Butyrate and Metformin Affect Energy Metabolism Independently of the Metabolic Phenotype in the Tumor Therapy Model
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Trypan Blue Cell Count
2.3. BALB/c Tumor Therapy Model
2.4. Glucose Measurement
2.5. Generation of Monoclonal Cell Lines
2.6. Giemsa Staining of Monoclonal Cell Clones
2.7. Growth Curves
2.8. Seahorse Analysis of Cells
2.9. Statistical Analysis
3. Results
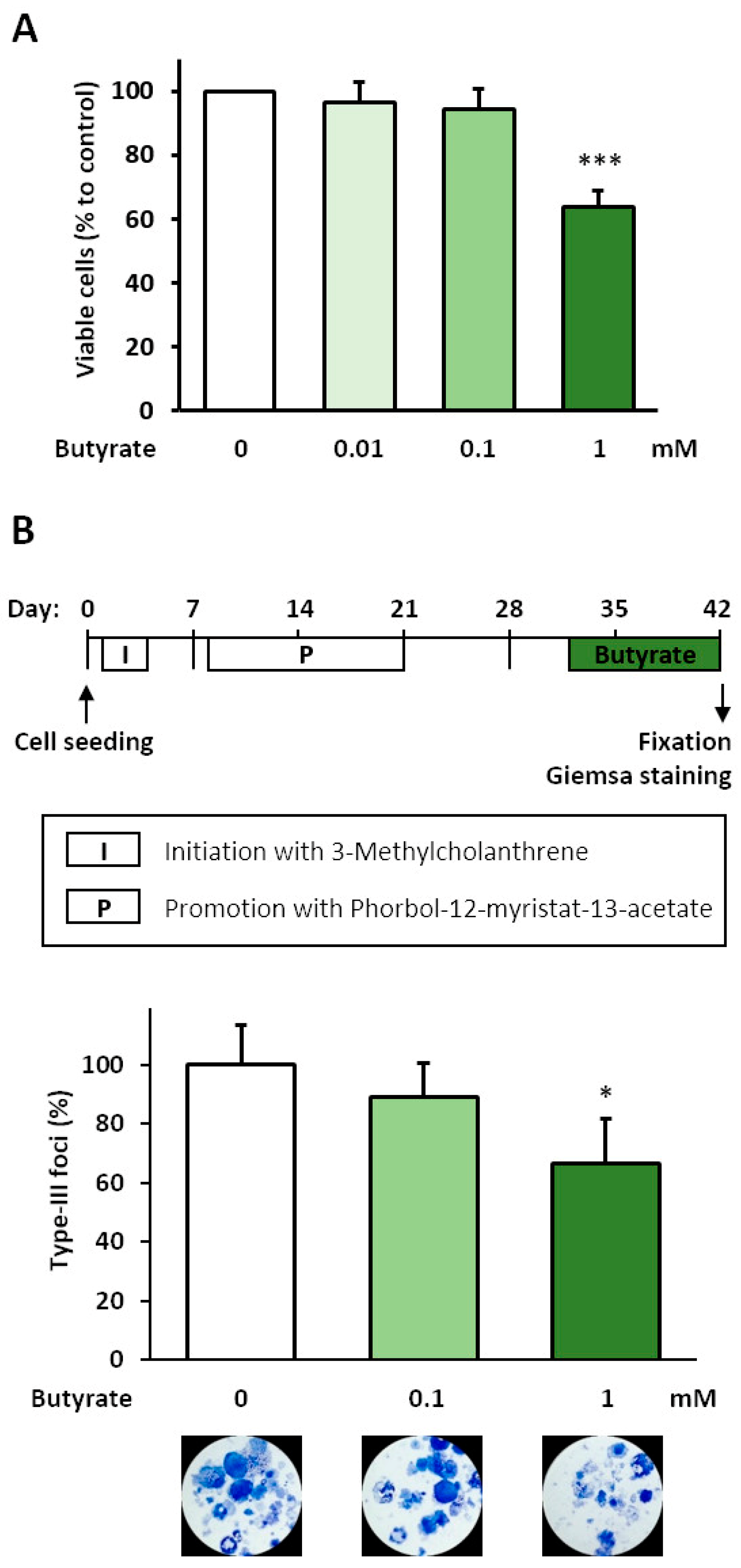
3.1. Analysis of Anti-Carcinogenic Effects of Butyrate in the BALB-TTM
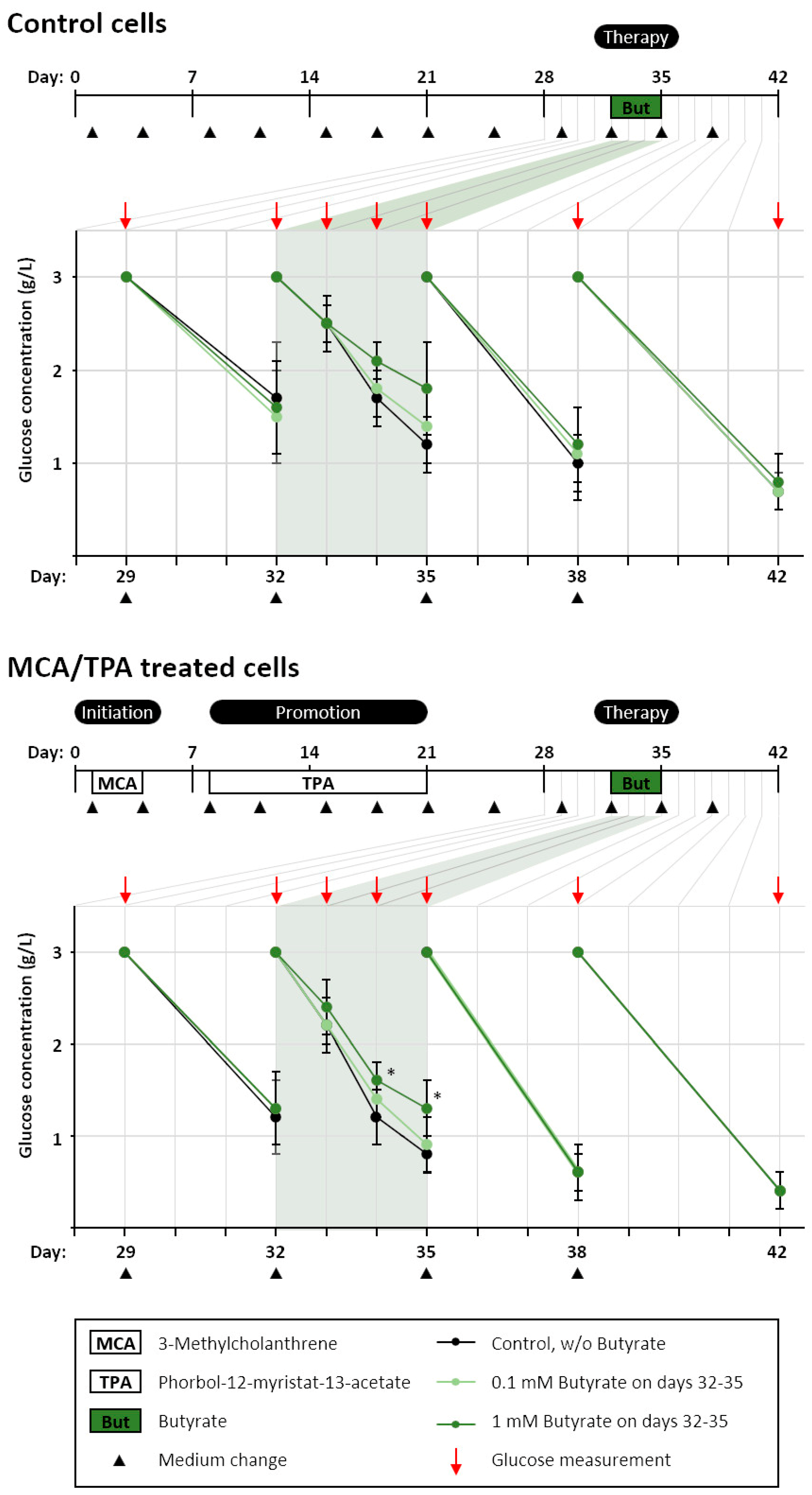
3.2. Glucose Consumption after Butyrate Treatment in the BALB-TTM
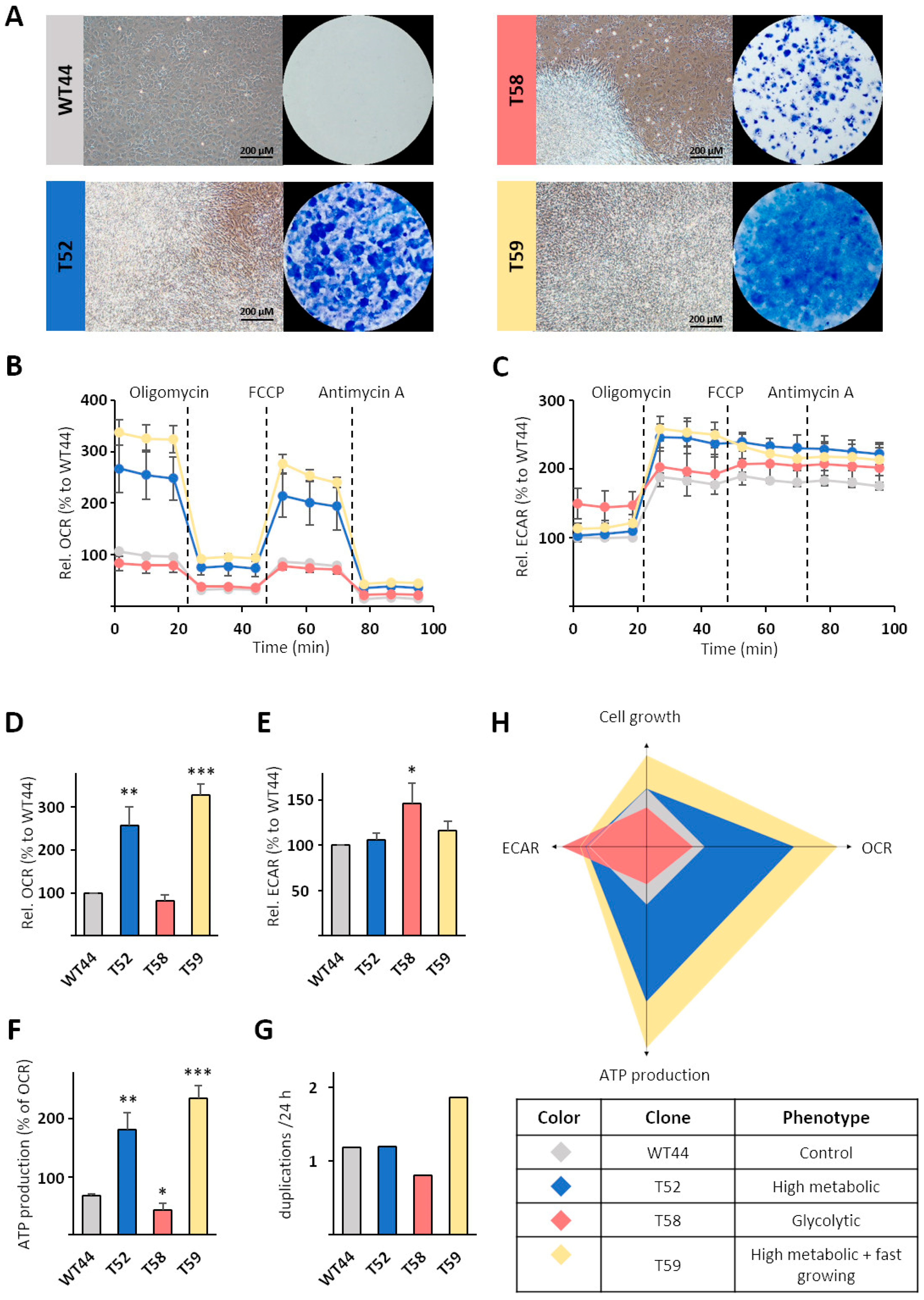
3.3. Generation of Monoclonal Cell Lines from the BALB-CTA
3.4. Metabolic Characterization of Selected Cell Clones
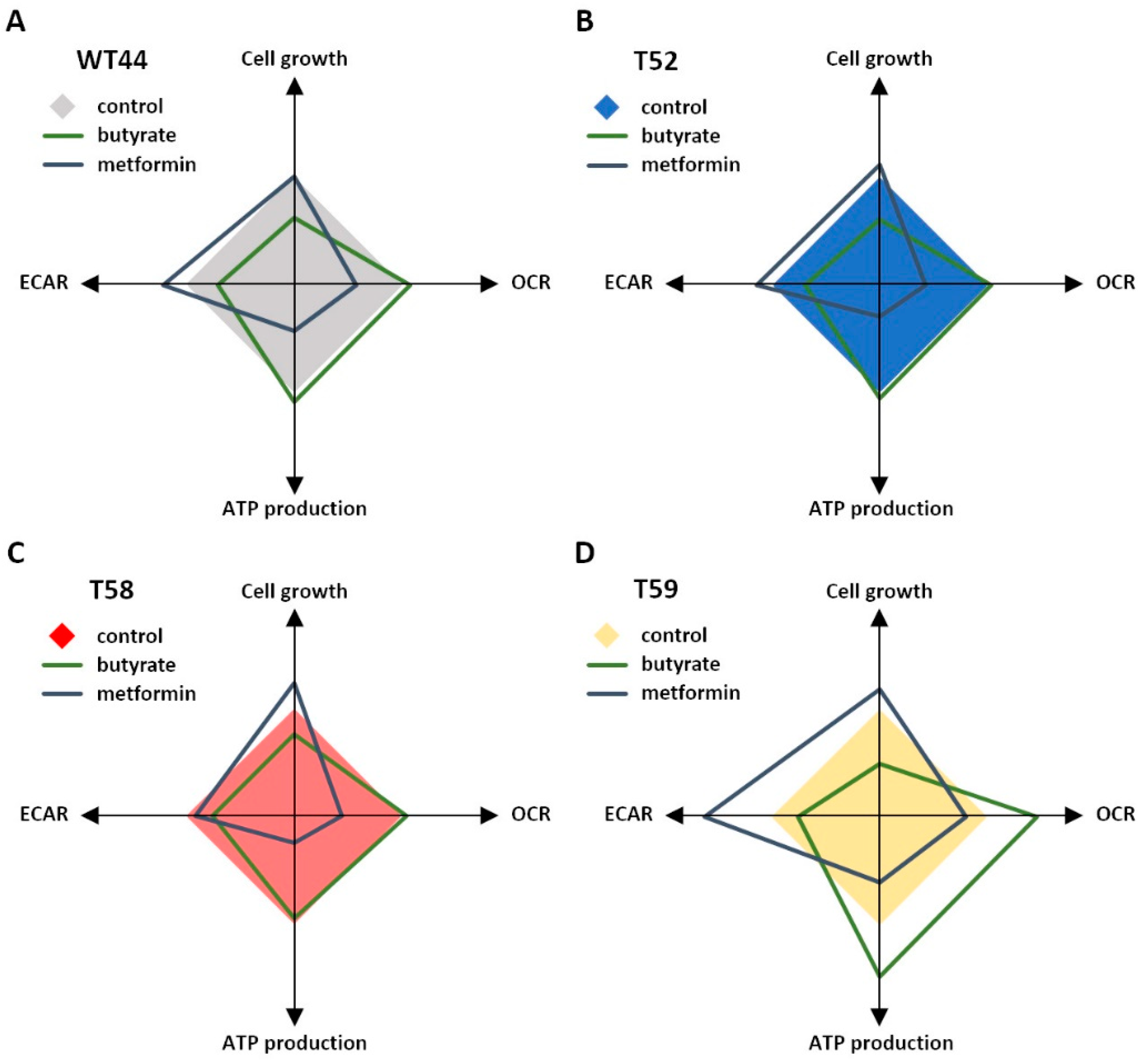
3.5. Treatment of Cell Clones with Butyrate and Metformin
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Warburg, O. On the origin of cancer cells. Science 1956, 123, 309–314. [Google Scholar] [CrossRef]
- Warburg, O. Über den Stoffwechsel der Carcinomzelle. Naturwissenschaften 1924, 12, 1131–1137. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- DeBerardinis, R.J.; Chandel, N.S. Fundamentals of cancer metabolism. Sci. Adv. 2016, 2, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Vander Heiden, M.G.; DeBerardinis, R.J. Understanding the Intersections between Metabolism and Cancer Biology. Cell 2017, 168, 657–669. [Google Scholar] [CrossRef] [PubMed]
