Salting-Out Approach Is Worthy of Comparison with Ultracentrifugation for Extracellular Vesicle Isolation from Tumor and Healthy Models
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Cultures and Conditioned Medium
2.2. Human Fluids
2.3. EVs Isolation
2.4. Nanoparticle Tracking Analysis (NTA)
2.5. Dynamic Light Scattering (DLS)
2.6. Transmission Electron Microscopy (TEM) Imaging
2.7. Flow Cytometry: Sample Preparation and Data Acquisition
2.8. Colorimetric Nanoplasmonic (CONAN) Assay
2.9. Western Blot Analysis
2.10. Statistical Analyses
3. Results
3.1. Physical Characteristics of EVs
3.2. Biochemical Characteristics of EVs
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Sáenz-Cuesta, M.; Arbelaiz, A.; Oregi, A.; Irizar, H.; Osorio-Querejeta, I.; Muñoz-Culla, M.; Banales, J.M.; Falcón-Pérez, J.M.; Olascoaga, J.; Otaegui, D. Methods for extracellular vesicles isolation in a hospital setting. Front. Immunol. 2015, 6, 50. [Google Scholar] [CrossRef]
- Mittal, S.; Gupta, P.; Chaluvally-Raghavan, P.; Pradeep, S. Emerging role of extracellular vesicles in immune regulation and cancer progression. Cancers 2020, 12, 3563. [Google Scholar] [CrossRef] [PubMed]
- Hosseinkhani, B.; Kuypers, S.; van den Akker, N.M.S.; Molin, D.G.M.; Michiels, L. Extracellular vesicles work as a functional inflammatory mediator between vascular endothelial cells and immune cells. Front. Immunol. 2018, 9, 1789. [Google Scholar] [CrossRef] [Green Version]
- Yang, E.; Wang, X.; Gong, Z.; Yu, M.; Wu, H.; Zhang, D. Exosome-mediated metabolic reprogramming: The emerging role in tumor microenvironment remodeling and its influence on cancer progression. Signal Transduct. Target. Ther. 2020, 5, 242. [Google Scholar] [CrossRef] [PubMed]
- Fu, S.; Zhang, Y.; Li, Y.; Luo, L.; Zhao, Y.; Yao, Y. Extracellular vesicles in cardiovascular diseases. Cell Death Discov. 2020, 6, 68. [Google Scholar] [CrossRef]
- György, B.; Szabó, T.G.; Pásztói, M.; Pál, Z.; Misják, P.; Aradi, B.; László, V.; Pállinger, É.; Pap, E.; Kittel, Á.; et al. Membrane vesicles, current state-of-the-art: Emerging role of extracellular vesicles. Cell. Mol. Life Sci. 2011, 68, 2667–2688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef] [Green Version]
