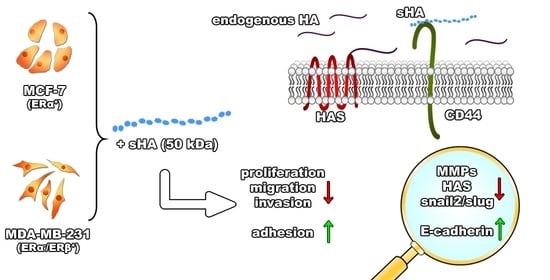
Sulfated Hyaluronan Modulates the Functional Properties and Matrix Effectors Expression of Breast Cancer Cells with Different Estrogen Receptor Status
Abstract
:1. Introduction
2. Materials and Methods
2.1. HA and sHA Fragments
2.2. Cell Cultures and Reagents
2.3. Proliferation Assay
2.4. Wound Healing Assay
2.5. Collagen Type I Cell Adhesion Assay
2.6. Collagen Type I Invasion Assay
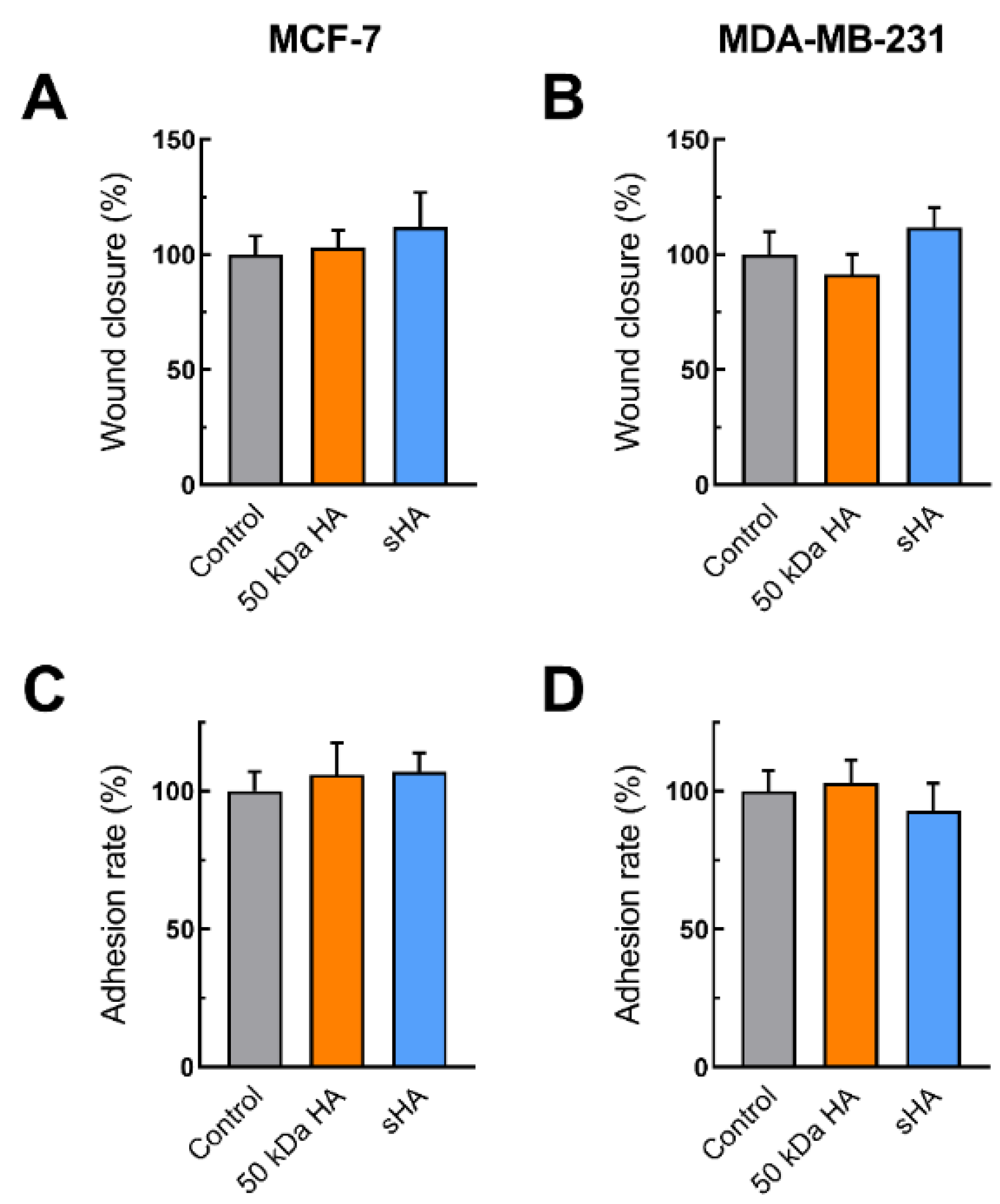
2.7. Wound Healing and Adhesion Assays Using Hyaluronidase Pre-Treatment
2.8. RNA Isolation, cDNA Synthesis and Real-Time PCR
2.9. Immunofluorescence
2.10. Western Blot Analysis
2.11. Statistical Analysis
3. Results
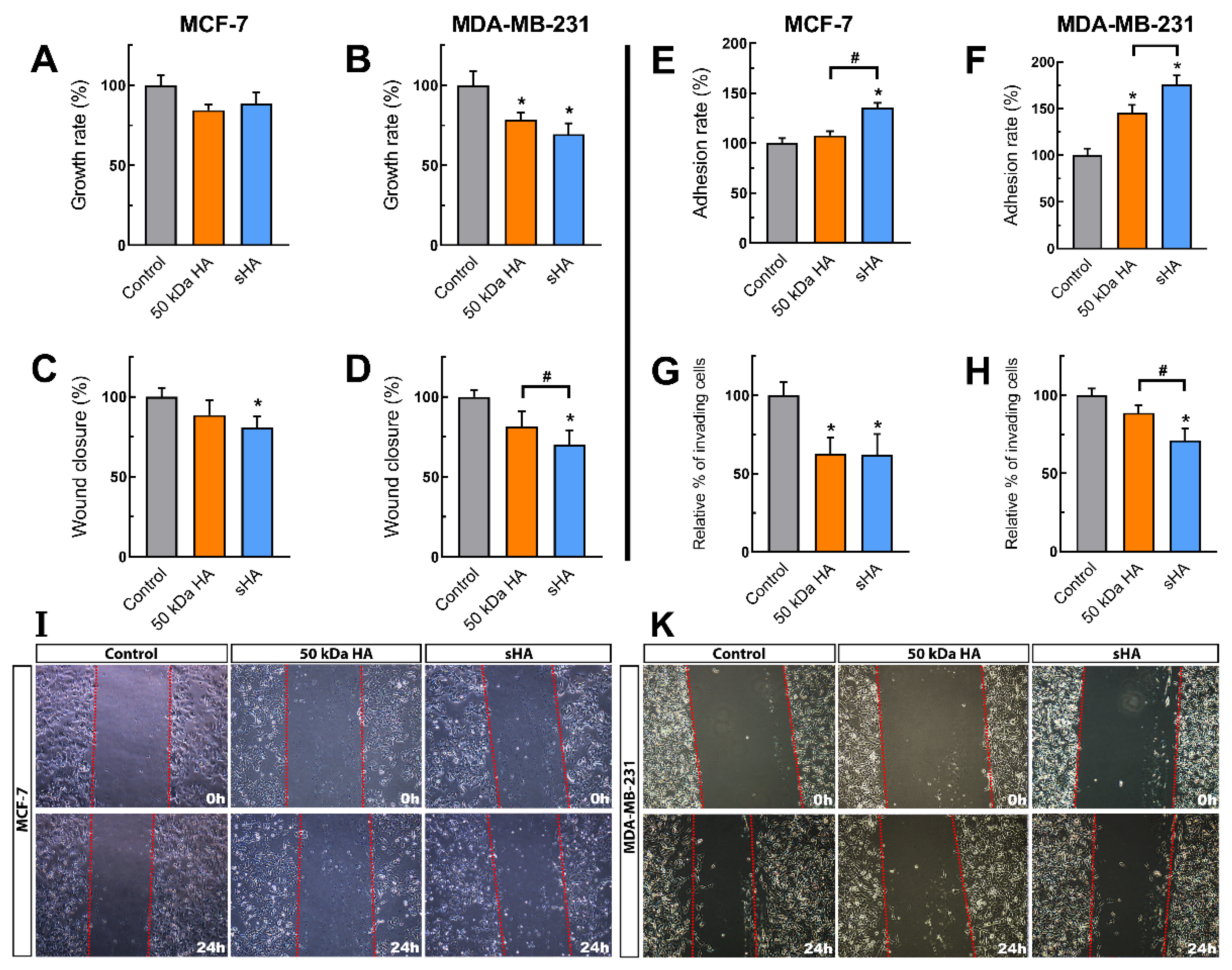
3.1. Sulfated HA Affects Breast Cancer Cells’ Functional Properties
3.2. Sulfated HA Competition with Endogenous HA Affects Cell Migration and Adhesion
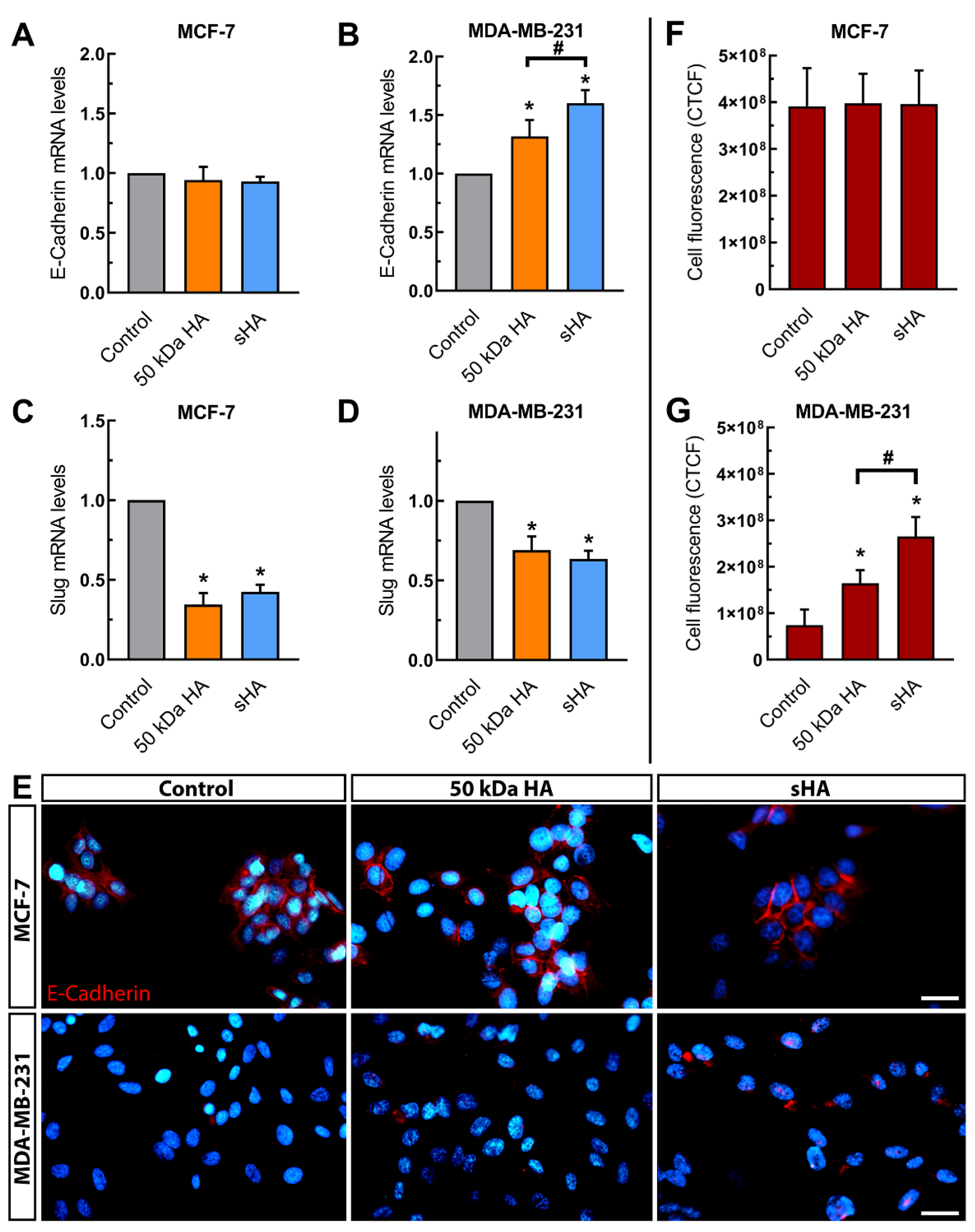
3.3. Sulfated HA Modulates the Expression of EMT Markers
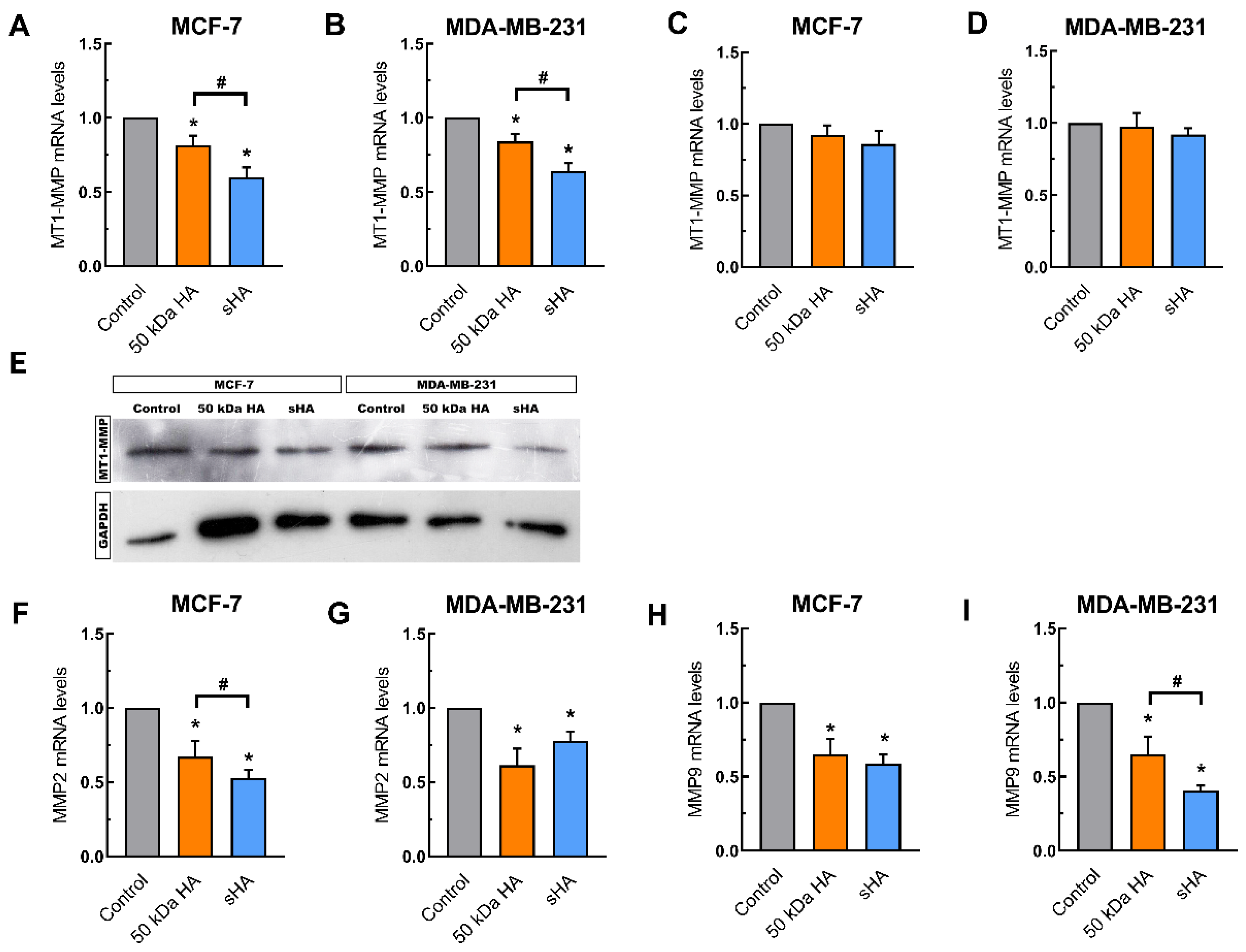
3.4. Sulfated HA Downregulates the Expression of Matrix Metalloproteinases
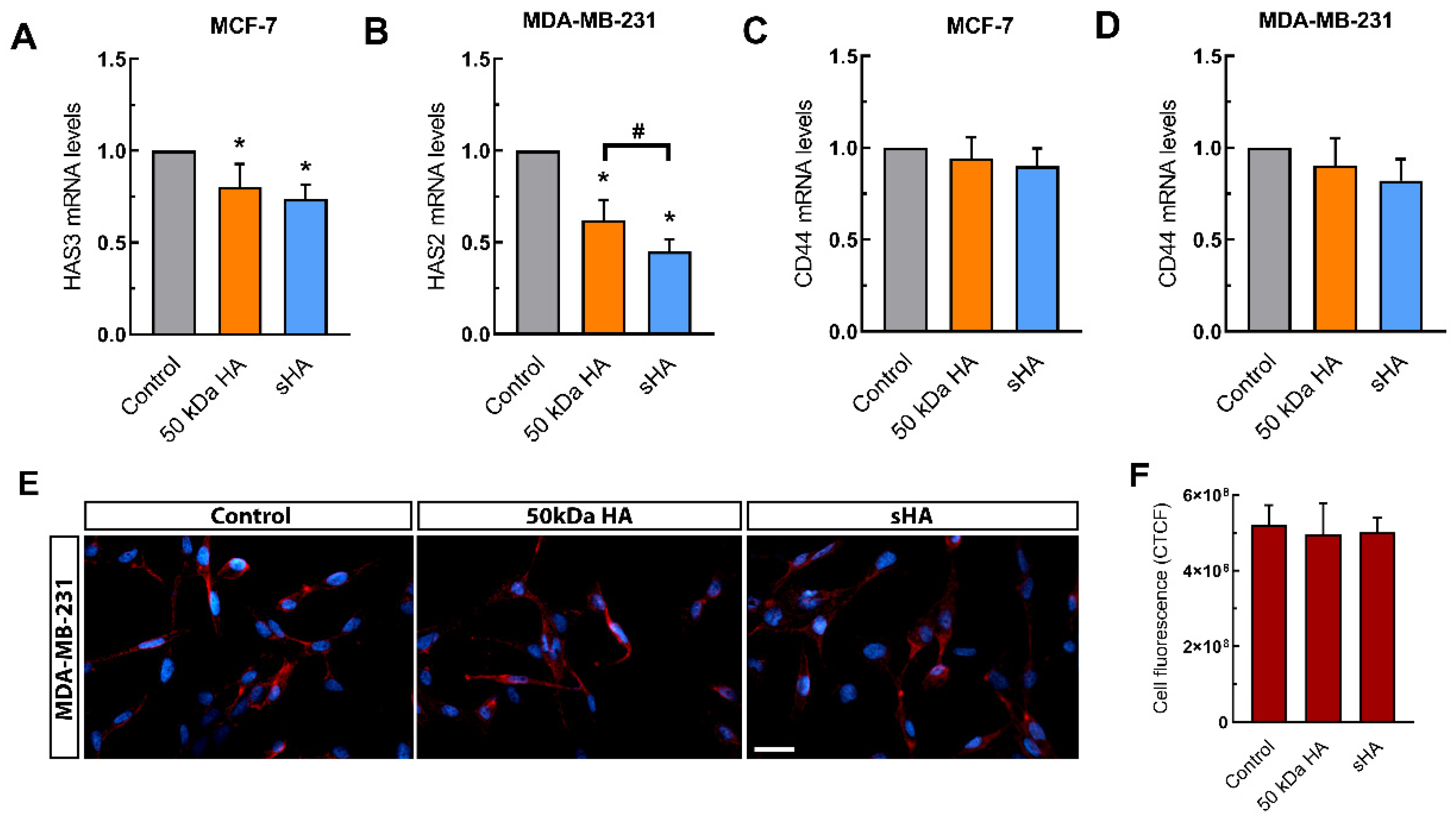
3.5. Sulfated HA Modulates the Expression of HAS but Not CD44
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Piperigkou, Z.; Karamanos, N.K. Estrogen Receptor-Mediated Targeting of the Extracellular Matrix Network in Cancer. Semin. Cancer Biol. 2020, 62, 116–124. [Google Scholar] [CrossRef] [PubMed]
