, C.H. The effects of the molecular weights of hyaluronic acid on the immune responses. Biomater. Res. 2021, 25, 27. [Google Scholar] [CrossRef]
- Shi, Q.; Zhao, L.; Xu, C.; Zhang, L.; Zhao, H. High Molecular Weight Hyaluronan Suppresses Macrophage M1 Polarization and Enhances IL–10 Production in PM2.5–Induced Lung Inflammation. Molecules 2019, 24, 1766. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vistejnova, L.; Safrankova, B.; Nesporova, K.; Slavkovsky, R.; Hermannova, M.; Hosek, P.; Velebny, V.; Kubala, L. Low molecular weight hyaluronan mediated CD44 dependent induction of IL–6 and chemokines in human dermal fibroblasts potentiates innate immune response. Cytokine 2014, 70, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Cha, J.; Jang, M.; Kim, P. Hyaluronic acid–based extracellular matrix triggers spontaneous M2–like polarity of monocyte/macrophage. Biomater. Sci. 2019, 7, 2264–7771. [Google Scholar] [CrossRef] [PubMed]
- Meyer–Siegler, K.L.; Leifheit, E.C.; Vera, P.L. Inhibition of macrophage migration inhibitory factor decreases proliferation and cytokine expression in bladder cancer cells. BMC Cancer 2004, 4, 34. [Google Scholar] [CrossRef] [Green Version]
- Tamoto, K.; Nochi, H.; Tada, M.; Shimada, S.; Mori, Y.; Kataoka, S.; Suzuki, Y.; Nakamura, T. High–molecular–weight hyaluronic acids inhibit chemotaxis and phagocytosis but not lysosomal enzyme release induced by receptor–mediated stimulations in guinea pig phagocytes. Microbiol. Immunol. 1994, 38, 73–80. [Google Scholar] [CrossRef]
- Schimizzi, A.L.; Massie, J.B.; Murphy, M.; Perry, A.; Kim, C.W.; Garfin, S.R.; Akeson, W.H. High–molecular–weight hyaluronan inhibits macrophage proliferation and cytokine release in the early wound of a preclinical postlaminectomy rat model. Spine J. 2006, 6, 550–556. [Google Scholar] [CrossRef]
- Sheehan, K.M.; DeLott, L.B.; West, R.A.; Bonnema, J.D.; DeHeer, D.H. Hyaluronic acid of high molecular weight inhibits proliferation and induces cell death in U937 macrophage cells. Life Sci. 2004, 75, 3087–3102. [Google Scholar] [CrossRef]
- Huang, L.; Gu, H.; Burd, A. A reappraisal of the biological effects of hyaluronan on human dermal fibroblast. J. Biomed. Mater. Res. A 2009, 90, 1177–1785. [Google Scholar] [CrossRef]
- Evanko, S.P.; Potter–Perigo, S.; Petty, L.J.; Workman, G.A.; Wight, T.N. Hyaluronan Controls the Deposition of Fibronectin and Collagen and Modulates TGF–beta1 Induction of Lung Myofibroblasts. Matrix Biol. 2015, 42, 74–92. [Google Scholar] [CrossRef] [Green Version]
- Kawano, Y.; Patrulea, V.; Sublet, E.; Borchard, G.; Iyoda, T.; Kageyama, R.; Morita, A.; Seino, S.; Yoshida, H.; Jordan, O.; et al. Wound Healing Promotion by Hyaluronic Acid: Effect of Molecular Weight on Gene Expression and In Vivo Wound Closure. Pharmaceuticals 2021, 14, 301. [Google Scholar] [CrossRef]
- Hoffmann, A.; Hoing, J.L.; Newman, M.; Simman, R. Role of Hyaluronic Acid Treatment in the Prevention of Keloid Scarring. J. Am. Coll. Clin. Wound Spec. 2012, 4, 23–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- D’Agostino, A.; Stellavato, A.; Corsuto, L.; Diana, P.; Filosa, R.; La Gatta, A.; De Rosa, M.; Schiraldi, C. Is molecular size a discriminating factor in hyaluronan interaction with human cells? Carbohydr. Polym. 2017, 157, 21–30. [Google Scholar] [CrossRef]