- Stoorvogel, W.; Kleijmeer, M.J.; Geuze, H.J.; Raposo, G. The biogenesis and functions of exosomes. Traffic 2002, 3, 321–330. [Google Scholar] [CrossRef]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurian, T.K.; Banik, S.; Gopal, D.; Chakrabarti, S.; Mazumder, N. Elucidating Methods for Isolation and Quantification of Exosomes: A Review. Mol. Biotechnol. 2021, 63, 249–266. [Google Scholar] [CrossRef]
- Muralidharan-Chari, V.; Clancy, J.W.; Sedgwick, A.; D’Souza-Schorey, C. Microvesicles: Mediators of extracellular communication during cancer progression. J. Cell Sci. 2010, 123, 1603–1611. [Google Scholar] [CrossRef] [Green Version]
- Kakarla, R.; Hur, J.; Kim, Y.J.; Kim, J.; Chwae, Y.J. Apoptotic cell-derived exosomes: Messages from dying cells. Exp. Mol. Med. 2020, 52, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Kalluri, R. The biology and function of exosomes in cancer. J. Clin. Investig. 2016, 126, 1208–1215. [Google Scholar] [CrossRef]
- Skotland, T.; Hessvik, N.P.; Sandvig, K.; Llorente, A. Exosomal lipid composition and the role of ether lipids and phosphoinositides in exosome biology. J. Lipid Res. 2019, 60, 9–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lydic, T.A.; Townsend, S.; Adda, C.G.; Collins, C.; Mathivanan, S.; Reid, G.E. Rapid and comprehensive “shotgun” lipidome profiling of colorectal cancer cell derived exosomes. Methods 2015, 87, 83–95. [Google Scholar] [CrossRef] [Green Version]
- Deatheragea, B.L.; Cooksona, B.T. Membrane vesicle release in bacteria, eukaryotes, and archaea: A conserved yet underappreciated aspect of microbial life. Infect. Immun. 2012, 80, 1948–1957. [Google Scholar] [CrossRef] [Green Version]
- Schwechheimer, C.; Kuehn, M.J. Outer-membrane vesicles from Gram-negative bacteria: Biogenesis and functions. Nat. Rev. Microbiol. 2015, 13, 605–619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tannetta, D.; Dragovic, R.; Alyahyaei, Z.; Southcombe, J. Extracellular vesicles and reproduction-promotion of successful pregnancy. Cell. Mol. Immunol. 2014, 11, 548–563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Winters, C.M.; Chiang, H.-L. Yeast as a Model System to Study Trafficking of Small Vesicles Carrying Signal-less Proteins In and Out of the Cell. Curr. Protein Pept. Sci. 2016, 17, 808–820. [Google Scholar] [CrossRef] [PubMed]