- Farhadi, P.; Yarani, R.; Dokaneheifard, S.; Mansouri, K. The emerging role of targeting cancer metabolism for cancer therapy. Tumor Biol. 2020, 42, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Locasale, J.W.; Bielas, J.H.; O’Sullivan, J.; Sheahan, K.; Cantley, L.C.; Vander Heiden, M.G.; Vitkup, D. Heterogeneity of tumor-induced gene expression changes in the human metabolic network. Nat. Biotechnol. 2013, 31, 522–529. [Google Scholar] [CrossRef] [PubMed]
- Faubert, B.; Solmonson, A.; DeBerardinis, R.J. Metabolic reprogramming and cancer progression. Science 2020, 368, 1–12. [Google Scholar] [CrossRef]
- Zaal, E.A.; Berkers, C.R. The Influence of Metabolism on Drug Response in Cancer. Front. Oncol. 2018, 8, 1–15. [Google Scholar] [CrossRef]
- Desbats, M.A.; Giacomini, I.; Prayer-Galetti, T.; Montopoli, M. Metabolic Plasticity in Chemotherapy Resistance. Front. Oncol. 2020, 10, 1–20. [Google Scholar] [CrossRef]
- Poburski, D.; Thierbach, R. Improvement of the BALB/c-3T3 cell transformation assay: A tool for investigating cancer mechanisms and therapies. Sci. Rep. 2016, 6, 1–8. [Google Scholar] [CrossRef]
- Poburski, D.; Leovsky, C.; Boerner, J.B.; Szimmtenings, L.; Ristow, M.; Glei, M.; Thierbach, R. Insulin-IGF signaling affects cell transformation in the BALB/c 3T3 cell model. Sci. Rep. 2016, 6, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Meyer, F.B.; Goebel, S.; Spangel, S.B.; Leovsky, C.; Hoelzer, D.; Thierbach, R. Metformin alters therapeutic effects in the BALB/c tumor therapy model. BMC Cancer 2021, 21, 629. [Google Scholar] [CrossRef] [PubMed]
- Donohoe, D.R.; Collins, L.B.; Wali, A.; Bigler, R.; Sun, W.; Bultman, S.J. The Warburg effect dictates the mechanism of butyrate-mediated histone acetylation and cell proliferation. Mol. Cell 2012, 48, 612–626. [Google Scholar] [CrossRef] [PubMed]