- Bouris, P.; Skandalis, S.S.; Piperigkou, Z.; Afratis, N.; Karamanou, K.; Aletras, A.J.; Moustakas, A.; Theocharis, A.D.; Karamanos, N.K. Estrogen Receptor Alpha Mediates Epithelial to Mesenchymal Transition, Expression of Specific Matrix Effectors and Functional Properties of Breast Cancer Cells. Matrix Biol. 2015, 43, 42–60. [Google Scholar] [CrossRef] [PubMed]
- Chekhun, S.; Bezdenezhnykh, N.; Shvets, J.; Lukianova, N. Expression of Biomarkers Related to Cell Adhesion, Metastasis and Invasion of Breast Cancer Cell Lines of Different Molecular Subtype. Exp. Oncol. 2013, 35, 174–179. [Google Scholar]
- Nicolini, A.; Ferrari, P.; Duffy, M.J. Prognostic and Predictive Biomarkers in Breast Cancer: Past, Present and Future. Semin. Cancer Biol. 2018, 52, 56–73. [Google Scholar] [CrossRef]
- Gritsenko, P.G.; Ilina, O.; Friedl, P. Interstitial Guidance of Cancer Invasion. J. Pathol. 2012, 226, 185–199. [Google Scholar] [CrossRef]
- Jo, S.J.; Park, P.-G.; Cha, H.-R.; Ahn, S.G.; Kim, M.J.; Kim, H.; Koo, J.S.; Jeong, J.; Park, J.H.; Dong, S.M.; et al. Cellular Inhibitor of Apoptosis Protein 2 Promotes the Epithelial-Mesenchymal Transition in Triple-Negative Breast Cancer Cells through Activation of the AKT Signaling Pathway. Oncotarget 2017, 8, 78781–78795. [Google Scholar] [CrossRef] [Green Version]
- Medici, D.; Hay, E.D.; Olsen, B.R. Snail and Slug Promote Epithelial-Mesenchymal Transition through β-Catenin–T-Cell Factor-4-Dependent Expression of Transforming Growth Factor-Β3. MBoC 2008, 19, 4875–4887. [Google Scholar] [CrossRef] [Green Version]
- Thiery, J.P. Epithelial–Mesenchymal Transitions in Tumour Progression. Nat. Rev. Cancer 2002, 2, 442–454. [Google Scholar] [CrossRef]