- Fronza, M.; Caetano, G.F.; Leite, M.N.; Bitencourt, C.S.; Paula–Silva, F.W.; Andrade, T.A.; Frade, M.A.; Merfort, I.; Faccioli, L.H. Hyaluronidase modulates inflammatory response and accelerates the cutaneous wound healing. PLoS ONE 2014, 9, e112297. [Google Scholar] [CrossRef] [PubMed]
- Tolg, C.; Telmer, P.; Turley, E. Specific sizes of hyaluronan oligosaccharides stimulate fibroblast migration and excisional wound repair. PLoS ONE 2014, 9, e88479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Damodarasamy, M.; Johnson, R.S.; Bentov, I.; MacCoss, M.J.; Vernon, R.B.; Reed, M.J. Hyaluronan enhances wound repair and increases collagen III in aged dermal wounds. Wound Repair Regen. 2014, 22, 521–526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Radrezza, S.; Baron, G.; Nukala, S.B.; Depta, G.; Aldini, G.; Carini, M.; D’Amato, A. Advanced quantitative proteomics to evaluate molecular effects of low–molecular–weight hyaluronic acid in human dermal fibroblasts. J. Pharm. Biomed. Anal. 2020, 185, 113199. [Google Scholar] [CrossRef] [PubMed]
- Nagy, N.; Kuipers, H.F.; Marshall, P.L.; Wang, E.; Kaber, G.; Bollyky, P.L. Hyaluronan in immune dysregulation and autoimmune diseases. Matrix Biol. 2019, 78, 292–313. [Google Scholar] [CrossRef]
- Jin, J.; Li, Y.; Zhao, Q.; Chen, Y.; Fu, S.; Wu, J. Coordinated regulation of immune contexture: Crosstalk between STAT3 and immune cells during breast cancer progression. Cell Commun. Signal. 2021, 19, 50. [Google Scholar] [CrossRef]
- Jenkins, S.; Wesolowski, R.; Gatti–Mays, M.E. Improving Breast Cancer Responses to Immunotherapy–a Search for the Achilles Heel of the Tumor Microenvironment. Curr. Oncol. Rep. 2021, 23, 55. [Google Scholar] [CrossRef] [PubMed]
- Deligne, C.; Midwood, K.S. Macrophages and Extracellular Matrix in Breast Cancer: Partners in Crime or Protective Allies? Front. Oncol. 2021, 11, 620773. [Google Scholar] [CrossRef] [PubMed]
- Joshi, R.S.; Kanugula, S.S.; Sudhir, S.; Pereira, M.P.; Jain, S.; Aghi, M.K. The Role of Cancer–Associated Fibroblasts in Tumor Progression. Cancers 2021, 13, 1399. [Google Scholar] [CrossRef] [PubMed]
- Deepak, K.G.K.; Vempati, R.; Nagaraju, G.P.; Dasari, V.R.; Nagini, S.; Rao, D.N.; Malla, R.R. Tumor microenvironment: Challenges and opportunities in targeting metastasis of triple negative breast cancer. Pharmacol. Res. 2020, 153, 104683. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Chen, L.; Wang, Y.; Jin, J.; Xie, X.; Zhang, J. Hyaluronic acid predicts poor prognosis in breast cancer patients: A protocol for systematic review and meta analysis. Medicine 2020, 99, e20438. [Google Scholar] [CrossRef]
- Wu, M.; Cao, M.; He, Y.; Liu, Y.; Yang, C.; Du, Y.; Wang, W.; Gao, F. A novel role of low molecular weight hyaluronan in breast cancer metastasis. FASEB J. 2015, 29, 1290–2198. [Google Scholar] [CrossRef]
- Bohrer, L.R.; Chuntova, P.; Bade, L.K.; Beadnell, T.C.; Leon, R.P.; Brady, N.J.; Ryu, Y.; Goldberg, J.E.; Schmechel, S.C.; Koopmeiners, J.S. Activation of the FGFR–STAT3 pathway in breast cancer cells induces a hyaluronan–rich microenvironment that licenses tumor formation. Cancer Res. 2014, 74, 374–386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Whiteside, T.L. Regulatory T cell subsets in human cancer: Are they regulating for or against tumor progression? Cancer Immunol. Immunother. 2014, 63, 67–72. [Google Scholar] [CrossRef] [Green Version]
