Inhibition of LPMOs by Fermented Persimmon Juice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Chemicals
2.2. Characterization
2.3. Enzyme Production and Purification
2.4. Activity and Inhibition Assays
2.4.1. Kakishibu-Impregnated Filter Paper
2.4.2. Reducing-End Quantification
2.4.3. Chromogenic AZCL-HEC Substrate
2.4.4. LPMO Inhibition Susceptibility
2.5. Data Analysis
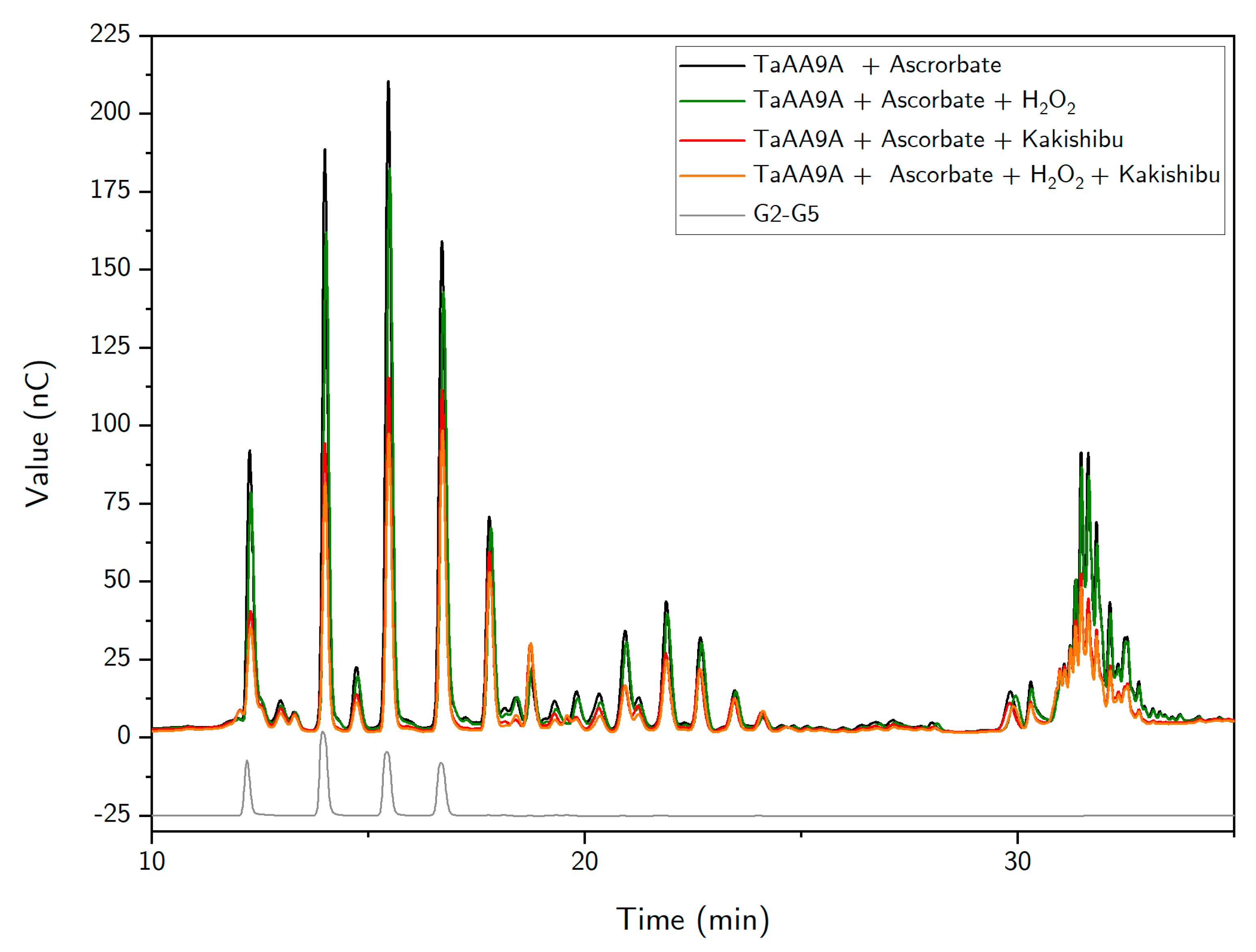
3. Results
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| Compound | Molecular Formula | RT (min) | Accurate Mass [M − H]− | Mass Error (ppm) | Fragments m/z (Relative Abundance) |
|---|---|---|---|---|---|
| p-hydroxybenzoic acid | C7H6O3 | 6.2 | 137.0234 | 7 | 137.02 (100), 93.06 (6), 108.93 (1) |
| Coumaric acid | C9H8O3 | 8.86 | 163.0396 | 3 | 118.99 (100), 142.99 (3), 162 (2), 163.03 (1) |
| Gallic acid | C7H6O5 | 2.49 | 169.0139 | 2 | 125.02 (100), 169.02 (80), 81.032 (2) |
| Kaempferol | C15H10O6 | 15.79 | 285.0411 | 2 | 285.05 (100), 151.10 (1) |
| Luteolin | C15H10O6 | 13.12 | 285.0412 | 3 | 285.05 (100), 257.07 (24), 244.04 (2), 167.53 (2) |
| Eriodictyol | C15H12O6 | 13.40 | 287.0566 | 2 | 151.00 (100), 287.05 (20), 181.61 (13) |
| Catechin | C15H14O6 | 6.51 | 289.0719 | 0 | 174.95 (100), 158.97 (52), 289.07 (30), 245.08 (28) |
| Quercetin | C15H10O7 | 13.71 | 301.0357 | 1 | 301.03 (100), 151.00 (19), 178.99 (10), 255.05 (6) |
| Taxifolin | C15H12O7 | 10.11 | 303.0517 | 2 | 285.04 (100), 125.02 (30), 303.05(20) |
| Myricetin | C15H10O8 | 11.65 | 317.0307 | 1 | 317.03 (100), 178.99 (10), 141.06 (8), 227.54 (7) |
References
- Yaqub, S.; Farooq, U.; Shafi, A.; Akram, K.; Murtaza, M.A.; Kausar, T.; Siddique, F. Chemistry and Functionality of Bioactive Compounds Present in Persimmon. J. Chem. 2016, 2016, 3424025. [Google Scholar] [CrossRef] [Green Version]
- Perez-Burillo, S.; Oliveras, M.; Quesada, J.; Rufián-Henares, J.; Pastoriza, S. Relationship between composition and bioactivity of persimmon and kiwifruit. Food Res. Int. 2018, 105, 461–472. [Google Scholar] [CrossRef]
- Ito, S.; Oshima, Y. Studies on the Tannin of Japanese Persimmon (Diospyros Kaki L.). Agric. Biol. Chem. 1962, 26, 156–161. [Google Scholar] [CrossRef]
- Okonogi, T.; Hattori, Z.; Ogiso, A.; Mitsui, S. Detoxification by persimmon tannin of snake venoms and bacterial toxins. Toxicon 1979, 17, 524–527. [Google Scholar] [CrossRef]
- McRae, J.M.; Falconer, R.J.; Kennedy, J.A. Thermodynamics of Grape and Wine Tannin Interaction with Polyproline: Implications for Red Wine Astringency. J. Agric. Food Chem. 2010, 58, 12510–12518. [Google Scholar] [CrossRef]
- Mandels, M.; Reese, E.T. Inhibition of Cellulases. Annu. Rev. Phytopathol. 1965, 3, 85–102. [Google Scholar] [CrossRef]