- Kutralam-Muniasamy, G.; Flores-Cotera, L.B.; Perez-Guevara, F. Potential of yeast secretory vesicles in biodelivery systems. Drug Discov. Today 2015, 20, 659–666. [Google Scholar] [CrossRef]
- Beer, K.B.; Wehman, A.M. Mechanisms and functions of extracellular vesicle release in vivo—What we can learn from flies and worms. Cell Adhes. Migr. 2017, 11, 135–150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernández-Lázaro, D.; Hernández, J.L.G.; García, A.C.; Martínez, A.C.; Mielgo-Ayuso, J.; Cruz-Hernández, J.J. Liquid biopsy as novel tool in precision medicine: Origins, properties, identification and clinical perspective of cancer’s biomarkers. Diagnostics 2020, 10, 215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Konoshenko, M.Y.; Lekchnov, E.A.; Vlassov, A.V.; Laktionov, P.P. Isolation of Extracellular Vesicles: General Methodologies and Latest Trends. BioMed Res. Int. 2018, 2018, 8545347. [Google Scholar] [CrossRef]
- Hornung, S.; Dutta, S.; Bitan, G. CNS-Derived Blood Exosomes as a Promising Source of Biomarkers: Opportunities and Challenges. Front. Mol. Neurosci. 2020, 13, 38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hood, J.L.; Wickline, S.A. A systematic approach to exosome-based translational nanomedicine. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2012, 4, 458–467. [Google Scholar] [CrossRef]
- Kholafazad Kordasht, H.; Hasanzadeh, M. Biomedical analysis of exosomes using biosensing methods: Recent progress. Anal. Methods 2020, 12, 2795–2811. [Google Scholar] [CrossRef]
- Yamamoto, T.; Kosaka, N.; Ochiya, T. Latest advances in extracellular vesicles: From bench to bedside. Sci. Technol. Adv. Mater. 2019, 20, 746–757. [Google Scholar] [CrossRef] [Green Version]
- Pang, B.; Zhu, Y.; Ni, J.; Thompson, J.; Malouf, D.; Bucci, J.; Graham, P.; Li, Y. Extracellular vesicles: The next generation of biomarkers for liquid biopsy-based prostate cancer diagnosis. Theranostics 2020, 10, 2309–2326. [Google Scholar] [CrossRef]
- Zhao, Z.; Fan, J.; Hsu, Y.M.S.; Lyon, C.J.; Ning, B.; Hu, T.Y. Extracellular vesicles as cancer liquid biopsies: From discovery, validation, to clinical application. Lab Chip 2019, 19, 1114–1140. [Google Scholar] [CrossRef]