- Urakawa, H.; Nishida, Y.; Wasa, J.; Arai, E.; Zhuo, L.; Kimata, K.; Kozawa, E.; Futamura, N.; Ishiguro, N. Inhibition of hyaluronan synthesis in breast cancer cells by 4–methylumbelliferone suppresses tumorigenicity in vitro and metastatic lesions of bone in vivo. Int. J. Cancer 2012, 130, 454–466. [Google Scholar] [CrossRef]
- Spinelli, F.M.; Vitale, D.L.; Icardi, A.; Caon, I.; Brandone, A.; Giannoni, P.; Saturno, V.; Passi, A.; García, M.; Sevic, I.; et al. Hyaluronan preconditioning of monocytes/macrophages affects their angiogenic behavior and regulation of TSG–6 expression in a tumor type–specific manner. FEBS J. 2019, 286, 3433–3449. [Google Scholar] [CrossRef]
- Spinelli, F.M.; Vitale, D.L.; Sevic, I.; Alaniz, L. Hyaluronan in the Tumor Microenvironment. Adv. Exp. Med. Biol. 2020, 1245, 67–83. [Google Scholar]
- Witschen, P.M.; Chaffee, T.S.; Brady, N.J.; Huggins, D.N.; Knutson, T.P.; LaRue, R.S.; Munro, S.A.; Tiegs, L.; McCarthy, J.B.; Nelson, A.C.; et al. Tumor Cell Associated Hyaluronan–CD44 Signaling Promotes Pro–Tumor Inflammation in Breast Cancer. Cancers 2020, 12, 1325. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, J.B.; El–Ashry, D.; Turley, E.A. Hyaluronan, Cancer–Associated Fibroblasts and the Tumor Microenvironment in Malignant Progression. Front. Cell Dev. Biol. 2018, 6, 48. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Thompson, B.J.; Chen, K.; Gao, F.; Blouw, B.; Marella, M.; Zimmerman, S.; Kimbler, T.; Garrovillo, S.; Bahn, J.; et al. The growth of a xenograft breast cancer tumor model with engineered hyaluronan–accumulating stroma is dependent on hyaluronan and independent of CD44. Oncotarget 2019, 10, 6561–6576. [Google Scholar] [CrossRef] [PubMed]
- Lompardia, S.; Diaz, M.; Pibuel, M.; Papademetrio, D.; Poodts, D.; Mihalez, C.; Álvarez, É.; Hajos, S. Hyaluronan abrogates imatinib–induced senescence in chronic myeloid leukemia cell lines. Sci. Rep. 2019, 9, 10930. [Google Scholar] [CrossRef] [PubMed]
- Tansi, F.L.; Frobel, F.; Maduabuchi, W.O.; Steiniger, F.; Westermann, M.; Quaas, R.; Teichgräber, U.K.; Hilger, I. Effect of Matrix–Modulating Enzymes on The Cellular Uptake of Magnetic Nanoparticles and on Magnetic Hyperthermia Treatment of Pancreatic Cancer Models In Vivo. Nanomaterials 2021, 11, 438. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Du, Y.; Liu, Y.; He, Y.; Zhang, G.; Yang, C.; Gao, F. Hyaluronan arrests human breast cancer cell growth by prolonging the G0/G1 phase of the cell cycle. Acta Biochim. Biophys. Sin. 2018, 50, 1181–1189. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Qiao, S.; Hou, X.; Tian, H.; Deng, S.; Ye, K.; Nie, Y.; Chen, X.; Yan, H.; Tian, W. Bioengineered tumor microenvironments with naked mole rats high–molecular–weight hyaluronan induces apoptosis in breast cancer cells. Oncogene 2019, 38, 4297–4309. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.F.; Qiao, S.P.; Shi, S.L.; Yao, L.F.; Hou, X.L.; Li, C.F.; Lin, F.H.; Guo, K.; Acharya, A.; Chen, X.B.; et al. Modulating Three–Dimensional Microenvironment with Hyaluronan of Different Molecular Weights Alters Breast Cancer Cell Invasion Behavior. ACS Appl. Mater. Interfaces 2017, 9, 9327–9338. [Google Scholar] [CrossRef] [PubMed]