- Polianskyte-Prause, Z.; Tolvanen, T.A.; Lindfors, S.; Dumont, V.; Van, M.; Wang, H.; Dash, S.N.; Berg, M.; Naams, J.B.; Hautala, L.C.; et al. Metformin increases glucose uptake and acts renoprotectively by reducing SHIP2 activity. FASEB J. 2019, 33, 2858–2869. [Google Scholar] [CrossRef]
- Sajan, M.P.; Bandyopadhyay, G.; Miura, A.; Standaert, M.L.; Nimal, S.; Longnus, S.L.; Van Obberghen, E.; Hainault, I.; Foufelle, F.; Kahn, R.; et al. AICAR and metformin, but not exercise, increase muscle glucose transport through AMPK-, ERK-, and PDK1-dependent activation of atypical PKC. Am. J. Physiol.-Endoc. Metab. 2010, 298, E179–E192. [Google Scholar] [CrossRef]
- Harada, K.; Ferdous, T.; Harada, T.; Ueyama, Y. Metformin in combination with 5-fluorouracil suppresses tumor growth by inhibiting the Warburg effect in human oral squamous cell carcinoma. Int. J. Oncol. 2016, 49, 276–284. [Google Scholar] [CrossRef]
- Jia, Y.L.; Ma, Z.Y.; Liu, X.F.; Zhou, W.J.; He, S.; Xu, X.; Ren, G.J.; Xu, G.; Tian, K.L. Metformin prevents DMH-induced colorectal cancer in diabetic rats by reversing the warburg effect. Cancer Med. 2015, 4, 1730–1741. [Google Scholar] [CrossRef] [PubMed]
- Marini, C.; Salani, B.; Massollo, M.; Amaro, A.; Esposito, A.I.; Orengo, A.M.; Capitanio, S.; Emionite, L.; Riondato, M.; Bottoni, G.; et al. Direct inhibition of hexokinase activity by metformin at least partially impairs glucose metabolism and tumor growth in experimental breast cancer. Cell Cycle 2013, 12, 3490–3499. [Google Scholar] [CrossRef]
- Salani, B.; Marini, C.; Rio, A.D.; Ravera, S.; Massollo, M.; Orengo, A.M.; Amaro, A.; Passalacqua, M.; Maffioli, S.; Pfeffer, U.; et al. Metformin impairs glucose consumption and survival in Calu-1 cells by direct inhibition of hexokinase-II. Sci. Rep. 2013, 3, 1–8. [Google Scholar] [CrossRef]