- Xue, Y.-L.; Miyakawa, T.; Hayashi, Y.; Okamoto, K.; Hu, F.; Mitani, N.; Furihata, K.; Sawano, Y.; Tanokura, M. Isolation and Tyrosinase Inhibitory Effects of Polyphenols from the Leaves of Persimmon, Diospyros kaki. J. Agric. Food Chem. 2011, 59, 6011–6017. [Google Scholar] [CrossRef]
- Kracher, D.; Andlar, M.; Furtmüller, P.G.; Ludwig, R. Active-site copper reduction promotes substrate binding of fungal lytic polysaccharide monooxygenase and reduces stability. J. Biol. Chem. 2018, 293, 1676–1687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tokin, R.; Frandsen, K.E.H.; Ipsen, J.Ø.; Leggio, L.L.; Poojary, M.M.; Berrin, J.; Grisel, S.; Brander, S.; Jensen, P.E.; Johansen, K.S. Inhibition of lytic polysaccharide monooxygenase by natural plant extracts. New Phytol. 2021, 232, 1337–1349. [Google Scholar] [CrossRef]
- Harris, P.V.; Welner, D.; McFarland, K.C.; Re, E.; Poulsen, J.-C.N.; Brown, K.; Salbo, R.; Ding, H.; Vlasenko, E.; Merino, S.; et al. Stimulation of Lignocellulosic Biomass Hydrolysis by Proteins of Glycoside Hydrolase Family 61: Structure and Function of a Large, Enigmatic Family. Biochemistry 2010, 49, 3305–3316. [Google Scholar] [CrossRef]
- Beeson, W.T.; Phillips, C.M.; Cate, J.H.D.; Marletta, M.A. Oxidative Cleavage of Cellulose by Fungal Copper-Dependent Polysaccharide Monooxygenases. J. Am. Chem. Soc. 2012, 134, 890–892. [Google Scholar] [CrossRef]
- Phillips, C.M.; Beeson, W.T.; Cate, J.H.; Marletta, M.A. Cellobiose Dehydrogenase and a Copper-Dependent Polysaccharide Monooxygenase Potentiate Cellulose Degradation by Neurospora crassa. ACS Chem. Biol. 2011, 6, 1399–1406. [Google Scholar] [CrossRef]
- Nierman, W.C.; Pain, A.; Anderson, M.J.; Wortman, J.; Kim, H.; Arroyo, J.; Berriman, M.; Abe, K.; Archer, D.B.; Bermejo, C.; et al. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nat. Cell Biol. 2005, 438, 1151–1156. [Google Scholar] [CrossRef]
- Simmons, T.J.; Frandsen, K.E.H.; Ciano, L.; Tryfona, T.; Lenfant, N.; Poulsen, J.C.; Wilson, L.F.L.; Tandrup, T.; Tovborg, M.; Schnorr, K.; et al. Structural and electronic determinants of lytic polysaccharide monooxygenase reactivity on polysaccharide substrates. Nat. Commun. 2017, 8, 1064. [Google Scholar] [CrossRef]
- Tokin, R.; Ipsen, J.Ø.; Westh, P.; Johansen, K.S. The synergy between LPMOs and cellulases in enzymatic saccharification of cellulose is both enzyme- and substrate-dependent. Biotechnol. Lett. 2020, 42, 1975–1984. [Google Scholar] [CrossRef] [PubMed]
- Teter, S.; Ward, C.; Cherry, J.; Jones, A.; Harris, P.; Yi, J. Variants of Glycoside Hydrolases. U.S. Patent 8383385, 26 February 2013. [Google Scholar]
