Identification and Validation of PLOD2 as an Adverse Prognostic Biomarker for Oral Squamous Cell Carcinoma
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients and Samples
2.2. GC–MS-Based Untargeted Analysis
2.3. UHPLC-MS/MS Targeted Quantitative Analysis
2.4. Public Databases Analysis
2.5. Western Blot Assay
2.6. IHC Analysis
2.7. Quantification of IHC
2.8. Gene Set Enrichment Analysis
2.9. Statistical Analysis
3. Results
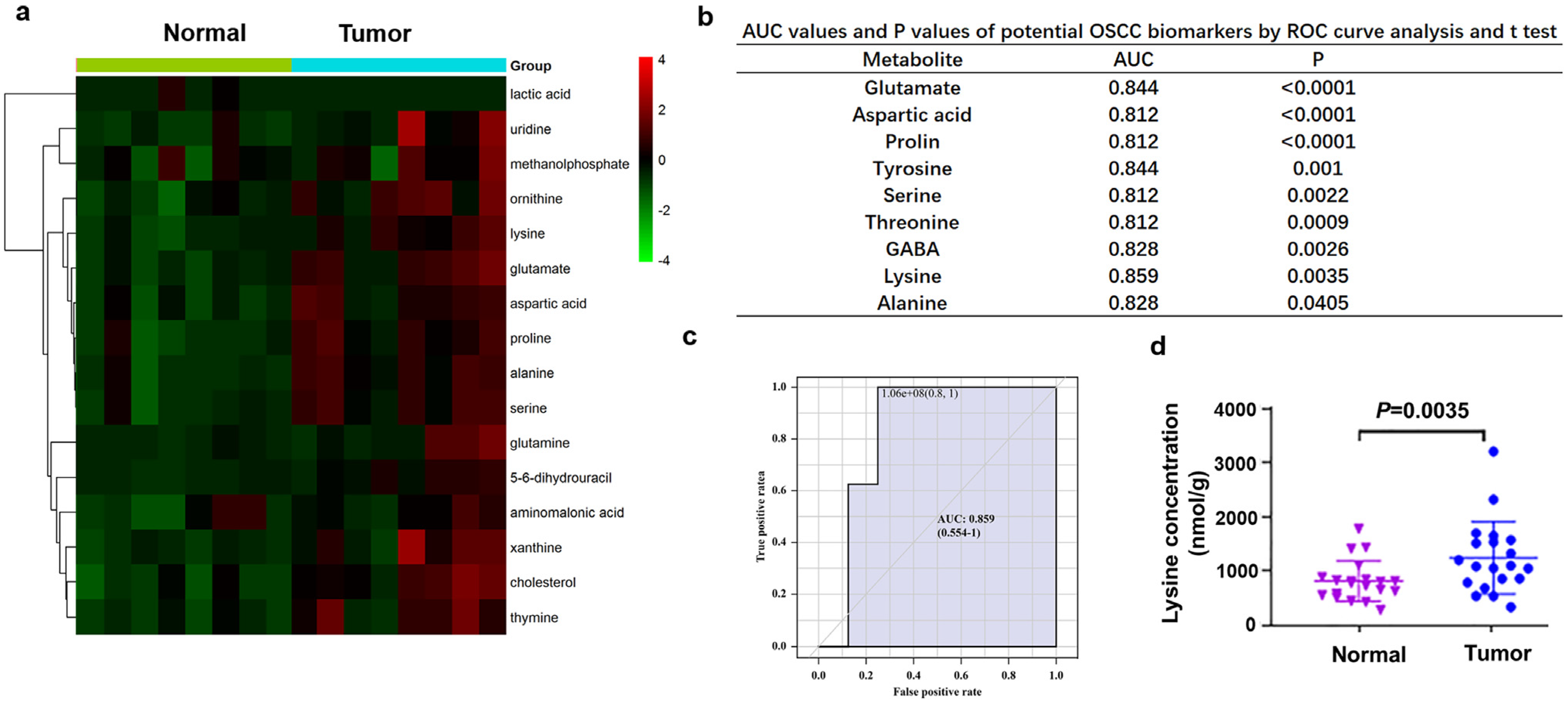
3.1. Lysine was Significantly Elevated in OSCC Tissues
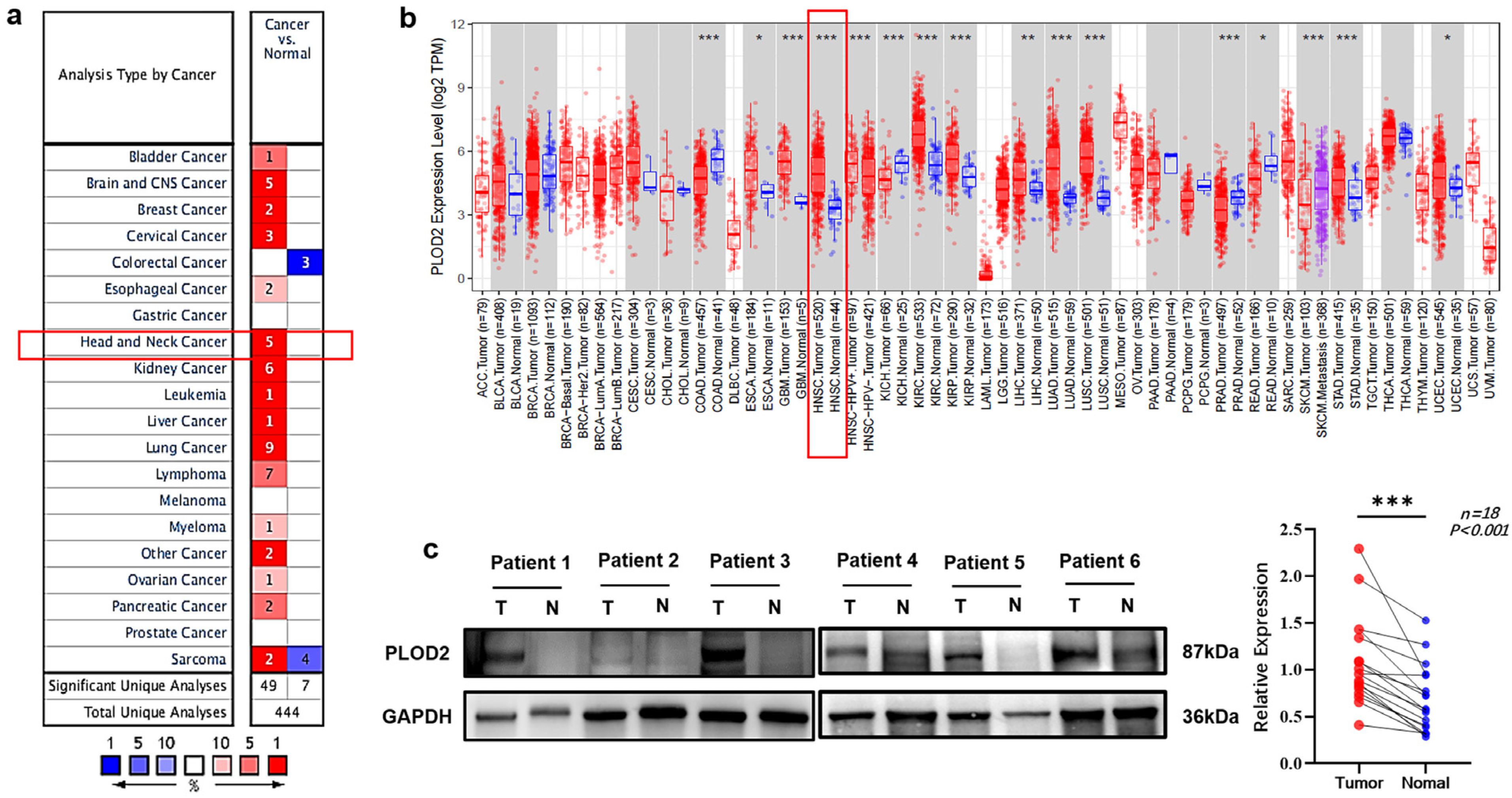
3.2. PLOD2 mRNA and Protein Levels Were Upregulated in OSCC Patients
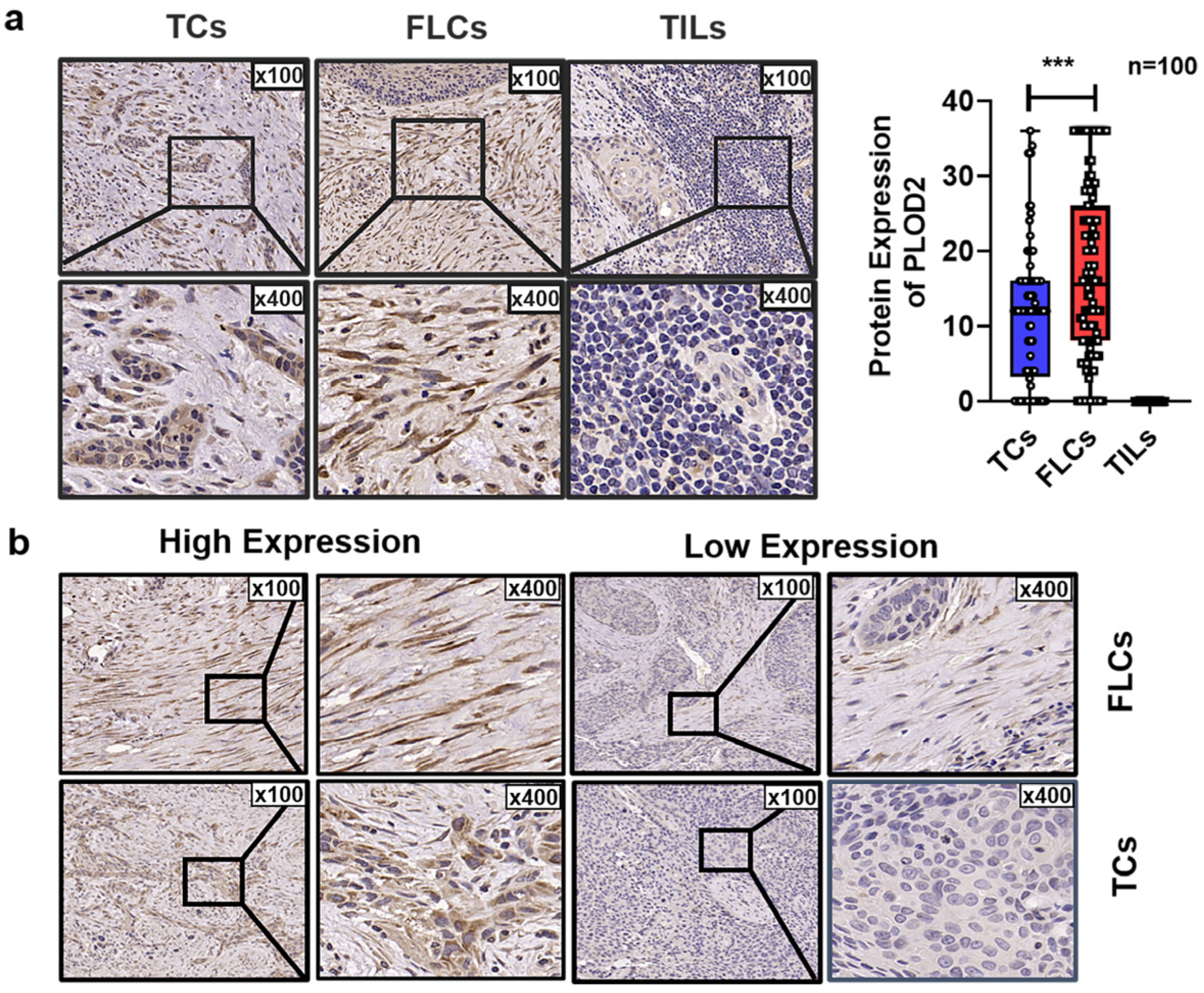
3.3. Spatial Distribution of PLOD2 and Its Correlation with Clinicopathological Characteristics of OSCC Patients
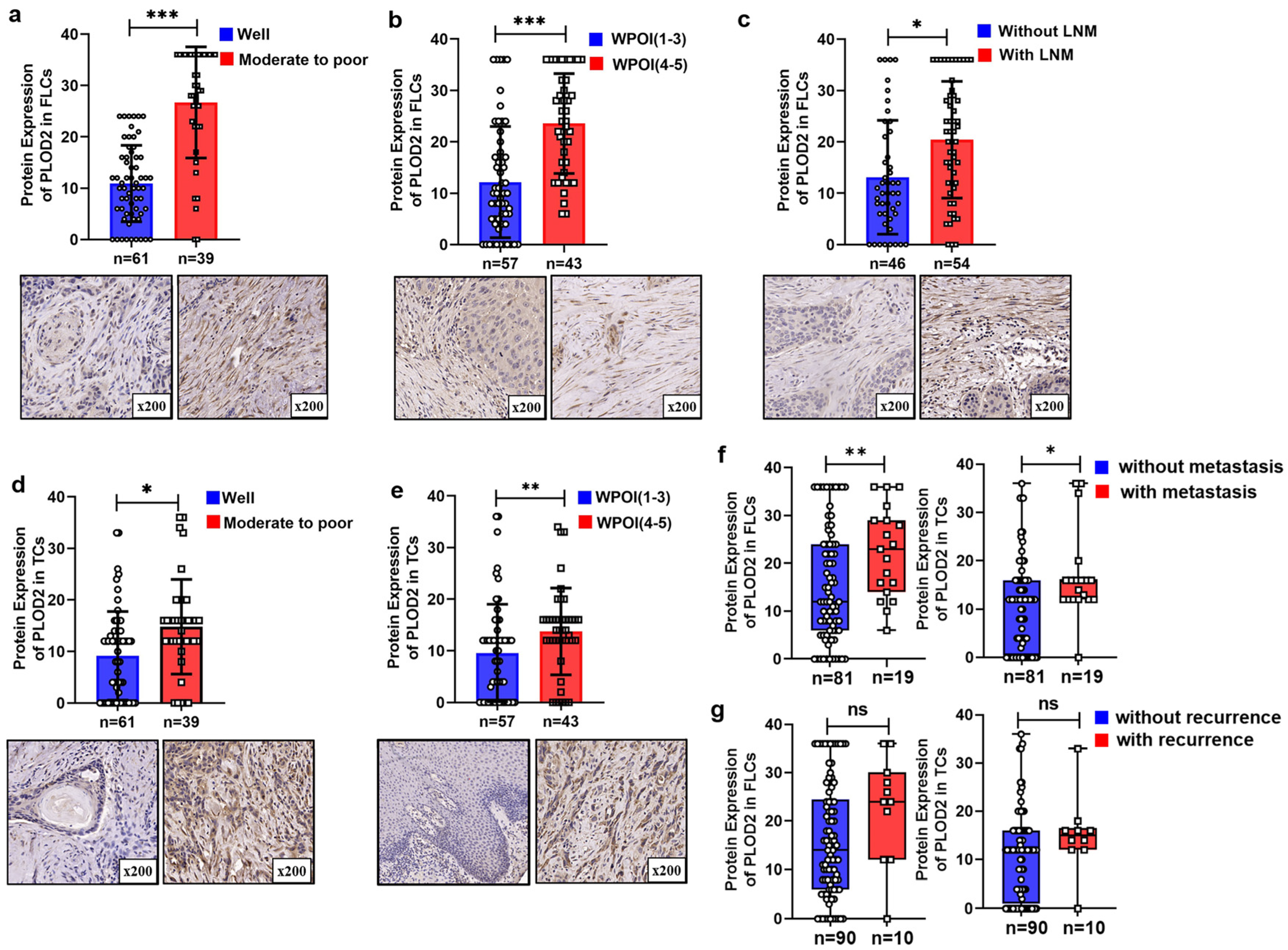
3.4. Highly Expressed PLOD2 Was Associated with Higher Postoperative Metastasis Risk and Poor Survival Time
3.5. PLOD2FLCs Was an Independent Prognostic Factor for OSCC
3.6. Signaling Pathways Involved in PLOD2 Regulation in HNSCC
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Torre, L.A.; Bray, F.; Siegel, R.L.; Ferlay, J.; Lortet-Tieulent, J.; Jemal, A. Global cancer statistics, 2012. CA Cancer J. Clin. 2015, 65, 87–108. [Google Scholar] [CrossRef] [Green Version]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Faubert, B.; Solmonson, A.; DeBerardinis, R.J. Metabolic reprogramming and cancer progression. Science 2020, 368. [Google Scholar] [CrossRef]
- Schmidt, D.R.; Patel, R.; Kirsch, D.G.; Lewis, C.A.; Vander Heiden, M.G.; Locasale, J.W. Metabolomics in cancer research and emerging applications in clinical oncology. CA Cancer J. Clin. 2021, 71, 333–358. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.H.; Zhang, X.X.; Jing, Y.; Ding, L.; Fu, Y.; Wang, S.; Hu, S.Q.; Zhang, L.; Huang, X.F.; Ni, Y.H.; et al. Amino acids signatures of distance-related surgical margins of oral squamous cell carcinoma. EBioMedicine 2019, 48, 81–91. [Google Scholar] [CrossRef] [Green Version]
- Qi, Y.; Xu, R. Roles of PLODs in Collagen Synthesis and Cancer Progression. Front. Cell Dev. Biol. 2018, 6, 66. [Google Scholar] [CrossRef] [PubMed]
- Quail, D.F.; Joyce, J.A. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 2013, 19, 1423–1437. [Google Scholar] [CrossRef] [PubMed]