- Tang, D.H.; Xu, L.; Zhang, M.M.; Dorfman, R.G.; Pan, Y.D.; Zhou, Q.; Zhou, L.X.; Wang, Y.M.; Li, Y.; Yin, Y.Y.; et al. Metformin facilitates BG45-induced apoptosis via an anti-Warburg effect in cholangiocarcinoma cells. Oncol. Rep. 2018, 39, 1957–1965. [Google Scholar] [CrossRef]
- Sasaki, K.; Bohnenberger, S.; Hayashi, K.; Kunkelmann, T.; Muramatsu, D.; Poth, A.; Sakai, A.; Salovaara, S.; Tanaka, N.; Thomas, B.C.; et al. Photo catalogue for the classification of foci in the BALB/c 3T3 cell transformation assay. Mutat. Res. 2012, 744, 42–53. [Google Scholar] [CrossRef] [PubMed]
- McFarland, D.C. Preparation of pure cell cultures by cloning. Methods Cell Sci. 2000, 22, 63–66. [Google Scholar] [CrossRef] [PubMed]
- Skehan, P.; Storeng, R.; Scudiero, D.; Monks, A.; McMahon, J.; Vistica, D.; Warren, J.T.; Bokesch, H.; Kenney, S.; Boyd, M.R. New colorimetric cytotoxicity assay for anticancer-drug screening. J. Natl. Cancer Inst. 1990, 82, 1107–1112. [Google Scholar] [CrossRef]
- Lajqi, T.; Marx, C.; Hudalla, H.; Haas, F.; Grosse, S.; Wang, Z.Q.; Heller, R.; Bauer, M.; Wetzker, R.; Bauer, R. The Role of the Pathogen Dose and PI3Kgamma in Immunometabolic Reprogramming of Microglia for Innate Immune Memory. Int. J. Mol. Sci. 2021, 22, 2578. [Google Scholar] [CrossRef]
- Glass, G.V.; Peckham, P.D.; Sanders, J.R. Consequences of Failure to Meet Assumptions Underlying Fixed Effects Analyses of Variance and Covariance. Rev. Educ. Res. 1972, 42, 237–288. [Google Scholar] [CrossRef]
- Harwell, M.R.; Rubinstein, E.N.; Hayes, W.S.; Olds, C.C. Summarizing Monte-Carlo Results in Methodological Research - the 1-Factor and 2-Factor Fixed Effects Anova Cases. J. Educ. Stat. 1992, 17, 315–339. [Google Scholar] [CrossRef]
