LPS Tolerance Inhibits Cellular Respiration and Induces Global Changes in the Macrophage Secretome
Abstract
:1. Introduction
2. Materials and Methods
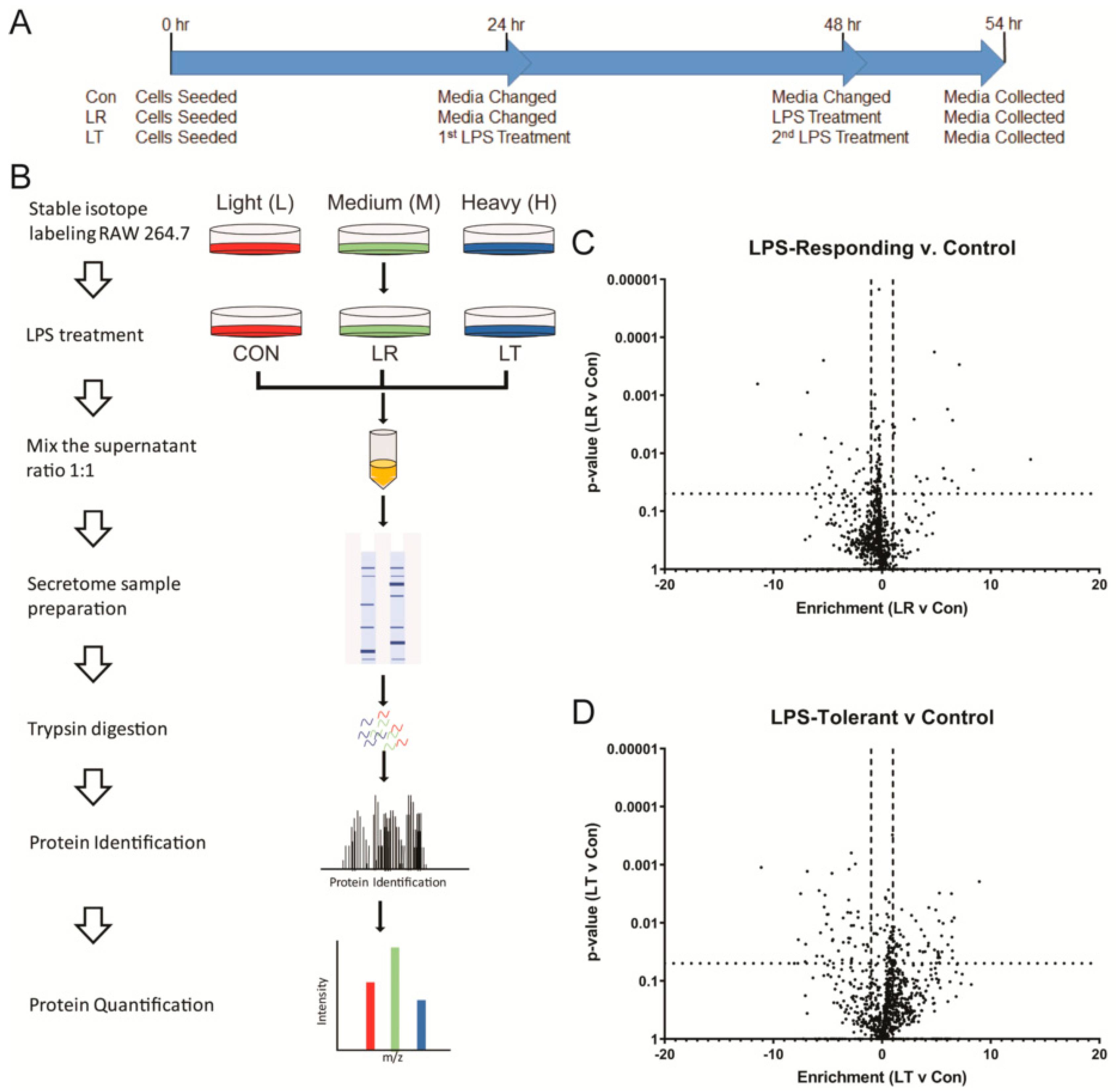
2.1. Cell Culture and Reagents
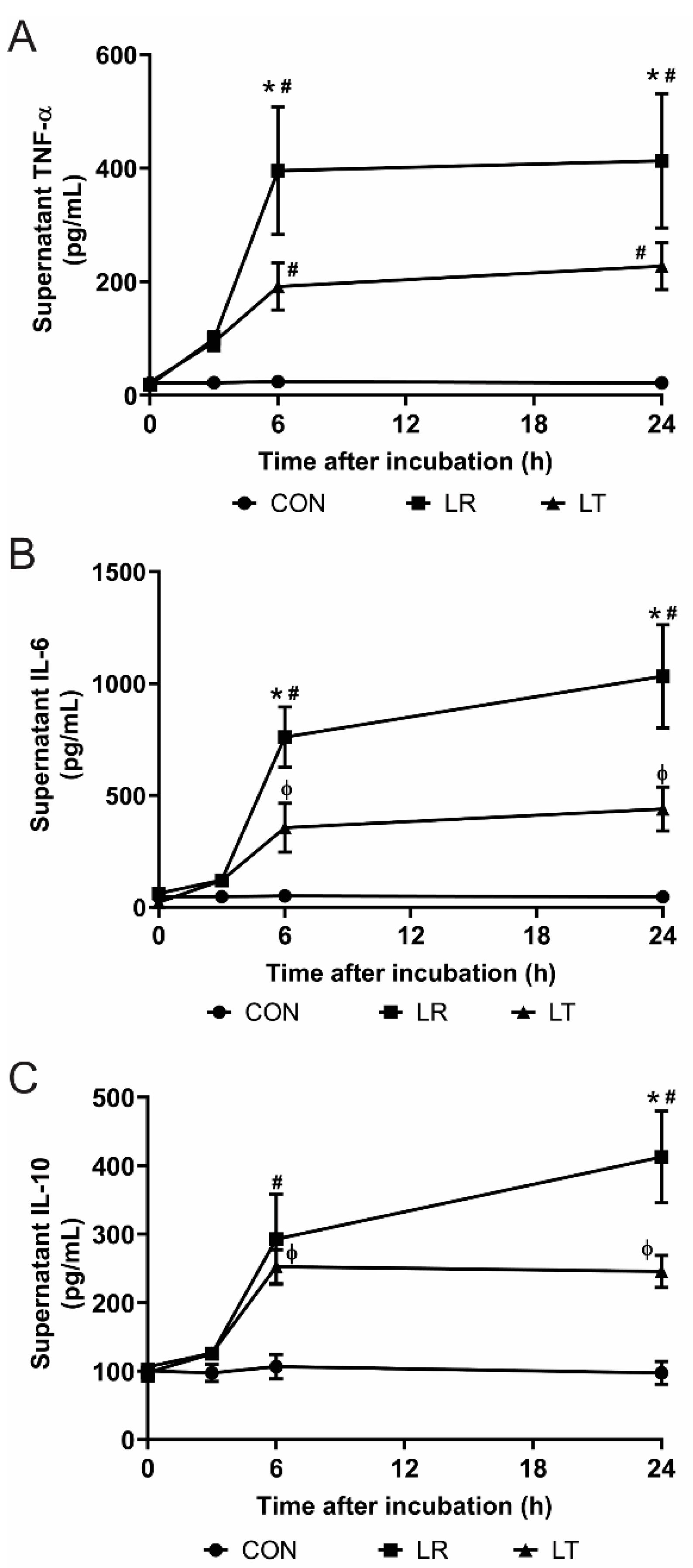
2.2. Quantification of Secreted Cytokines
2.3. Extracellular Flux Analysis
2.4. Collection of Secreted Proteins
2.5. In-Gel Digestion of Secreted Proteins
2.6. Mass Spectrometry
2.7. Analysis of MS Results
3. Results
3.1. LPS-Tolerant Macrophages Decrease Cellular Respiration
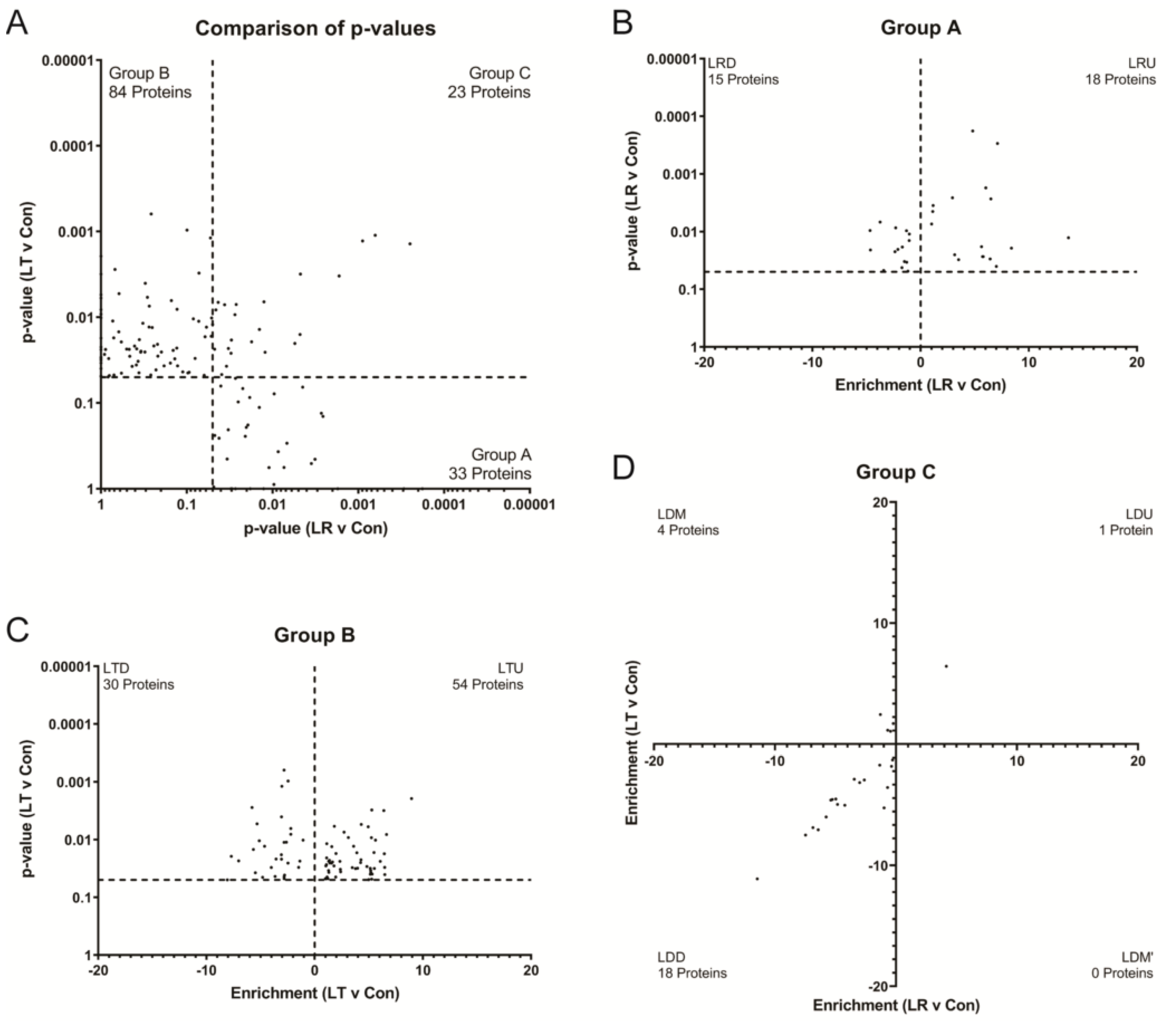
3.2. Variations in LPS Treatment Lead to Variations in the Secretome
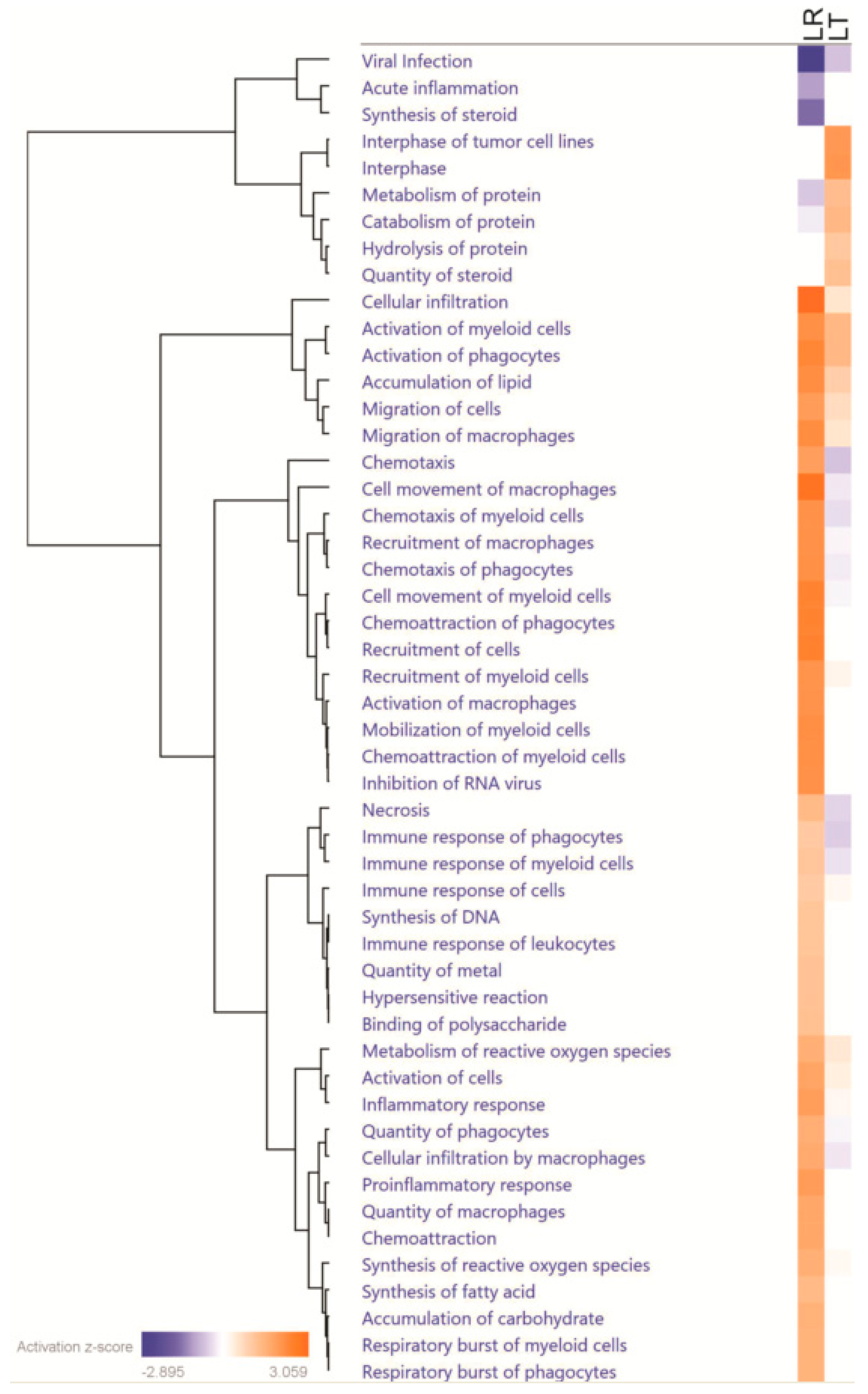
3.3. Pathway Analysis of Critical Groups
3.4. Relationship between Repeated LPS Stimulation and Cellular Exhaustion
4. Discussion