- Thone, M.N.; Kwon, Y.J. Extracellular blebs: Artificially-induced extracellular vesicles for facile production and clinical translation. Methods 2020, 177, 135–145. [Google Scholar] [CrossRef]
- Brennan, K.; Martin, K.; FitzGerald, S.P.; O’Sullivan, J.; Wu, Y.; Blanco, A.; Richardson, C.; Mc Gee, M.M. A comparison of methods for the isolation and separation of extracellular vesicles from protein and lipid particles in human serum. Sci. Rep. 2020, 10, 1039. [Google Scholar] [CrossRef] [Green Version]
- Ayala-Mar, S.; Donoso-Quezada, J.; Gallo-Villanueva, R.C.; Perez-Gonzalez, V.H.; González-Valdez, J. Recent advances and challenges in the recovery and purification of cellular exosomes. Electrophoresis 2019, 40, 3036–3049. [Google Scholar] [CrossRef]
- Brownlee, Z.; Lynn, K.D.; Thorpe, P.E.; Schroit, A.J. A novel “salting-out” procedure for the isolation of tumor-derived exosomes. J. Immunol. Methods 2014, 407, 120–126. [Google Scholar] [CrossRef] [Green Version]
- Bouvy, C.; Gheldof, D.; Chatelain, C.; Mullier, F.; Dogné, J.M. Contributing role of extracellular vesicles on vascular endothelium haemostatic balance in cancer. J. Extracell. Vesicles 2014, 3, 24400. [Google Scholar] [CrossRef]
- Mallardi, A.; Nuzziello, N.; Liguori, M.; Avolio, C.; Palazzo, G. Counting of peripheral extracellular vesicles in Multiple Sclerosis patients by an improved nanoplasmonic assay and dynamic light scattering. Colloids Surf. B Biointerfaces 2018, 168, 134–142. [Google Scholar] [CrossRef] [PubMed]
- Minafra, S.; Morello, V.; Glorioso, F.; La Fiura, A.M.; Tomasino, R.M.; Feo, S.; McIntosh, D.; Woolley, D.E. A new cell line (8701-BC) from primary ductal infiltrating carcinoma of human breast. Br. J. Cancer 1989, 60, 185–192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Serratì, S.; Margheri, F.; Fibbi, G.; Di Cara, G.; Minafra, L.; Pucci-Minafra, I.; Liotta, F.; Annunziato, F.; Pucci, D.M.; Del Rosso, M. Endothelial cells and normal breast epithelial cells enhance invasion of breast carcinoma cells by CXCR-4-dependent up-regulation of urokinase-type plasminogen activator receptor (uPAR, CD87) expression. J. Pathol. 2008, 214, 545–554. [Google Scholar] [CrossRef] [PubMed]