- Xu, S.; Liu, C.X.; Xu, W.; Huang, L.; Zhao, J.Y.; Zhao, S.M. Butyrate induces apoptosis by activating PDC and inhibiting complex I through SIRT3 inactivation. Sig. Transduct. Target Ther. 2017, 2, 1–9. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhang, W.; Zhang, S.L.; Hu, X.; Tam, K.Y. Phenyl butyrate inhibits pyruvate dehydrogenase kinase 1 and contributes to its anti-cancer effect. Eur. J. Pharm. Sci. 2017, 110, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Stanta, G.; Bonin, S. Overview on Clinical Relevance of Intra-Tumor Heterogeneity. Front. Med. 2018, 5, 1–10. [Google Scholar] [CrossRef]
- Schwaederle, M.; Zhao, M.; Lee, J.J.; Eggermont, A.M.; Schilsky, R.L.; Mendelsohn, J.; Lazar, V.; Kurzrock, R. Impact of Precision Medicine in Diverse Cancers: A Meta-Analysis of Phase II Clinical Trials. J. Clin. Oncol. 2015, 33, 3817–3825. [Google Scholar] [CrossRef] [PubMed]
- Stockley, T.L.; Oza, A.M.; Berman, H.K.; Leighl, N.B.; Knox, J.J.; Shepherd, F.A.; Chen, E.X.; Krzyzanowska, M.K.; Dhani, N.; Joshua, A.M.; et al. Molecular profiling of advanced solid tumors and patient outcomes with genotype-matched clinical trials: The Princess Margaret IMPACT/COMPACT trial. Genome Med. 2016, 8, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Jardim, D.L.; Schwaederle, M.; Wei, C.; Lee, J.J.; Hong, D.S.; Eggermont, A.M.; Schilsky, R.L.; Mendelsohn, J.; Lazar, V.; Kurzrock, R. Impact of a Biomarker-Based Strategy on Oncology Drug Development: A Meta-analysis of Clinical Trials Leading to FDA Approval. J. Natl. Cancer Inst. 2015, 107, 1–11. [Google Scholar] [CrossRef]