- Myllyharju, J.; Kivirikko, K.I. Collagens, modifying enzymes and their mutations in humans, flies and worms. Trends Genet. 2004, 20, 33–43. [Google Scholar] [CrossRef]
- Gilkes, D.M.; Bajpai, S.; Chaturvedi, P.; Wirtz, D.; Semenza, G.L. Hypoxia-inducible factor 1 (HIF-1) promotes extracellular matrix remodeling under hypoxic conditions by inducing P4HA1, P4HA2, and PLOD2 expression in fibroblasts. J. Biol. Chem. 2013, 288, 10819–10829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piersma, B.; Bank, R.A. Collagen cross-linking mediated by lysyl hydroxylase 2: An enzymatic battlefield to combat fibrosis. Essays Biochem. 2019, 63, 377–387. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Zhao, Y.; Wang, L.; Zhao, Y.; Wei, L.; Chen, D.; Chen, Z. Identification of PLOD Family Genes as Novel Prognostic Biomarkers for Hepatocellular Carcinoma. Front. Oncol. 2020, 10, 1695. [Google Scholar] [CrossRef]
- Sada, M.; Ohuchida, K.; Horioka, K.; Okumura, T.; Moriyama, T.; Miyasaka, Y.; Ohtsuka, T.; Mizumoto, K.; Oda, Y.; Nakamura, M. Hypoxic stellate cells of pancreatic cancer stroma regulate extracellular matrix fiber organization and cancer cell motility. Cancer Lett. 2016, 372, 210–218. [Google Scholar] [CrossRef] [PubMed]
- Eisinger-Mathason, T.S.; Zhang, M.; Qiu, Q.; Skuli, N.; Nakazawa, M.S.; Karakasheva, T.; Mucaj, V.; Shay, J.E.; Stangenberg, L.; Sadri, N.; et al. Hypoxia-dependent modification of collagen networks promotes sarcoma metastasis. Cancer Discov. 2013, 3, 1190–1205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sato, K.; Parag-Sharma, K.; Terajima, M.; Musicant, A.M.; Murphy, R.M.; Ramsey, M.R.; Hibi, H.; Yamauchi, M.; Amelio, A.L. Lysyl hydroxylase 2-induced collagen cross-link switching promotes metastasis in head and neck squamous cell carcinomas. Neoplasia 2021, 23, 594–606. [Google Scholar] [CrossRef] [PubMed]
- Ueki, Y.; Saito, K.; Iioka, H.; Sakamoto, I.; Kanda, Y.; Sakaguchi, M.; Horii, A.; Kondo, E. PLOD2 Is Essential to Functional Activation of Integrin beta1 for Invasion/Metastasis in Head and Neck Squamous Cell Carcinomas. iScience 2020, 23, 100850. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saito, K.; Mitsui, A.; Sumardika, I.W.; Yokoyama, Y.; Sakaguchi, M.; Kondo, E. PLOD2-driven IL-6/STAT3 signaling promotes the invasion and metastasis of oral squamous cell carcinoma via activation of integrin beta1. Int. J. Oncol. 2021, 58. [Google Scholar] [CrossRef] [PubMed]