- Brachmann, C.B.; Davies, A.; Cost, G.J.; Caputo, E.; Li, J.; Hieter, P.; Boeke, J.D. Designer deletion strains derived from Saccharomyces cerevisiae S288C: A useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 1998, 14, 115–132. [Google Scholar] [CrossRef]
- Biagioni, A.; Laurenzana, A.; Menicacci, B.; Peppicelli, S.; Andreucci, E.; Bianchini, F.; Guasti, D.; Paoli, P.; Serratì, S.; Mocali, A.; et al. uPAR-expressing melanoma exosomes promote angiogenesis by VE-Cadherin, EGFR and uPAR overexpression and rise of ERK1,2 signaling in endothelial cells. Cell. Mol. Life Sci. 2021, 78, 3057–3072. [Google Scholar] [CrossRef]
- Théry, C.; Amigorena, S.; Raposo, G.; Clayton, A. Isolation and Characterization of Exosomes from Cell Culture Supernatants and Biological Fluids. Curr. Protoc. Cell Biol. 2006, 30, 3–22. [Google Scholar] [CrossRef]
- Gorgun, C.; Reverberi, D.; Rotta, G.; Villa, F.; Quarto, R.; Tasso, R. Isolation and Flow Cytometry Characterization of Extracellular-Vesicle Subpopulations Derived from Human Mesenchymal Stromal Cells. Curr. Protoc. Stem Cell Biol. 2019, 48, e76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Serratì, S.; Porcelli, L.; Fragassi, F.; Garofoli, M.; Di Fonte, R.; Fucci, L.; Iacobazzi, R.M.; Palazzo, A.; Margheri, F.; Cristiani, G.; et al. The interaction between reactive peritoneal mesothelial cells and tumor cells via extracellular vesicles facilitates colorectal cancer dissemination. Cancers 2021, 13, 2505. [Google Scholar] [CrossRef]
- Porcelli, L.; Guida, M.; De Summa, S.; Di Fonte, R.; De Risi, I.; Garofoli, M.; Caputo, M.; Negri, A.; Strippoli, S.; Serratì, S.; et al. UPAR + extracellular vesicles: A robust biomarker of resistance to checkpoint inhibitor immunotherapy in metastatic melanoma patients. J. Immunother. Cancer 2021, 9, e002372. [Google Scholar] [CrossRef] [PubMed]
- Maiolo, D.; Paolini, L.; Di Noto, G.; Zendrini, A.; Berti, D.; Bergese, P.; Ricotta, D. Colorimetric nanoplasmonic assay to determine purity and titrate extracellular vesicles. Anal. Chem. 2015, 87, 4168–4176. [Google Scholar] [CrossRef]
- Turkevich, J.; Stevenson, P.C.; Hillier, J. A study of the nucleation and growth processes in the synthesis of colloidal gold. Discuss. Faraday Soc. 1951, 11, 55–75. [Google Scholar] [CrossRef]