- Bourguignon, L.Y.; Wong, G.; Earle, C.A.; Xia, W. Interaction of low molecular weight hyaluronan with CD44 and toll–like receptors promotes the actin filament–associated protein 110–actin binding and MyD88–NFkappaB signaling leading to proinflammatory cytokine/chemokine production and breast tumor invasion. Cytoskeleton 2011, 68, 671–693. [Google Scholar]
- Koyama, H.; Hibi, T.; Isogai, Z.; Yoneda, M.; Fujimori, M.; Amano, J.; Kawakubo, M.; Kannagi, R.; Kimata, K.; Taniguchi, S.; et al. Hyperproduction of hyaluronan in neu–induced mammary tumor accelerates angiogenesis through stromal cell recruitment: Possible involvement of versican/PG–M. Am. J. Pathol. 2007, 170, 1086–1099. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koyama, H.; Kobayashi, N.; Harada, M.; Takeoka, M.; Kawai, Y.; Sano, K.; Fujimori, M.; Amano, J.; Ohhashi, T.; Kannagi, R.; et al. Significance of tumor–associated stroma in promotion of intratumoral lymphangiogenesis: Pivotal role of a hyaluronan–rich tumor microenvironment. Am. J. Pathol. 2008, 172, 179–193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tavianatou, A.G.; Piperigkou, Z.; Barbera, C.; Beninatto, R.; Masola, V.; Caon, I.; Onisto, M.; Franchi, M.; Galesso, D.; Karamanos, N.K. Molecular size–dependent specificity of hyaluronan on functional properties, morphology and matrix composition of mammary cancer cells. Matrix Biol. Plus 2019, 3, 100008. [Google Scholar] [CrossRef] [PubMed]
- Lien, H.C.; Lee, Y.H.; Jeng, Y.M.; Lin, C.H.; Lu, Y.S.; Yao, Y.T. Differential expression of hyaluronan synthase 2 in breast carcinoma and its biological significance. Histopathology 2014, 65, 328–339. [Google Scholar] [CrossRef]
- Hamilton, S.R.; Fard, S.F.; Paiwand, F.F.; Tolg, C.; Veiseh, M.; Wang, C.; McCarthy, J.B.; Bissell, M.J.; Koropatnick, J.; Turley, E.A. The hyaluronan receptors CD44 and Rhamm (CD168) form complexes with ERK1,2 that sustain high basal motility in breast cancer cells. J. Biol. Chem. 2007, 282, 16667–16680. [Google Scholar] [CrossRef] [Green Version]
- Veiseh, M.; Kwon, D.H.; Borowsky, A.D.; Tolg, C.; Leong, H.S.; Lewis, J.D.; Turley, E.A.; Bissell, M.J. Cellular heterogeneity profiling by hyaluronan probes reveals an invasive but slow–growing breast tumor subset. Proc. Natl. Acad. Sci. USA 2014, 111, 1731–1739. [Google Scholar] [CrossRef] [Green Version]
- Glasner, A.; Plitas, G. Tumor resident regulatory T cells. Semin. Immunol. 2021, 101476, in press. [Google Scholar] [CrossRef]
- Govindaraju, P.; Todd, L.; Shetye, S.; Monslow, J.; Pure, E. CD44–dependent inflammation, fibrogenesis, and collagenolysis regulates extracellular matrix remodeling and tensile strength during cutaneous wound healing. Matrix Biol. 2019, 75, 314–330. [Google Scholar] [CrossRef]
- Shatirishvili, M.; Burk, A.S.; Franz, C.M.; Pace, G.; Kastilan, T.; Breuhahn, K.; Hinterseer, E.; Dierich, A.; Bakiri, L.; Wagner, E.F.; et al. Epidermal–specific deletion of CD44 reveals a function in keratinocytes in response to mechanical stress. Cell Death Dis. 2016, 7, e2461. [Google Scholar] [CrossRef] [PubMed]