- Saito, T.; Uzawa, K.; Terajima, M.; Shiiba, M.; Amelio, A.L.; Tanzawa, H.; Yamauchi, M. Aberrant Collagen Cross-linking in Human Oral Squamous Cell Carcinoma. J. Dent. Res. 2019, 98, 517–525. [Google Scholar] [CrossRef]
- Zhu, N.; Ding, L.; Fu, Y.; Yang, Y.; Chen, S.; Chen, W.; Zhao, M.; Zhao, X.; Lu, Z.; Ni, Y.; et al. Tumor-infiltrating lymphocyte-derived MLL2 independently predicts disease-free survival for patients with early-stage oral squamous cell carcinoma. J. Oral. Pathol. Med. 2020, 49, 126–136. [Google Scholar] [CrossRef]
- Zhao, M.; Ding, L.; Yang, Y.; Chen, S.; Zhu, N.; Fu, Y.; Ni, Y.; Wang, Z. Aberrant Expression Of PDCD4/eIF4A1 Signal Predicts Postoperative Recurrence For Early-Stage Oral Squamous Cell Carcinoma. Cancer Manag. Res. 2019, 11, 9553–9562. [Google Scholar] [CrossRef] [Green Version]
- Han, S.; Huang, T.; Wu, X.; Wang, X.; Li, W.; Liu, S.; Yang, W.; Shi, Q.; Li, H.; Shi, K.; et al. Prognostic value of ALDH1 and Nestin in advanced cancer: A systematic meta-analysis with trial sequential analysis. Ther. Adv. Med. Oncol. 2019, 11, 1758835919830831. [Google Scholar] [CrossRef] [Green Version]
- Miyamoto, K.; Seki, N.; Matsushita, R.; Yonemori, M.; Yoshino, H.; Nakagawa, M.; Enokida, H. Tumour-suppressive miRNA-26a-5p and miR-26b-5p inhibit cell aggressiveness by regulating PLOD2 in bladder cancer. Br. J. Cancer 2016, 115, 354–363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurozumi, A.; Kato, M.; Goto, Y.; Matsushita, R.; Nishikawa, R.; Okato, A.; Fukumoto, I.; Ichikawa, T.; Seki, N. Regulation of the collagen cross-linking enzymes LOXL2 and PLOD2 by tumor-suppressive microRNA-26a/b in renal cell carcinoma. Int. J. Oncol. 2016, 48, 1837–1846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rajkumar, T.; Sabitha, K.; Vijayalakshmi, N.; Shirley, S.; Bose, M.V.; Gopal, G.; Selvaluxmy, G. Identification and validation of genes involved in cervical tumourigenesis. BMC Cancer 2011, 11, 80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, L.; Wang, W.; Li, X.; Gao, T. Association of ECRG4 with PLK1, CDK4, PLOD1 and PLOD2 in esophageal squamous cell carcinoma. Am. J. Transl. Res. 2017, 9, 3741–3748. [Google Scholar]