- Palazzo, G.; Paduano, L. Diffusion measuring techniques. Colloid. Found. Nanosci. 2022, 10, 257–287. [Google Scholar] [CrossRef]
- Laurati, M.; Berti, D. Structure and dynamics: Static scattering of radiation and optical correlation techniques. Colloid. Found. Nanosci. 2022, 9, 227–256. [Google Scholar] [CrossRef]
- Doyle, L.; Wang, M. Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis. Cells 2019, 8, 727. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalishwaralal, K.; Kwon, W.Y.; Park, K.S. Exosomes for Non-Invasive Cancer Monitoring. Biotechnol. J. 2019, 14, e1800430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zendrini, A.; Paolini, L.; Busatto, S.; Radeghieri, A.; Romano, M.; Wauben, M.H.M.; van Herwijnen, M.J.C.; Nejsum, P.; Borup, A.; Ridolfi, A.; et al. Augmented COlorimetric NANoplasmonic (CONAN) Method for Grading Purity and Determine Concentration of EV Microliter Volume Solutions. Front. Bioeng. Biotechnol. 2020, 7, 452. [Google Scholar] [CrossRef]
- Lener, T.; Gimona, M.; Aigner, L.; Börger, V.; Buzas, E.; Camussi, G.; Chaput, N.; Chatterjee, D.; Court, F.A.; del Portillo, H.A.; et al. Applying extracellular vesicles based therapeutics in clinical trials—An ISEV position paper. J. Extracell. Vesicles 2015, 4, 30087. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, A.; Xu, F.; Lee, S. Human saliva and model saliva at bulk to adsorbed phases – similarities and differences. Adv. Colloid Interface Sci. 2019, 273, 102034. [Google Scholar] [CrossRef] [PubMed]
- Zlotogorski-Hurvitz, A.; Dayan, D.; Chaushu, G.; Korvala, J.; Salo, T.; Sormunen, R.; Vered, M. Human Saliva-Derived Exosomes: Comparing Methods of Isolation. J. Histochem. Cytochem. 2015, 63, 181–189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moschetti, R.; Palazzo, A.; Lorusso, P.; Viggiano, L.; Marsano, R.M. “What You Need, Baby, I Got It”: Transposable Elements as Suppliers of Cis-Operating Sequences in Drosophila. Biology 2020, 9, 25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
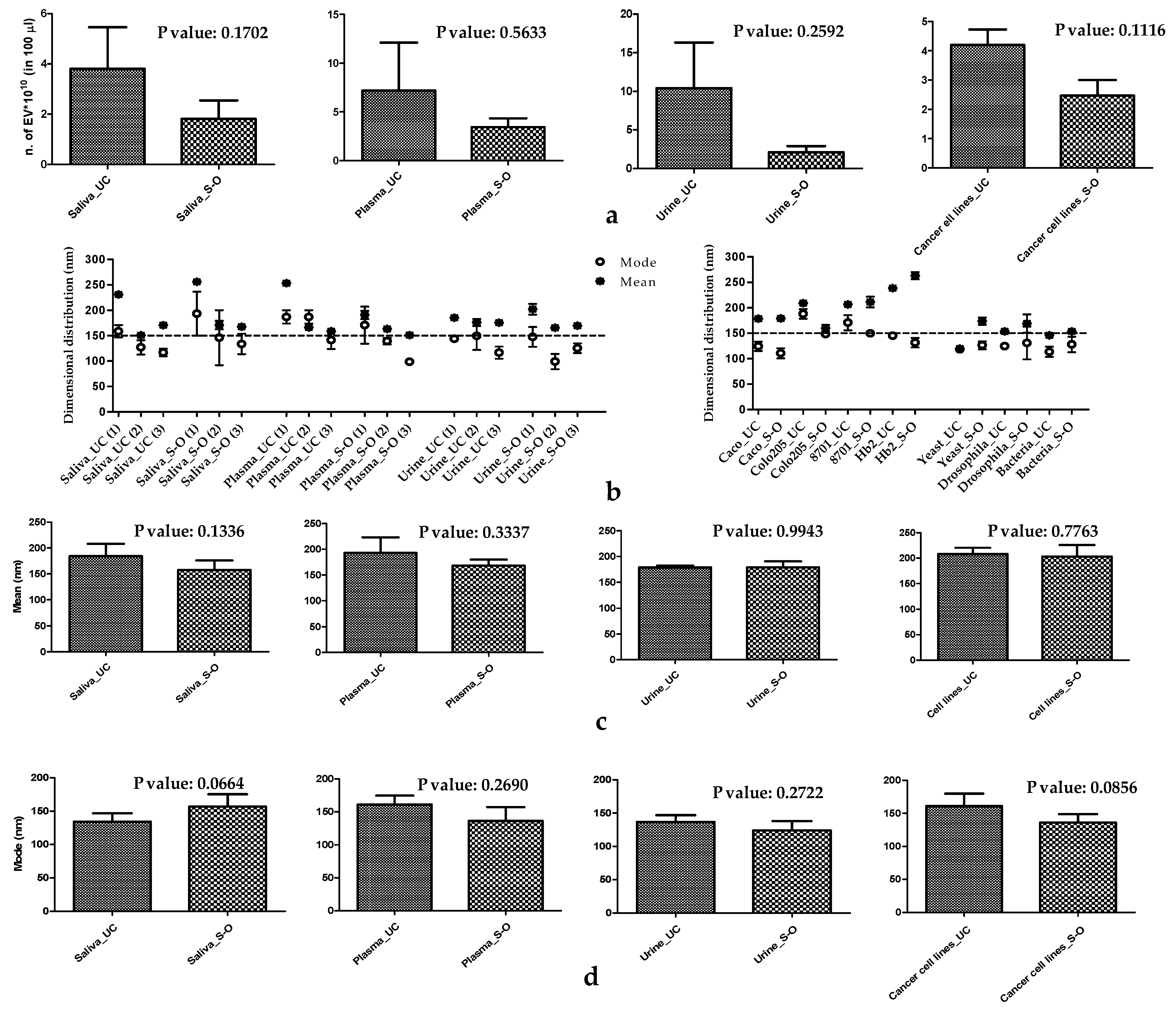

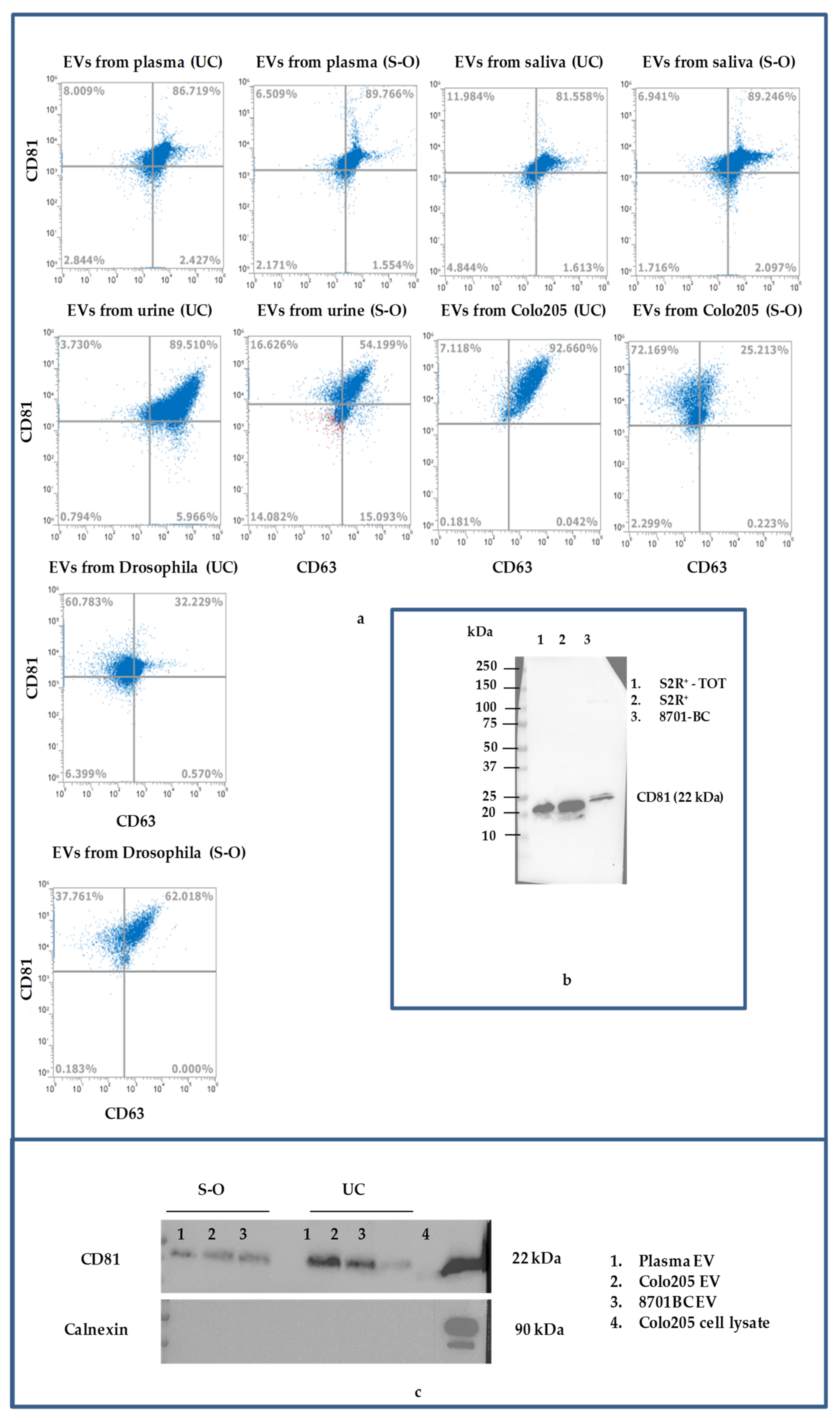
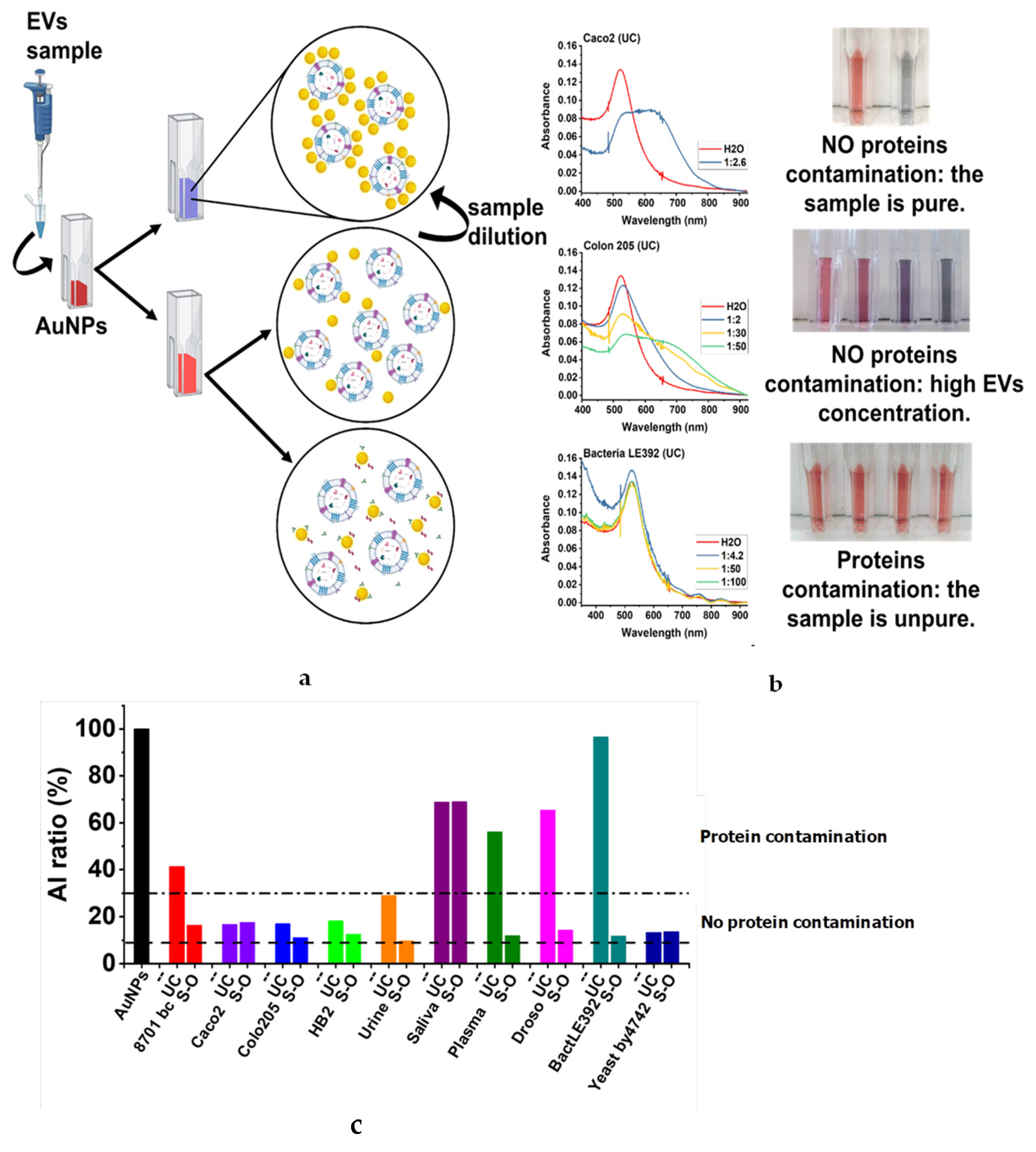
| Sample | NTA | DLS | |||
|---|---|---|---|---|---|