- Lovvorn, H.N., III; Cass, D.L.; Sylvester, K.G.; Yang, E.Y.; Crombleholme, T.M.; Adzick, N.S.; Savani, R.C. Hyaluronan receptor expression increases in fetal excisional skin wounds and correlates with fibroplasia. J. Pediatr. Surg. 1998, 33, 1062–1069. [Google Scholar] [CrossRef]
- Tolg, C.; Hamilton, S.R.; Nakrieko, K.A.; Kooshesh, F.; Walton, P.; McCarthy, J.B.; Bissell, M.J.; Turley, E.A. Rhamm–/– fibroblasts are defective in CD44–mediated ERK1,2 motogenic signaling, leading to defective skin wound repair. J. Cell Biol. 2006, 175, 1017–1028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kouvidi, K.; Berdiaki, A.; Nikitovic, D.; Katonis, P.; Afratis, N.; Hascall, V.C.; Karamanos, N.K.; Tzanakakis, G.N. Role of receptor for hyaluronic acid–mediated motility (RHAMM) in low molecular weight hyaluronan (LMWHA)–mediated fibrosarcoma cell adhesion. J. Biol. Chem. 2011, 286, 38509–38520. [Google Scholar] [CrossRef] [Green Version]
- Wu, K.; Kim, S.; Liu, V.M.; Sabino, A.; Minkhorst, K.; Yazdani, A.; Turley, E.A. Function–blocking RHAMM peptides attenuate fibrosis and promote anti–fibrotic adipokines in a bleomycin–induced murine model of systemic sclerosis. J. Investig. Dermatol. 2020, 141, 1482–1492. [Google Scholar] [CrossRef]
- Gao, F.; Yang, C.X.; Mo, W.; Liu, Y.W.; He, Y.Q. Hyaluronan oligosaccharides are potential stimulators to angiogenesis via RHAMM mediated signal pathway in wound healing. Clin. Investig. Med. 2008, 31, 106–116. [Google Scholar] [CrossRef] [Green Version]
- Seifert, A.W.; Monaghan, J.R.; Voss, S.R.; Maden, M. Skin regeneration in adult axolotls: A blueprint for scar–free healing in vertebrates. PLoS ONE 2012, 7, e32875. [Google Scholar]
- Imanaka–Yoshida, K.; Tawara, I.; Yoshida, T. Tenascin–C in cardiac disease: A sophisticated controller of inflammation, repair, and fibrosis. Am. J. Physiol. Cell Physiol. 2020, 319, 781–796. [Google Scholar] [CrossRef]
- Walsh, H.R.; Cruickshank, B.M.; Brown, J.M.; Marcato, P. The Flick of a Switch: Conferring Survival Advantage to Breast Cancer Stem Cells Through Metabolic Plasticity. Front. Oncol. 2019, 9, 753. [Google Scholar] [CrossRef]
- Saeg, F.; Anbalagan, M. Breast cancer stem cells and the challenges of eradication: A review of novel therapies. Stem Cell Investig. 2018, 5, 39. [Google Scholar] [CrossRef]
- Louderbough, J.M.; Schroeder, J.A. Understanding the dual nature of CD44 in breast cancer progression. Mol. Cancer Res. 2011, 9, 1573–1586. [Google Scholar] [CrossRef] [Green Version]
- Huang, P.; Chen, A.; He, W.; Li, Z.; Zhang, G.; Liu, Z.; Liu, G.; Liu, X.; He, S.; Xiao, G.; et al. BMP–2 induces EMT and breast cancer stemness through Rb and CD44. Cell Death Discov. 2017, 3, 17039. [Google Scholar] [CrossRef]
- Muller, S.; Sindikubwabo, F.; Caneque, T.; Lafon, A.; Versini, A.; Lombard, B.; Loew, D.; Wu, T.D.; Ginestier, C.; Charafe–Jauffret, E.; et al. CD44 regulates epigenetic plasticity by mediating iron endocytosis. Nat. Chem. 2020, 12, 92–938. [Google Scholar] [CrossRef]
- Lopez, J.I.; Camenisch, T.D.; Stevens, M.V.; Sands, B.J.; McDonald, J.; Schroeder, J.A. CD44 attenuates metastatic invasion during breast cancer progression. Cancer Res. 2005, 65, 6755–6763. [Google Scholar] [CrossRef] [Green Version]