- Bennati-Granier, C.; Garajova, S.; Champion, C.; Grisel, S.; Haon, M.; Zhou, S.; Fanuel, M.; Ropartz, D.; Rogniaux, H.; Gimbert, I.; et al. Substrate specificity and regioselectivity of fungal AA9 lytic polysaccharide monooxygenases secreted by Podospora anserina. Biotechnol. Biofuels 2015, 8, 90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Filiatrault-Chastel, C.; Navarro, D.; Haon, M.; Grisel, S.; Herpoël-Gimbert, I.; Chevret, D.; Fanuel, M.; Henrissat, B.; Heiss-Blanquet, S.; Margeot, A.; et al. AA16, a new lytic polysaccharide monooxygenase family identified in fungal secretomes. Biotechnol. Biofuels 2019, 12, 55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kruer-Zerhusen, N.; Alahuhta, M.; Lunin, V.V.; Himmel, M.E.; Bomble, Y.J.; Wilson, D.B. Structure of a Thermobifida fusca lytic polysaccharide monooxygenase and mutagenesis of key residues. Biotechnol. Biofuels 2017, 10, 243. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Rollán, C.; Falkenberg, K.B.; Rennig, M.; Bertelsen, A.B.; Ipsen, J.Ø.; Brander, S.; Daley, D.O.; Johansen, K.S.; Nørholm, M.H.H. LyGo: A Platform for Rapid Screening of Lytic Polysaccharide Monooxygenase Production. ACS Synth. Biol. 2021, 10, 897–906. [Google Scholar] [CrossRef] [PubMed]
- Barkholt, V.; Jensen, A.L. Amino acid analysis: Determination of cysteine plus half-cystine in proteins after hydrochloric acid hydrolysis with a disulfide compound as additive. Anal. Biochem. 1989, 177, 318–322. [Google Scholar] [CrossRef]
- Brander, S.; Horvath, I.; Ipsen, J.Ø.; Peciulyte, A.; Olsson, L.; Hernández-Rollán, C.; Nørholm, M.H.H.; Mossin, S.; Leggio, L.L.; Probst, C.; et al. Biochemical evidence of both copper chelation and oxygenase activity at the histidine brace. Sci. Rep. 2020, 10, 16369. [Google Scholar] [CrossRef]
- Lever, M. Colorimetric and fluorometric carbohydrate determination with p-hydroxybenzoic acid hydrazide. Biochem. Med. 1973, 7, 274–281. [Google Scholar] [CrossRef]
- Quinlan, R.J.; Sweeney, M.D.; Leggio, L.L.; Otten, H.; Poulsen, J.-C.N.; Johansen, K.; Krogh, K.B.R.M.; Jørgensen, C.I.; Tovborg, M.; Anthonsen, A.; et al. Insights into the oxidative degradation of cellulose by a copper metalloenzyme that exploits biomass components. Proc. Natl. Acad. Sci. USA 2011, 108, 15079–15084. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Besharati, M.; Taghizadeh, A. Effect of Tannin-Binding Agents (Polyethylene Glycol and Polyvinylpyrrolidone) Supplementation on In Vitro Gas Production Kinetics of Some Grape Yield Byproducts. Int. Sch. Res. Not. 2011, 2011, 780540. [Google Scholar] [CrossRef] [Green Version]
- Tanner, G.J.; Moore, A.E.; Larkin, P.J. Proanthocyanidins inhibit hydrolysis of leaf proteins by rumen microflora in vitro. Br. J. Nutr. 1994, 71, 947–958. [Google Scholar] [CrossRef] [Green Version]
- Goldstein, J.L.; Swain, T. The inhibition of enzymes by tannins. Phytochemistry 1965, 4, 185–192. [Google Scholar] [CrossRef]