4.1. Most Evident Protein Level Changes in the Secretome
4.2. Potential Protein Regulators of LPS Tolerance
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Akira, S.; Uematsu, S.; Takeuchi, O. Pathogen recognition and innate immunity. Cell 2006, 124, 783–801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawai, T.; Akira, S. The role of pattern-recognition receptors in innate immunity: Update on Toll-like receptors. Nat. Immunol. 2010, 11, 373–384. [Google Scholar] [CrossRef] [PubMed]
- Salomao, R.; Brunialti, M.K.; Rapozo, M.M.; Baggio-Zappia, G.L.; Galanos, C.; Freudenberg, M. Bacterial sensing, cell signaling, and modulation of the immune response during sepsis. Shock 2012, 38, 227–242. [Google Scholar] [CrossRef] [Green Version]
- Seeley, J.J.; Ghosh, S. Molecular mechanisms of innate memory and tolerance to LPS. J. Leukoc. Biol. 2017, 101, 107–119. [Google Scholar] [CrossRef] [PubMed]
- Hotchkiss, R.S.; Monneret, G.; Payen, D. Sepsis-induced immunosuppression: From cellular dysfunctions to immunotherapy. Nat. Rev. Immunol. 2013, 13, 862–874. [Google Scholar] [CrossRef] [PubMed]
- Foster, S.L.; Medzhitov, R. Gene-specific control of the TLR-induced inflammatory response. Clin. Immunol. 2009, 130, 7–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Netea, M.G.; Quintin, J.; van der Meer, J.W. Trained immunity: A memory for innate host defense. Cell Host Microbe 2011, 9, 355–361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biswas, S.K.; Lopez-Collazo, E. Endotoxin tolerance: New mechanisms, molecules and clinical significance. Trends Immunol. 2009, 30, 475–487. [Google Scholar] [CrossRef]