- Iorio, F.; Knijnenburg, T.A.; Vis, D.J.; Bignell, G.R.; Menden, M.P.; Schubert, M.; Aben, N.; Goncalves, E.; Barthorpe, S.; Lightfoot, H.; et al. A Landscape of Pharmacogenomic Interactions in Cancer. Cell 2016, 166, 740–754. [Google Scholar] [CrossRef] [PubMed]
- Gillet, J.P.; Varma, S.; Gottesman, M.M. The clinical relevance of cancer cell lines. J. Natl. Cancer Inst. 2013, 105, 452–458. [Google Scholar] [CrossRef]
- Doctor, A.; Seifert, V.; Ullrich, M.; Hauser, S.; Pietzsch, J. Three-Dimensional Cell Culture Systems in Radiopharmaceutical Cancer Research. Cancers 2020, 12, 765. [Google Scholar] [CrossRef]
- Forcella, M.; Callegaro, G.; Melchioretto, P.; Gribaldo, L.; Frattini, M.; Stefanini, F.M.; Fusi, P.; Urani, C. Cadmium-transformed cells in the in vitro cell transformation assay reveal different proliferative behaviours and activated pathways. Toxicol. In Vitro 2016, 36, 71–80. [Google Scholar] [CrossRef] [PubMed]
- Reznikoff, C.A.; Bertram, J.S.; Brankow, D.W.; Heidelberger, C. Quantitative and qualitative studies of chemical transformation of cloned C3H mouse embryo cells sensitive to postconfluence inhibition of cell division. Cancer Res. 1973, 33, 3239–3249. [Google Scholar] [PubMed]
- Male, R.; Bjerkvig, R.; Lillehaug, J.R. Biological and biochemical characterization of cell lines derived from initiation-promotion transformed C3H/10T1/2 cells. Carcinogenesis 1987, 8, 1375–1383. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R. The biology and function of fibroblasts in cancer. Nat. Rev. Cancer 2016, 16, 582–598. [Google Scholar] [CrossRef]
- Hamer, H.M.; Jonkers, D.; Venema, K.; Vanhoutvin, S.; Troost, F.J.; Brummer, R.J. Review article: The role of butyrate on colonic function. Aliment. Pharmacol. Ther. 2008, 27, 104–119. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Wu, Y.; He, L.; Wu, L.; Wang, X.; Liu, Z. Effects of the intestinal microbial metabolite butyrate on the development of colorectal cancer. J. Cancer 2018, 9, 2510–2517. [Google Scholar] [CrossRef] [PubMed]