- Liu J, C.S.; Wang, W.; Ning, B.F.; Chen, F.; Shen, W.; Ding, J.; Chen, W.; Xie, W.F.; Zhang, X. Cancer-associated fibroblasts promote hepatocellular carcinoma metastasis through chemokine-activated hedgehog and TGF-β pathways. Cancer Lett. 2016, 379, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, J.; Liu, Y.; Wu, W.; Li, X.; Wu, Y.; Chen, H.; Zhang, K.; Gu, L. miR-101 represses lung cancer by inhibiting interaction of fibroblasts and cancer cells by down-regulating CXCL12. Biomed. Pharmacother. 2015, 74, 215–221. [Google Scholar] [CrossRef] [PubMed]
- Gaggioli, C.; Hooper, S.; Hidalgo-Carcedo, C.; Grosse, R.; Marshall, J.F.; Harrington, K.; Sahai, E. Fibroblast-led collective invasion of carcinoma cells with differing roles for RhoGTPases in leading and following cells. Nat. Cell Biol. 2007, 9, 1392–1400. [Google Scholar] [CrossRef] [PubMed]
- Slany, A.; Meshcheryakova, A.; Beer, A.; Ankersmit, H.J.; Paulitschke, V.; Gerner, C. Plasticity of fibroblasts demonstrated by tissue-specific and function-related proteome profiling. Clin. Proteom. 2014, 11, 41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bozoky, B.; Savchenko, A.; Csermely, P.; Korcsmaros, T.; Dul, Z.; Ponten, F.; Szekely, L.; Klein, G. Novel signatures of cancer-associated fibroblasts. Int. J. Cancer 2013, 133, 286–293. [Google Scholar] [CrossRef] [Green Version]
- Pankova, D.; Chen, Y.; Terajima, M.; Schliekelman, M.J.; Baird, B.N.; Fahrenholtz, M.; Sun, L.; Gill, B.J.; Vadakkan, T.J.; Kim, M.P.; et al. Cancer-Associated Fibroblasts Induce a Collagen Cross-link Switch in Tumor Stroma. Mol. Cancer Res. 2016, 14, 287–295. [Google Scholar] [CrossRef] [Green Version]
- Du, W.; Liu, N.; Zhang, Y.; Liu, X.; Yang, Y.; Chen, W.; He, Y. PLOD2 promotes aerobic glycolysis and cell progression in colorectal cancer by upregulating HK2. Biochem. Cell Biol. 2020, 98, 386–395. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Zhang, J.; Hu, G.; Liu, L.; Liang, W. Hypoxia and TGF-beta1 induced PLOD2 expression improve the migration and invasion of cervical cancer cells by promoting epithelial-to-mesenchymal transition (EMT) and focal adhesion formation. Cancer Cell Int. 2017, 17, 54. [Google Scholar] [CrossRef]