- Sciacovelli, M.; Frezza, C. Metabolic Reprogramming and Epithelial-to-Mesenchymal Transition in Cancer. FEBS J. 2017, 284, 3132–3144. [Google Scholar] [CrossRef]
- Gialeli, C.; Theocharis, A.D.; Karamanos, N.K. Roles of Matrix Metalloproteinases in Cancer Progression and Their Pharmacological Targeting: MMPs as Potential Targets in Malignancy. FEBS J. 2011, 278, 16–27. [Google Scholar] [CrossRef]
- Karamanos, N.K.; Theocharis, A.D.; Neill, T.; Iozzo, R.V. Matrix Modeling and Remodeling: A Biological Interplay Regulating Tissue Homeostasis and Diseases. Matrix Biol. 2019, 75–76, 1–11. [Google Scholar] [CrossRef]
- Theocharis, A.D.; Skandalis, S.S.; Gialeli, C.; Karamanos, N.K. Extracellular Matrix Structure. Adv. Drug Deliv. Rev. 2016, 97, 4–27. [Google Scholar] [CrossRef]
- Oikari, S.; Kettunen, T.; Tiainen, S.; Häyrinen, J.; Masarwah, A.; Sudah, M.; Sutela, A.; Vanninen, R.; Tammi, M.; Auvinen, P. UDP-Sugar Accumulation Drives Hyaluronan Synthesis in Breast Cancer. Matrix Biol. 2018, 67, 63–74. [Google Scholar] [CrossRef] [Green Version]
- Heldin, P.; Lin, C.-Y.; Kolliopoulos, C.; Chen, Y.-H.; Skandalis, S.S. Regulation of Hyaluronan Biosynthesis and Clinical Impact of Excessive Hyaluronan Production. Matrix Biol. 2019, 78–79, 100–117. [Google Scholar] [CrossRef]
- Passi, A.; Vigetti, D.; Buraschi, S.; Iozzo, R.V. Dissecting the Role of Hyaluronan Synthases in the Tumor Microenvironment. FEBS J. 2019, 286, 2937–2949. [Google Scholar] [CrossRef]
- Ouhtit, A.; Rizeq, B.; Saleh, H.A.; Rahman, M.M.; Zayed, H. Novel CD44-Downstream Signaling Pathways Mediating Breast Tumor Invasion. Int. J. Biol. Sci. 2018, 14, 1782–1790. [Google Scholar] [CrossRef] [Green Version]