| Human Cell Lines | n. of EVs ± S.D./μL | Mean ± S.D. (d. nm) | Mode ± S.D. (d. nm) | Z-Average ± S.D. | Mode by Number (FWHH *) |
| Caco-2_UC | (5.2 ± 1.3) × 10⁸ | 178.6 ± 9.5 | 124.2 ± 9.1 | 440.9 ± 27.7 | 122 (80) |
| Caco-2_S-O | (3.1 ± 0.2) × 10⁸ | 179 ± 5 | 110.5 ± 9.9 | 552.6 ± 28.7 | 100 (50) |
| Colo 205_UC | (4.0 ± 0.4) × 10⁸ | 209 ± 8 | 187.9 ± 9.5 | 374 ± 95 | 106 (56) |
| Colo 205_S-O | (1.4 ± 0.1) × 10⁸ | 159.8 ± 11.2 | 148.2 ± 3.9 | 944.5 ± 76.2 | 142 (50) |
| 8701-BC_UC | (3.4 ± 0.2) × 10⁸ | 206.7 ± 4.8 | 170.6 ± 15.1 | 663.4 ± 35.6 | 122 (66) |
| 8701-BC_S-O | (2.9 ± 0.6) × 10⁸ | 211.4 ± 18.3 | 149.7 ± 3.8 | 267.1 ± 26.8 | 113 (58) |
| HB2_UC | (6.8 ± 0.4) × 10⁸ | 238.5 ± 2.2 | 144.9 ± 1.3 | 547.3 ± 32.5 | 130 (40) |
| HB2_S-O | (3.3 ± 0.1) × 10⁸ | 263.1 ± 12.1 | 131.3 ± 9.3 | 381.1 ± 3.7 | 143 (64) |
| Body Fluids | |||||
| Saliva_UC (1) | (6.7 ± 0.3) × 10⁸ | 231.1 ± 3.1 | 158.7 ± 12.2 | 195 ± 16 | 143 (66) |
| Saliva_UC (2) | (9.80 ± 0.9) × 107 | 150.9 ± 6.2 | 127.0 ± 14.1 | 212 ± 32 | 142 (62) |
| Saliva_UC (3) | (3.74 ± 0.2) × 108 | 170.5 ± 3.0 | 116.9 ± 7.4 | 237 ± 40 | 105 (61) |
| Saliva_S-O (1) | (2.9 ± 0.4) × 10⁸ | 256.1 ± 3.9 | 193.4 ± 43.2 | 255 ± 14 | 121 (54) |
| Saliva_S-O (2) | (4.10 ± 0.2) × 107 | 170.8 ± 14.8 | 146.0 ± 54.1 | 574 ± 44 | 134 (47) |
| Saliva_S-O (3) | (2.13 ± 0.2) × 108 | 167.6 ± 3.3 | 133.7 ± 20.1 | 1022 ± 200 | 165 (45) |
| Plasma_UC (1) | (1.7 ± 0.1) × 10⁹ | 253.4 ± 7.3 | 186.9 ± 13.1 | 96.7 ± 8.3 | 68 (35) |
| Plasma_UC (2) | (2.71 ± 0.2) × 108 | 166.0 ± 2.6 | 141.3 ± 17.5 | 132 ± 9 | 93 (47) |
| Plasma_UC (3) | (1.93 ± 0.1) × 108 | 158.8 ± 11.1 | 155.8 ± 35.6 | 147 ± 12 | 81 (43) |
| Plasma_S-O (1) | (2.3 ± 0.3) × 10⁸ | 190.4 ± 14.2 | 170.8 ± 36.6 | not suitable | not suitable |
| Plasma_S-O(2) | (5.24 ± 1.1) × 108 | 163.3 ± 3.7 | 139.3 ± 6.6 | 268 ± 70 | 106 (43) |
| Plasma_S-O (3) | (2.82 ± 0.3) × 108 | 151.2 ± 2.2 | 98.5 ± 3.0 | 850 ± 350 | 61 (35) |
| Urine_UC (1) | (2.2 ± 0.1) × 10⁹ | 185.4 ± 2.8 | 143.8 ± 4.0 | 144.4 ± 0.7 | 60 (22) |
| Urine_UC (2) | (6.65 ± 0.1) × 108 | 175.7 ± 1.1 | 116.7 ± 12.0 | 247 ± 9 | 105 (45) |
| Urine_UC (3) | (2.59 ± 0.2) × 108 | 176.0 ± 12.2 | 149.7 ± 27.8 | 275 ± 14 | 108 (42) |
| Urine_S-O (1) | (3.5 ± 0.1) × 10⁸ | 202.0 ± 19 | 147.6 ± 19.5 | 332.5 ± 48.9 | 152 (60) |