- Ding, Z.; He, Y.; Fu, Y.; Zhu, N.; Zhao, M.; Song, Y.; Huang, X.; Chen, S.; Yang, Y.; Zhang, C.; et al. CD38 Multi-Functionality in Oral Squamous Cell Carcinoma: Prognostic Implications, Immune Balance, and Immune Checkpoint. Front. Oncol. 2021, 11, 687430. [Google Scholar] [CrossRef] [PubMed]
- Noda, T.Y.H.; Takemasa, I.; Yamada, D.; Uemura, M.; Wada, H.; Kobayashi, S.; Marubashi, S.; Eguchi, H.; Tanemura, M.; Umeshita, K.; et al. PLOD2 induced under hypoxia is a novel prognostic factor for hepatocellular carcinoma after curative resection. Liver Int. 2012, 32, 110–118. [Google Scholar] [CrossRef] [PubMed]
- He, J.Y.; Wei, X.H.; Li, S.J.; Liu, Y.; Hu, H.L.; Li, Z.Z.; Kuang, X.H.; Wang, L.; Shi, X.; Yuan, S.T.; et al. Adipocyte-derived IL-6 and leptin promote breast Cancer metastasis via upregulation of Lysyl Hydroxylase-2 expression. Cell Commun. Signal. 2018, 16, 100. [Google Scholar] [CrossRef] [Green Version]
- Wan, J.; Qin, J.; Cao, Q.; Hu, P.; Zhong, C.; Tu, C. Hypoxia-induced PLOD2 regulates invasion and epithelial-mesenchymal transition in endometrial carcinoma cells. Genes Genom. 2020, 42, 317–324. [Google Scholar] [CrossRef] [PubMed]


| FILs | TCs | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Characteristics | N | Low (%) | High (%) | χ2 | p Value | N | Low (%) | High (%) | χ2 | p Value |
| Gender | ||||||||||
| Male | 58 | 31 (53.4%) | 27 (46.6%) | 0.369 | 0.543 | 58 | 32 (51.7%) | 26 (48.3%) | 0.095 | 0.758 |
| Female | 42 | 19 (45.2%) | 23 (54.8%) | 42 | 21 (65.9%) | 21 (34.1%) | ||||
| Age (year) | ||||||||||
| <60 | 43 | 22 (51.2%) | 21 (48.8%) | 0 | 1 | 43 | 26 (58.1%) | 17 (41.9%) | 1.203 | 0.273 |
| ≥60 | 57 | 28 (49.1% | 29 (50.9%) | 57 | 27 (57.1%) | 30 (42.9%) | ||||
| Smoking | ||||||||||
| NO | 60 | 26 (43.3%) | 34 (56.7%) | 2.042 | 0.153 | 60 | 27 (50.8%) | 33 (49.1%) | 3.093 | 0.079 |
| YES | 40 | 24 (60%) | 16 (40%) | 40 | 19 (57.5%) | 22 (42.5%) | ||||
| T stage | ||||||||||
| Ⅰ-Ⅱ | 59 | 31 (52.5%) | 28 (47.5%) | 0.165 | 0.684 | 59 | 34 (57.6%) | 25 (42.4%) | 0.825 | 0.364 |
| Ⅲ-Ⅳ | 41 | 19 (46.3%) | 22 (53.7%) | 41 | 19 (57.5%) | 22 (42.5%) | ||||
| LNM | ||||||||||
| NO | 46 | 34 (73.9%) | 12 (26.1%) | 17.754 | <0.001 * | 46 | 28 (66.7%) | 18 (33.3%) | 1.573 | 0.21 |