- Misra, S.; Heldin, P.; Hascall, V.C.; Karamanos, N.K.; Skandalis, S.S.; Markwald, R.R.; Ghatak, S. Hyaluronan-CD44 Interactions as Potential Targets for Cancer Therapy. FEBS J. 2011, 278, 1429–1443. [Google Scholar] [CrossRef] [Green Version]
- Skandalis, S.S.; Karalis, T.T.; Chatzopoulos, A.; Karamanos, N.K. Hyaluronan-CD44 Axis Orchestrates Cancer Stem Cell Functions. Cell. Signal. 2019, 63, 109377. [Google Scholar] [CrossRef]
- Garantziotis, S.; Savani, R.C. Hyaluronan Biology: A Complex Balancing Act of Structure, Function, Location and Context. Matrix Biol. 2019, 78–79, 1–10. [Google Scholar] [CrossRef]
- Velesiotis, C.; Vasileiou, S.; Vynios, D.H. A Guide to Hyaluronan and Related Enzymes in Breast Cancer: Biological Significance and Diagnostic Value. FEBS J. 2019, 286, 3057–3074. [Google Scholar] [CrossRef]
- Tammi, M.I.; Oikari, S.; Pasonen-Seppänen, S.; Rilla, K.; Auvinen, P.; Tammi, R.H. Activated Hyaluronan Metabolism in the Tumor Matrix—Causes and Consequences. Matrix Biol. 2019, 78–79, 147–164. [Google Scholar] [CrossRef]
- Piperigkou, Z.; Kyriakopoulou, K.; Koutsakis, C.; Mastronikolis, S.; Karamanos, N.K. Key Matrix Remodeling Enzymes: Functions and Targeting in Cancer. Cancers 2021, 13, 1441. [Google Scholar] [CrossRef]
- Tavianatou, A.; Piperigkou, Z.; Koutsakis, C.; Barbera, C.; Beninatto, R.; Franchi, M.; Karamanos, N.K. The Action of Hyaluronan in Functional Properties, Morphology and Expression of Matrix Effectors in Mammary Cancer Cells Depends on Its Molecular Size. FEBS J. 2021. [Google Scholar] [CrossRef]