- Olsen, S.; Bohlin, C.; Murphy, L.; Borch, K.; McFarland, K.; Sweeny, M.; Westh, P. Effects of non-ionic surfactants on the interactions between cellulases and tannic acid: A model system for cellulase–poly-phenol interactions. Enzym. Microb. Technol. 2011, 49, 353–359. [Google Scholar] [CrossRef] [PubMed]
- Chávez-González, M.; Rodríguez-Durán, L.V.; Balagurusamy, N.; Prado-Barragán, A.; Rodríguez, R.; Contreras, J.C.; Aguilar, C.N. Biotechnological Advances and Challenges of Tannase: An Overview. Food Bioprocess. Technol. 2011, 5, 445–459. [Google Scholar] [CrossRef]
- Bissaro, B.; Røhr, Å.K.; Müller, G.; Chylenski, P.; Skaugen, M.; Forsberg, Z.; Horn, S.J.; Vaaje-Kolstad, G.; Eijsink, V.G.H. Oxidative cleavage of polysaccharides by monocopper enzymes depends on H2O2. Nat. Chem. Biol. 2017, 13, 1123–1128. [Google Scholar] [CrossRef] [PubMed]
- Calderaro, F.; Keser, M.; Akeroyd, M.; Bevers, L.E.; Eijsink, V.G.H.; Várnai, A.; Berg, M.A.V.D. Characterization of an AA9 LPMO from Thielavia australiensis, TausLPMO9B, under industrially relevant lignocellulose saccharification conditions. Biotechnol. Biofuels 2020, 13, 195. [Google Scholar] [CrossRef] [PubMed]
- Brander, S.; Tokin, R.; Ipse2, J.Ø.; Jensen, P.E.; Hernández-Rollán, C.; Nørholm, M.H.H.; Leggio, L.L.; Dupree, P.; Johansen, K.S. Scission of Glucosidic Bonds by a Lentinus similis LPMO is Strictly Dependent on H2O2 While Oxidation of Saccharide Products Depends on O2. ASC Catal. 2021, 11, 13848–13859. [Google Scholar]
- Mandels, M.; Reese, E.T. Inhibition of cellulases and β-glucosidases. In Advances in Enzymic Hydrolysis of Cellulose and Related Materials; Elsevier BV: Amsterdam, The Netherlands, 1963; pp. 115–157. [Google Scholar]
- Lee, Y.A.; Cho, E.J.; Tanaka, T.; Yokozawa, T. Inhibitory Activities of Proanthocyanidins from Persimmon against Oxidative Stress and Digestive Enzymes Related to Diabetes. J. Nutr. Sci. Vitaminol. 2007, 53, 287–292. [Google Scholar] [CrossRef] [Green Version]
- Ximenes, E.; Kim, Y.; Mosier, N.; Dien, B.; Ladisch, M. Deactivation of cellulases by phenols. Enzym. Microb. Technol. 2011, 48, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Ximenes, E.; Kim, Y.; Mosier, N.; Dien, B.; Ladisch, M. Inhibition of cellulases by phenols. Enzym. Microb. Technol. 2010, 46, 170–176. [Google Scholar] [CrossRef]
- Ramawat, K.G.; Mérillon, J.M. Natural Products; Springer: Berlin/Heidelberg, Germany, 2013; ISBN 978-3-642-22143-9. [Google Scholar]
- Börjesson, J.; Engqvist, M.; Sipos, B.; Tjerneld, F. Effect of poly(ethylene glycol) on enzymatic hydrolysis and adsorption of cellulase enzymes to pretreated lignocellulose. Enzym. Microb. Technol. 2007, 41, 186–195. [Google Scholar] [CrossRef]
- Feng, X. Methods of Reducing the Inhibitory Effect of a Tannin on the Enzymatic Hydrolysis of Cellulosic Material. CN101910405A, 8 December 2010. [Google Scholar]