| YES | 54 | 16 (29.6%) | 38 (70.4%) | 54 | 25 (50%) | 29 (50%) | ||||
| Differentiation | ||||||||||
| Well | 61 | 41 (67.2%) | 20 (32.8%) | 16.814 | <0.001 * | 61 | 38 (67.8%) | 23 (32.2%) | 4.51 | 0.034 * |
| Moderate to poor | 39 | 9 (23.1%) | 30 (76.9%) | 39 | 15 (42.5%) | 24 (57.5%) | ||||
| WPOI | ||||||||||
| 1~3 | 57 | 40 (70.2%) | 17 (29.8%) | 19.747 | <0.001 * | 57 | 43 (75%) | 14 (25%) | 24.739 | <0.001 * |
| 4~5 | 43 | 10 (23.3%) | 33 (76.7%) | 43 | 10 (34.9% | 33 (65.1%) | ||||
| Variables | Univariate Analysis | p | Multivariate Analysis | p | ||
|---|---|---|---|---|---|---|
| HR | 95%CI | HR | 95%CI | |||
| OS | ||||||
| Gender: Male vs. Female | 1.647 | 0.779–3.482 | 0.192 | |||
| Age: ≥60 vs. <60 | 2.194 | 1.014–4.745 | 0.046 * | 2.219 | 1.005–4.903 | 0.049 * |
| Smoking: Yes vs. No | 0.751 | 0.362–1.560 | 0.443 | |||
| T Stage: III-IV vs. I-II | 0.881 | 0.430–1.802 | 0.728 | |||
| LNM: Yes vs. No | 1.119 | 0.556–2.252 | 0.752 | |||
| Differentiation: Moderate to poor vs. Well | 2.214 | 1.099–4.460 | 0.026 * | 1.528 | 0.736–3.169 | 0.255 |
| WPOI: 4–5 vs. 1–3 | 2.257 | 1.110–4.588 | 0.024 * | 0.833 | 0.370–1.876 | 0.659 |
| Metastasis: Yes vs. No | 0.951 | 0.391–2.314 | 0.913 | |||
| Recurrence: Yes vs. No | 0.862 | 0.262–2.839 | 0.807 | |||
| PLOD2 in FLCs: High vs. Low | 8.131 | 2.841–23.273 | 0.001 * | 6.127 | 2.046–18.347 | 0.001 * |
| PLOD2 in TCs: High vs. Low | 3.141 | 1.514–6.520 | 0.002 * | 2.202 | 0.975–4.974 | 0.057 |
| DFS | ||||||
| Gender: Male vs. Female | 1.336 | 0.648–2.756 | 0.433 | |||
| Age: ≥60 vs. <60 | 2.121 | 1.006–4.715 | 0.0048 * | 2.167 | 0.988–4.744 | 0.056 |
| Smoking: Yes vs. No | 0.604 | 0.292–1.250 | 0.174 | |||
| T Stage: III-IV vs. I-II | 0.813 | 0.399–1.657 | 0.569 | |||
| LNM: Yes vs. No | 1.158 | 0.582–2.301 | 0.676 | |||
| Differentiation: Moderate to poor vs. Well | 2.289 | 1.136–4.554 | 0.021 * | 1.367 | 0.685–2.868 | 0.45 |
| WPOI: 4–5 vs. 1–3 | 2.912 | 1.435–6.134 | 0.01 * | 1.168 | 0.548–2.865 | 0.654 |
| Metastasis: Yes vs. No | 1.428 | 0.712–3.865 | 0.189 | |||
| Recurrence: Yes vs. No | 1.452 | 0.654–4.658 | 0.356 | |||
| PLOD2 in FLCs: High vs. Low | 8.639 | 3.125–26.354 | 0.001 * | 6.425 | 2.068–19.267 | 0.002 * |
| PLOD2 in TCs: High vs. Low | 3.462 | 1.625–7.159 | 0.002 * | 2.065 | 0.954–4.769 | 0.068 |