| Urine_S-O (2) | (6.49 ± 0.6) × 107 | 165.6 ± 7.5 | 99.0 ± 15.1 | 756 ± 110 | 106 (36) |
| Urine_S-O (3) | (2.13 ± 0.2) × 108 | 169.7 ± 2.9 | 125.3 ± 9.5 | 1650 ± 215 | 174 (40) |
| Cell Cultures Models | |||||
| Yeast_UC | (8.8 ± 0.5) × 10⁸ | 119.9 ± 1.1 | 118.2 ± 4.4 | 187 ± 1 | 98 (50) |
| Yeast_S-O | (2.8 ± 0.5) × 10⁸ | 173.8 ± 12.6 | 126.4 ± 7.7 | not suitable | not suitable |
| Drosophila_UC | (3.8 ± 0.6) × 10⁸ | 153.7 ± 0.8 | 124.4 ± 5.4 | 188.5 ± 88.5 | 54 (22) |
| Drosophila_S-O | (1.3 ± 0.1) × 10⁸ | 169.1 ± 31.1 | 130.7 ± 32.2 | not suitable | not suitable |
| Bacteria_UC | (8.3 ± 5.9) × 10⁸ | 145.8 ± 9.3 | 113.4 ± 10.1 | 379.5 ± 70.1 | 84 (40) |
| Bacteria_S-O | (2.9 ± 1.5) × 10⁸ | 153.5 ± 5 | 127.9 ± 15.3 | not suitable | not suitable |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Serratì, S.; Palazzo, A.; Lapenna, A.; Mateos, H.; Mallardi, A.; Marsano, R.M.; Quarta, A.; Del Rosso, M.; Azzariti, A. Salting-Out Approach Is Worthy of Comparison with Ultracentrifugation for Extracellular Vesicle Isolation from Tumor and Healthy Models. Biomolecules 2021, 11, 1857. https://doi.org/10.3390/biom11121857
Serratì S, Palazzo A, Lapenna A, Mateos H, Mallardi A, Marsano RM, Quarta A, Del Rosso M, Azzariti A. Salting-Out Approach Is Worthy of Comparison with Ultracentrifugation for Extracellular Vesicle Isolation from Tumor and Healthy Models. Biomolecules. 2021; 11(12):1857. https://doi.org/10.3390/biom11121857
Chicago/Turabian StyleSerratì, Simona, Antonio Palazzo, Annamaria Lapenna, Helena Mateos, Antonia Mallardi, René Massimiliano Marsano, Alessandra Quarta, Mario Del Rosso, and Amalia Azzariti. 2021. "Salting-Out Approach Is Worthy of Comparison with Ultracentrifugation for Extracellular Vesicle Isolation from Tumor and Healthy Models" Biomolecules 11, no. 12: 1857. https://doi.org/10.3390/biom11121857
APA StyleSerratì, S., Palazzo, A., Lapenna, A., Mateos, H., Mallardi, A., Marsano, R. M., Quarta, A., Del Rosso, M., & Azzariti, A. (2021). Salting-Out Approach Is Worthy of Comparison with Ultracentrifugation for Extracellular Vesicle Isolation from Tumor and Healthy Models. Biomolecules, 11(12), 1857. https://doi.org/10.3390/biom11121857