- Frommhagen, M.; Mutte, S.K.; Westphal, A.H.; Koetsier, M.J.; Hinz, S.W.A.; Visser, J.; Vincken, J.-P.; Weijers, D.; Van Berkel, W.J.H.; Gruppen, H.; et al. Boosting LPMO-driven lignocellulose degradation by polyphenol oxidase-activated lignin building blocks. Biotechnol. Biofuels 2017, 10, 121. [Google Scholar] [CrossRef] [Green Version]
- Lu, M.-J.; Chu, S.-C.; Yan, L.; Chen, C. Effect of tannase treatment on protein–tannin aggregation and sensory attributes of green tea infusion. LWT 2009, 42, 338–342. [Google Scholar] [CrossRef]
- Adamczyk, B.; Simon, J.; Kitunen, V.; Adamczyk, S.; Smolander, A. Tannins and Their Complex Interaction with Different Organic Nitrogen Compounds and Enzymes: Old Paradigms versus Recent Advances. ChemistryOpen 2017, 6, 610–614. [Google Scholar] [CrossRef] [PubMed]
- Labuckas, D.; Maestri, D.; Lamarque, A. Molecular Characterization, Antioxidant and Protein Solubility-Related Properties of Polyphenolic Compounds from Walnut (Juglans regia). Nat. Prod. Commun. 2016, 11, 637–640. [Google Scholar] [CrossRef] [Green Version]
- Harbertson, J.F.; Kilmister, R.L.; Kelm, M.A.; Downey, M.O. Impact of condensed tannin size as individual and mixed polymers on bovine serum albumin precipitation. Food Chem. 2014, 160, 16–21. [Google Scholar] [CrossRef]
- Hagerman, A.; Butler, L. The specificity of proanthocyanidin-protein interactions. J. Biol. Chem. 1981, 256, 4494–4497. [Google Scholar] [CrossRef]
- Kont, R.; Pihlajaniemi, V.; Borisova, A.S.; Aro, N.; Marjamaa, K.; Loogen, J.; Büchs, J.; Eijsink, V.G.H.; Kruus, K.; Väljamäe, P. The liquid fraction from hydrothermal pretreatment of wheat straw provides lytic polysaccharide monooxygenases with both electrons and H2O2 co-substrate. Biotechnol. Biofuels 2019, 12, 235. [Google Scholar] [CrossRef] [PubMed] [Green Version]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tokin, R.; Ipsen, J.Ø.; Poojary, M.M.; Jensen, P.E.; Olsson, L.; Johansen, K.S. Inhibition of LPMOs by Fermented Persimmon Juice. Biomolecules 2021, 11, 1890. https://doi.org/10.3390/biom11121890
Tokin R, Ipsen JØ, Poojary MM, Jensen PE, Olsson L, Johansen KS. Inhibition of LPMOs by Fermented Persimmon Juice. Biomolecules. 2021; 11(12):1890. https://doi.org/10.3390/biom11121890
Chicago/Turabian StyleTokin, Radina, Johan Ørskov Ipsen, Mahesha M. Poojary, Poul Erik Jensen, Lisbeth Olsson, and Katja Salomon Johansen. 2021. "Inhibition of LPMOs by Fermented Persimmon Juice" Biomolecules 11, no. 12: 1890. https://doi.org/10.3390/biom11121890
APA StyleTokin, R., Ipsen, J. Ø., Poojary, M. M., Jensen, P. E., Olsson, L., & Johansen, K. S. (2021). Inhibition of LPMOs by Fermented Persimmon Juice. Biomolecules, 11(12), 1890. https://doi.org/10.3390/biom11121890