| Variables | Univariate Analysis | p | Multivariate Analysis | p | ||
|---|---|---|---|---|---|---|
| HR | 95%CI | HR | 95%CI | |||
| RFS | ||||||
| Gender: Male vs. Female | 1.703 | 0.805–3.604 | 0.164 | |||
| Age: ≥60 vs. <60 | 2.227 | 1.029–4.819 | 0.042 * | 2.149 | 0.973–4.475 | 0.058 |
| Smoking: Yes vs. No | 0.684 | 0.329–1.420 | 0.308 | |||
| T Stage: III-IV vs. I-II | 0.9 | 0.439–1.842 | 0.773 | |||
| LNM: Yes vs. No | 1.183 | 0.588–2.380 | 0.638 | |||
| Differentiation: Moderate to poor vs. Well | 2.269 | 1.126–4.574 | 0.022 * | 1.438 | 0.693–2.987 | 0.33 |
| WPOI: 4–5 vs. 1–3 | 2.547 | 1.253–5.180 | 0.01 * | 0.943 | 0.409–2.173 | 0.891 |
| Metastasis: Yes vs. No | 0.881 | 0.362–2.141 | 0.779 | |||
| Recurrence: Yes vs. No | 1.97 | 0.589–6.584 | 0.271 | |||
| PLOD2 in FLCs: High vs. Low | 8.559 | 2.990–24.501 | 0.001 * | 6.024 | 1.975–18.373 | 0.002 * |
| PLOD2 in TCs: High vs. Low | 3.248 | 1.564–6.745 | 0.002 * | 1.972 | 0.847–4.593 | 0.115 |
| MFS | ||||||
| Gender: Male vs. Female | 1.448 | 0.686–3.059 | 0.332 | |||
| Age: ≥60 vs. <60 | 2.207 | 1.021–4.773 | 0.044 * | 2.176 | 0.994–4.764 | 0.052 |
| Smoking: Yes vs. No | 0.721 | 0.347–1.500 | 0.381 | |||
| T Stage: III-IV vs. I-II | 0.839 | 0.410–1.720 | 0.632 | |||
| LNM: Yes vs. No | 1.175 | 0.583–2.366 | 0.652 | |||
| Differentiation: Moderate to poor vs. Well | 2.298 | 1.141–4.626 | 0.02 * | 1.287 | 0.609–2.720 | 0.5 |
| WPOI: 4–5 vs. 1–3 | 3.41 | 1.622–7.168 | 0.001 * | 1.365 | 0.593–3.143 | 0.465 |
| Metastasis: Yes vs. No | 2.326 | 0.910–5.944 | 0.078 | |||
| Recurrence: Yes vs. No | 0.762 | 0.231–2.512 | 0.655 | |||
| PLOD2 in FLCs: High vs. Low | 9.933 | 3.340–28.768 | 0.001 * | 6.796 | 2.244–20.582 | 0.001 * |
| PLOD2 in TCs: High vs. Low | 3.587 | 1.722–7.472 | 0.001 * | 2.222 | 0.990–4.986 | 0.053 |
| TERM | ES | NES | NOM p-val | FDR q-val | FWER p-val |
|---|---|---|---|---|---|
| HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION | 0.74 | 3.51 | <0.001 | <0.001 | <0.001 |
| HALLMARK_UV_RESPONSE_DN | 0.62 | 2.83 | <0.001 | <0.001 | <0.001 |
| HALLMARK_MITOTIC_SPINDLE | 0.54 | 2.55 | <0.001 | <0.001 | <0.001 |
| HALLMARK_G2M_CHECKPOINT | 0.52 | 2.47 | <0.001 | <0.001 | <0.001 |
| HALLMARK_TGF_BETA_SIGNALING | 0.61 | 2.36 | <0.001 | <0.001 | <0.001 |