- Xu, H.; Niu, M.; Yuan, X.; Wu, K.; Liu, A. CD44 as a tumor biomarker and therapeutic target. Exp. Hematol. Oncol. 2020, 9, 36. [Google Scholar] [CrossRef]
- Ma, L.; Dong, L.; Chang, P. CD44v6 engages in colorectal cancer progression. Cell Death Dis. 2019, 10, 30. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Thor, A.D.; Moore, D.H., II; Zhao, Y.; Kerschmann, R.; Stern, R.; Watson, P.H.; Turley, E.A. The overexpression of RHAMM, a hyaluronan–binding protein that regulates ras signaling, correlates with overexpression of mitogen–activated protein kinase and is a significant parameter in breast cancer progression. Clin. Cancer Res. 1998, 4, 567–576. [Google Scholar] [PubMed]
- Wang, J.; Li, D.; Shen, W.; Sun, W.; Gao, R.; Jiang, P.; Wang, L.; Liu, Y.; Chen, Y.; Zhou, W.; et al. RHAMM inhibits cell migration via the AKT/GSK3beta/Snail axis in luminal A subtype breast cancer. Anat. Rec. 2020, 303, 2344–2356. [Google Scholar] [CrossRef]
- Hall, C.L.; Yang, B.; Yang, X.; Zhang, S.; Turley, M.; Samuel, S.; Lange, L.A.; Wang, C.; Curpen, G.D.; Savani, R.C.; et al. Overexpression of the hyaluronan receptor RHAMM is transforming and is also required for H–ras transformation. Cell 1995, 82, 19–26. [Google Scholar] [CrossRef] [Green Version]
- Kotla, N.G.; Bonam, S.R.; Rasala, S.; Wankar, J.; Bohara, R.A.; Bayry, J.; Rochev, Y.; Pandit, A. Recent advances and prospects of hyaluronan as a multifunctional therapeutic system. J. Control. Release 2021, 336, 598–620. [Google Scholar] [CrossRef]
- Salari, N.; Mansouri, K.; Valipour, E.; Abam, F.; Jaymand, M.; Rasoulpoor, S.; Dokaneheifard, S.; Mohammadi, M. Hyaluronic acid–based drug nanocarriers as a novel drug delivery system for cancer chemotherapy: A systematic review. Daru 2021, 1–9. [Google Scholar] [CrossRef]
- Kultti, A.; Pasonen–Seppänen, S.; Jauhiainen, M.; Rilla, K.J.; Kärnä, R.; Pyöriä, E.; Tammi, R.H.; Tammi, M.I. 4–Methylumbelliferone inhibits hyaluronan synthesis by depletion of cellular UDP–glucuronic acid and downregulation of hyaluronan synthase 2 and 3. Exp. Cell Res. 2009, 315, 1914–1923. [Google Scholar] [CrossRef] [PubMed]
- Yates, T.J.; Lopez, L.E.; Lokeshwar, S.D.; Ortiz, N.; Kallifatidis, G.; Jordan, A.; Hoye, K.; Altman, N.; Lokeshwar, V.B. Dietary Supplement 4–Methylumbelliferone: An Effective Chemopreventive and Therapeutic Agent for Prostate Cancer. J. Natl. Cancer Inst. 2015, 107, djv085. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kudo, D.; Suto, A.; Hakamada, K. The Development of a Novel Therapeutic Strategy to Target Hyaluronan in the Extracellular Matrix of Pancreatic Ductal Adenocarcinoma. Int. J. Mol. Sci. 2017, 18, 600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagy, N.; Kuipers, H.F.; Frymoyer, A.R.; Ishak, H.D.; Bollyky, J.B.; Wight, T.N.; Bollyky, P.L. 4–methylumbelliferone treatment and hyaluronan inhibition as a therapeutic strategy in inflammation, autoimmunity, and cancer. Front. Immunol. 2015, 6, 123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pereira, P.M.R.; Ragupathi, A.; Shmuel, S.; Mandleywala, K.; Viola, N.T.; Lewis, J.S. HER2–Targeted PET Imaging and Therapy of Hyaluronan–Masked HER2–Overexpressing Breast Cancer. Mol. Pharm. 2020, 17, 327–337. [Google Scholar] [CrossRef]