- Tavianatou, A.-G.; Piperigkou, Z.; Barbera, C.; Beninatto, R.; Masola, V.; Caon, I.; Onisto, M.; Franchi, M.; Galesso, D.; Karamanos, N.K. Molecular Size-Dependent Specificity of Hyaluronan on Functional Properties, Morphology and Matrix Composition of Mammary Cancer Cells. Matrix Biol. Plus 2019, 3, 100008. [Google Scholar] [CrossRef]
- Tavianatou, A.G.; Caon, I.; Franchi, M.; Piperigkou, Z.; Galesso, D.; Karamanos, N.K. Hyaluronan: Molecular Size-dependent Signaling and Biological Functions in Inflammation and Cancer. FEBS J. 2019, 286, 2883–2908. [Google Scholar] [CrossRef]
- Toole, B.; Ghatak, S.; Misra, S. Hyaluronan Oligosaccharides as a Potential Anticancer Therapeutic. Curr. Pharm. Biotechnol. 2008, 9, 249–252. [Google Scholar] [CrossRef]
- Urakawa, H.; Nishida, Y.; Knudson, W.; Knudson, C.B.; Arai, E.; Kozawa, E.; Futamura, N.; Wasa, J.; Ishiguro, N. Therapeutic Potential of Hyaluronan Oligosaccharides for Bone Metastasis of Breast Cancer. J. Orthop. Res. 2012, 30, 662–672. [Google Scholar] [CrossRef]
- Zhao, Y.; Qiao, S.; Shi, S.; Yao, L.; Hou, X.; Li, C.; Lin, F.-H.; Guo, K.; Acharya, A.; Chen, X.; et al. Modulating Three-Dimensional Microenvironment with Hyaluronan of Different Molecular Weights Alters Breast Cancer Cell Invasion Behavior. ACS Appl. Mater. Interfaces 2017, 9, 9327–9338. [Google Scholar] [CrossRef]
- Benitez, A.; Yates, T.J.; Lopez, L.E.; Cerwinka, W.H.; Bakkar, A.; Lokeshwar, V.B. Targeting Hyaluronidase for Cancer Therapy: Antitumor Activity of Sulfated Hyaluronic Acid in Prostate Cancer Cells. Cancer Res. 2011, 71, 4085–4095. [Google Scholar] [CrossRef] [Green Version]