- Hajjar, R.; Richard, C.S.; Santos, M.M. The role of butyrate in surgical and oncological outcomes in colorectal cancer. Am J Physiol Gastrointest. Liver Physiol. 2021, 320, G601–G608. [Google Scholar] [CrossRef] [PubMed]
- Geng, H.W.; Yin, F.Y.; Zhang, Z.F.; Gong, X.; Yang, Y. Butyrate Suppresses Glucose Metabolism of Colorectal Cancer Cells via GPR109a-AKT Signaling Pathway and Enhances Chemotherapy. Front. Mol. Biosci. 2021, 8, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Reckzeh, E.S.; Karageorgis, G.; Schwalfenberg, M.; Ceballos, J.; Nowacki, J.; Stroet, M.C.M.; Binici, A.; Knauer, L.; Brand, S.; Choidas, A.; et al. Inhibition of Glucose Transporters and Glutaminase Synergistically Impairs Tumor Cell Growth. Cell Chem. Biol. 2019, 26, 1214–1228.e1225. [Google Scholar] [CrossRef]
- Hensley, C.T.; Faubert, B.; Yuan, Q.; Lev-Cohain, N.; Jin, E.; Kim, J.; Jiang, L.; Ko, B.; Skelton, R.; Loudat, L.; et al. Metabolic Heterogeneity in Human Lung Tumors. Cell 2016, 164, 681–694. [Google Scholar] [CrossRef]
- Terunuma, A.; Putluri, N.; Mishra, P.; Mathe, E.A.; Dorsey, T.H.; Yi, M.; Wallace, T.A.; Issaq, H.J.; Zhou, M.; Killian, J.K.; et al. MYC-driven accumulation of 2-hydroxyglutarate is associated with breast cancer prognosis. J. Clin. Investig. 2014, 124, 398–412. [Google Scholar] [CrossRef]
- Pecinova, A.; Brazdova, A.; Drahota, Z.; Houstek, J.; Mracek, T. Mitochondrial targets of metformin—Are they physiologically relevant? BioFactors 2019, 45, 703–711. [Google Scholar] [CrossRef] [PubMed]