| HALLMARK_HYPOXIA | 0.49 | 2.31 | <0.001 | <0.001 | <0.001 |
| HALLMARK_ANGIOGENESIS | 0.62 | 2.21 | <0.001 | <0.001 | <0.001 |
| HALLMARK_PROTEIN_SECRETION | 0.51 | 2.19 | <0.001 | <0.001 | <0.001 |
| HALLMARK_HEDGEHOG_SIGNALING | 0.6 | 2.16 | <0.001 | <0.001 | <0.001 |
| HALLMARK_INFLAMMATORY_RESPONSE | 0.45 | 2.13 | <0.001 | <0.001 | <0.001 |
| HALLMARK_E2F_TARGETS | 0.44 | 2.11 | <0.001 | <0.001 | <0.001 |
| HALLMARK_KRAS_SIGNALING_UP | 0.44 | 2.08 | <0.001 | <0.001 | <0.001 |
| HALLMARK_TNFA_SIGNALING_VIA_NFKB | 0.42 | 2.01 | <0.001 | <0.001 | <0.001 |
| HALLMARK_ANDROGEN_RESPONSE | 0.46 | 1.99 | <0.001 | <0.001 | 0.001 |
| HALLMARK_IL6_JAK_STAT3_SIGNALING | 0.45 | 1.91 | <0.001 | <0.001 | 0.003 |
| HALLMARK_APICAL_JUNCTION | 0.4 | 1.91 | <0.001 | <0.001 | 0.003 |
| HALLMARK_GLYCOLYSIS | 0.4 | 1.9 | <0.001 | <0.001 | 0.003 |
| HALLMARK_COAGULATION | 0.41 | 1.87 | <0.001 | <0.001 | 0.004 |
| HALLMARK_IL2_STAT5_SIGNALING | 0.36 | 1.7 | <0.001 | 0.002 | 0.035 |
| HALLMARK_COMPLEMENT | 0.35 | 1.66 | <0.001 | 0.003 | 0.054 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, Y.; Wang, S.; Zhang, X.; Wu, Z.; Li, Z.; Ding, Z.; Huang, X.; Chen, S.; Jing, Y.; Zhang, X.; et al. Identification and Validation of PLOD2 as an Adverse Prognostic Biomarker for Oral Squamous Cell Carcinoma. Biomolecules 2021, 11, 1842. https://doi.org/10.3390/biom11121842
Sun Y, Wang S, Zhang X, Wu Z, Li Z, Ding Z, Huang X, Chen S, Jing Y, Zhang X, et al. Identification and Validation of PLOD2 as an Adverse Prognostic Biomarker for Oral Squamous Cell Carcinoma. Biomolecules. 2021; 11(12):1842. https://doi.org/10.3390/biom11121842
Chicago/Turabian StyleSun, Yawei, Shuai Wang, Xingwei Zhang, Zhuhao Wu, Zihui Li, Zhuang Ding, Xiaofeng Huang, Sheng Chen, Yue Jing, Xiaoxin Zhang, and et al. 2021. "Identification and Validation of PLOD2 as an Adverse Prognostic Biomarker for Oral Squamous Cell Carcinoma" Biomolecules 11, no. 12: 1842. https://doi.org/10.3390/biom11121842
APA StyleSun, Y., Wang, S., Zhang, X., Wu, Z., Li, Z., Ding, Z., Huang, X., Chen, S., Jing, Y., Zhang, X., Ding, L., Song, Y., Sun, G., & Ni, Y. (2021). Identification and Validation of PLOD2 as an Adverse Prognostic Biomarker for Oral Squamous Cell Carcinoma. Biomolecules, 11(12), 1842. https://doi.org/10.3390/biom11121842