- Jordan, A.R.; Lokeshwar, S.D.; Lopez, L.E.; Hennig, M.; Chipollini, J.; Yates, T.; Hupe, M.C.; Merseburger, A.S.; Shiedlin, A.; Cerwinka, W.H.; et al. Antitumor Activity of Sulfated Hyaluronic Acid Fragments in Pre-Clinical Models of Bladder Cancer. Oncotarget 2017, 8, 24262–24274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feoktistova, M.; Geserick, P.; Leverkus, M. Crystal Violet Assay for Determining Viability of Cultured Cells. Cold Spring Harb. Protoc. 2016, 2016. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An Open-Source Platform for Biological-Image Analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Wever, O.; Hendrix, A.; De Boeck, A.; Westbroek, W.; Braems, G.; Emami, S.; Sabbah, M.; Gespach, C.; Bracke, M. Modeling and Quantification of Cancer Cell Invasion through Collagen Type I Matrices. Int. J. Dev. Biol. 2010, 54, 887–896. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [Green Version]
- Kolliopoulos, C.; Lin, C.-Y.; Heldin, C.-H.; Moustakas, A.; Heldin, P. Has2 Natural Antisense RNA and Hmga2 Promote Has2 Expression during TGFβ-Induced EMT in Breast Cancer. Matrix Biol. 2019, 80, 29–45. [Google Scholar] [CrossRef]
- Heldin, P.; Basu, K.; Kozlova, I.; Porsch, H. HAS2 and CD44 in Breast Tumorigenesis. In Advances in Cancer Research; Elsevier: Amsterdam, The Netherlands, 2014; Volume 123, pp. 211–229. ISBN 978-0-12-800092-2. [Google Scholar]