- Birsoy, K.; Possemato, R.; Lorbeer, F.K.; Bayraktar, E.C.; Thiru, P.; Yucel, B.; Wang, T.; Chen, W.W.; Clish, C.B.; Sabatini, D.M. Metabolic determinants of cancer cell sensitivity to glucose limitation and biguanides. Nature 2014, 508, 108–112. [Google Scholar] [CrossRef]
- Alhourani, A.H.; Tidwell, T.R.; Bokil, A.A.; Rosland, G.V.; Tronstad, K.J.; Soreide, K.; Hagland, H.R. Metformin treatment response is dependent on glucose growth conditions and metabolic phenotype in colorectal cancer cells. Sci. Rep. 2021, 11, 1–10. [Google Scholar] [CrossRef]
- Kim, J.; DeBerardinis, R.J. Mechanisms and Implications of Metabolic Heterogeneity in Cancer. Cell Metab. 2019, 30, 434–446. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meyer, F.B.; Marx, C.; Spangel, S.B.; Thierbach, R. Butyrate and Metformin Affect Energy Metabolism Independently of the Metabolic Phenotype in the Tumor Therapy Model. Biomolecules 2021, 11, 1831. https://doi.org/10.3390/biom11121831
Meyer FB, Marx C, Spangel SB, Thierbach R. Butyrate and Metformin Affect Energy Metabolism Independently of the Metabolic Phenotype in the Tumor Therapy Model. Biomolecules. 2021; 11(12):1831. https://doi.org/10.3390/biom11121831
Chicago/Turabian StyleMeyer, Felix B., Christian Marx, Sonja B. Spangel, and René Thierbach. 2021. "Butyrate and Metformin Affect Energy Metabolism Independently of the Metabolic Phenotype in the Tumor Therapy Model" Biomolecules 11, no. 12: 1831. https://doi.org/10.3390/biom11121831
APA StyleMeyer, F. B., Marx, C., Spangel, S. B., & Thierbach, R. (2021). Butyrate and Metformin Affect Energy Metabolism Independently of the Metabolic Phenotype in the Tumor Therapy Model. Biomolecules, 11(12), 1831. https://doi.org/10.3390/biom11121831