- Palyi–Krekk, Z.; Barok, M.; Isola, J.; Tammi, M.; Szollosi, J.; Nagy, P. Hyaluronan–induced masking of ErbB2 and CD44–enhanced trastuzumab internalisation in trastuzumab resistant breast cancer. Eur. J. Cancer 2007, 43, 2423–2433. [Google Scholar] [CrossRef] [Green Version]
- Wong, K.M.; Horton, K.J.; Coveler, A.L.; Hingorani, S.R.; Harris, W.P. Targeting the Tumor Stroma: The Biology and Clinical Development of Pegylated Recombinant Human Hyaluronidase (PEGPH20). Curr. Oncol. Rep. 2017, 19, 47. [Google Scholar] [CrossRef]
- Adel, N. Current treatment landscape and emerging therapies for pancreatic cancer. Am. J. Manag. Care 2019, 25, 3–10. [Google Scholar]
- Verdaguer, H.; Arroyo, A.; Macarulla, T. New Horizons in the Treatment of Metastatic Pancreatic Cancer: A Review of the Key Biology Features and the Most Recent Advances to Treat Metastatic Pancreatic Cancer. Target Oncol. 2018, 13, 691–704. [Google Scholar] [CrossRef]
- Brundel, D.H.; Feeney, O.M.; Nowell, C.J.; Suys, E.J.; Gracia, G.; Kaminskas, L.M.; McIntosh, M.M.; Kang, D.W.; Porter, C.J. Depolymerization of hyaluronan using PEGylated human recombinant hyaluronidase promotes nanoparticle tumor penetration. Nanomedicine 2021, 16, 275–292. [Google Scholar] [CrossRef] [PubMed]
- Chaudhry, G.E.; Akim, A.; Naveed Zafar, M.; Safdar, N.; Sung, Y.Y.; Muhammad, T.S.T. Understanding Hyaluronan Receptor (CD44) Interaction, HA–CD44 Activated Potential Targets in Cancer Therapeutics. Adv. Pharm. Bull. 2021, 11, 426–438. [Google Scholar] [CrossRef] [PubMed]
- Diebolder, P.; Mpoy, C.; Scott, J.; Huynh, T.T.; Fields, R.; Spitzer, D.; Bandara, N.; Rogers, B.E. Preclinical Evaluation of an Engineered Single–Chain Fragment Variable–Fragment Crystallizable Targeting Human CD44. J. Nucl. Med. 2021, 62, 137–143. [Google Scholar] [CrossRef]
- Menke–van der Houven van Oordt, C.W.; Gomez–Roca, C.; van Herpen, C.; Coveler, A.L.; Mahalingam, D.; Verheul, H.M.; van der Graaf, W.T.; Christen, R.; Rüttinger, D.; Weigand, S.; et al. First–in–human phase I clinical trial of RG7356, an anti–CD44 humanized antibody, in patients with advanced, CD44–expressing solid tumors. Oncotarget 2016, 7, 80046–80058. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tolg, C.; Messam, B.J.-A.; McCarthy, J.B.; Nelson, A.C.; Turley, E.A. Hyaluronan Functions in Wound Repair That Are Captured to Fuel Breast Cancer Progression. Biomolecules 2021, 11, 1551. https://doi.org/10.3390/biom11111551
Tolg C, Messam BJ-A, McCarthy JB, Nelson AC, Turley EA. Hyaluronan Functions in Wound Repair That Are Captured to Fuel Breast Cancer Progression. Biomolecules. 2021; 11(11):1551. https://doi.org/10.3390/biom11111551
Chicago/Turabian StyleTolg, Cornelia, Britney Jodi-Ann Messam, James Benjamin McCarthy, Andrew Cook Nelson, and Eva Ann Turley. 2021. "Hyaluronan Functions in Wound Repair That Are Captured to Fuel Breast Cancer Progression" Biomolecules 11, no. 11: 1551. https://doi.org/10.3390/biom11111551
APA StyleTolg, C., Messam, B. J. -A., McCarthy, J. B., Nelson, A. C., & Turley, E. A. (2021). Hyaluronan Functions in Wound Repair That Are Captured to Fuel Breast Cancer Progression. Biomolecules, 11(11), 1551. https://doi.org/10.3390/biom11111551