- Bernert, B.; Porsch, H.; Heldin, P. Hyaluronan Synthase 2 (HAS2) Promotes Breast Cancer Cell Invasion by Suppression of Tissue Metalloproteinase Inhibitor 1 (TIMP-1). J. Biol. Chem. 2011, 286, 42349–42359. [Google Scholar] [CrossRef] [Green Version]
- Bohaumilitzky, L.; Huber, A.-K.; Stork, E.M.; Wengert, S.; Woelfl, F.; Boehm, H. A Trickster in Disguise: Hyaluronan’s Ambivalent Roles in the Matrix. Front. Oncol. 2017, 7, 242. [Google Scholar] [CrossRef]
- Hamilton, S.R.; Fard, S.F.; Paiwand, F.F.; Tolg, C.; Veiseh, M.; Wang, C.; McCarthy, J.B.; Bissell, M.J.; Koropatnick, J.; Turley, E.A. The Hyaluronan Receptors CD44 and Rhamm (CD168) Form Complexes with ERK1,2 That Sustain High Basal Motility in Breast Cancer Cells. J. Biol. Chem. 2007, 282, 16667–16680. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Qi, L.; Liang, Z.; Song, W.; Liu, Y.; Wang, Y.; Sun, B.; Zhang, B.; Cao, W. Transforming Growth Factor-Β1 Induces EMT by the Transactivation of Epidermal Growth Factor Signaling through HA/CD44 in Lung and Breast Cancer Cells. Int. J. Mol. Med. 2015, 36, 113–122. [Google Scholar] [CrossRef] [Green Version]
| Gene | Primer Sequence (5′–3′) | Annealing T (°C) | |
|---|---|---|---|
| HAS2 | F | TCGCAACACGTAACGCAAT | 60 °C |
| R | ACTTCTCTTTTTCCACCCCATTT | ||
| HAS3 | F | AACAAGTACGACTCATGGATTTCCT | 60 °C |
| R | GCCCGCTCCACGTTGA | ||
| e-cadherin | F | TACGCCTGGGACTCCACCTA | 60 °C |
| R | CCAGAAACGGAGGCCTGAT | ||
| snail2/slug | F | AGACCCTGGTTGCTTCAAGGA | 60 °C |
| R | CTCAGATTTGACCTGTCTGCAAA | ||
| MT1-MMP | F | CATGGGCAGCGATGAAGTCT | 60 °C |
| R | CCAGTATTTGTTCCCCTTGTAGAAGTA | ||
| MMP2 | F | CGTCTGTCCCAGGATGACATC | 62 °C |
| R | ATGTCAGGAGAGGCCCCATA | ||
| GAPDH | F | AGGCTGTTGTCATACTTCTCAT | 60 °C |
| R | GGAGTCCACTGGCGTCTT | ||
| MMP9 | F | TTCCAGTACCGAGAGAAAGCCTAT | 60 °C |
| R | GGTCACGTAGCCCACTTGGT | ||
| CD44 | F | ATAATAAAGGAGCAGCACTTCAGGA | 60 °C |
| R | ATAATTTGTGTCTTGGTCTCTGGTAGC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koutsakis, C.; Tavianatou, A.-G.; Kokoretsis, D.; Baroutas, G.; Karamanos, N.K. Sulfated Hyaluronan Modulates the Functional Properties and Matrix Effectors Expression of Breast Cancer Cells with Different Estrogen Receptor Status. Biomolecules 2021, 11, 1916. https://doi.org/10.3390/biom11121916
Koutsakis C, Tavianatou A-G, Kokoretsis D, Baroutas G, Karamanos NK. Sulfated Hyaluronan Modulates the Functional Properties and Matrix Effectors Expression of Breast Cancer Cells with Different Estrogen Receptor Status. Biomolecules. 2021; 11(12):1916. https://doi.org/10.3390/biom11121916
Chicago/Turabian StyleKoutsakis, Christos, Anastasia-Gerasimoula Tavianatou, Dimitris Kokoretsis, Georgios Baroutas, and Nikos K. Karamanos. 2021. "Sulfated Hyaluronan Modulates the Functional Properties and Matrix Effectors Expression of Breast Cancer Cells with Different Estrogen Receptor Status" Biomolecules 11, no. 12: 1916. https://doi.org/10.3390/biom11121916
APA StyleKoutsakis, C., Tavianatou, A. -G., Kokoretsis, D., Baroutas, G., & Karamanos, N. K. (2021). Sulfated Hyaluronan Modulates the Functional Properties and Matrix Effectors Expression of Breast Cancer Cells with Different Estrogen Receptor Status. Biomolecules, 11(12), 1916. https://doi.org/10.3390/biom11121916