- Quinn, E.M.; Wang, J.; Redmond, H.P. The emerging role of microRNA in regulation of endotoxin tolerance. J. Leukoc. Biol. 2012, 91, 721–727. [Google Scholar] [CrossRef]
- He, X.; Jing, Z.; Cheng, G. MicroRNAs: New regulators of Toll-like receptor signalling pathways. Biomed. Res. Int. 2014, 2014, 945169. [Google Scholar] [CrossRef] [Green Version]
- Pena, O.M.; Hancock, D.G.; Lyle, N.H.; Linder, A.; Russell, J.A.; Xia, J.; Fjell, C.D.; Boyd, J.H.; Hancock, R.E. An Endotoxin Tolerance Signature Predicts Sepsis and Organ Dysfunction at Initial Clinical Presentation. EBioMedicine 2014, 1, 64–71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shalova, I.N.; Lim, J.Y.; Chittezhath, M.; Zinkernagel, A.S.; Beasley, F.; Hernandez-Jimenez, E.; Toledano, V.; Cubillos-Zapata, C.; Rapisarda, A.; Chen, J.; et al. Human monocytes undergo functional re-programming during sepsis mediated by hypoxia-inducible factor-1alpha. Immunity 2015, 42, 484–498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vergadi, E.; Vaporidi, K.; Tsatsanis, C. Regulation of Endotoxin Tolerance and Compensatory Anti-inflammatory Response Syndrome by Non-coding RNAs. Front. Immunol. 2018, 9, 2705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lopez-Collazo, E.; del Fresno, C. Pathophysiology of endotoxin tolerance: Mechanisms and clinical consequences. Crit. Care 2013, 17, 242. [Google Scholar] [CrossRef] [Green Version]
- Ondee, T.; Gillen, J.; Visitchanakun, P.; Somparn, P.; Issara-Amphorn, J.; Dang Phi, C.; Chancharoenthana, W.; Gurusamy, D.; Nita-Lazar, A.; Leelahavanichkul, A. Lipocalin-2 (Lcn-2) Attenuates Polymicrobial Sepsis with LPS Preconditioning (LPS Tolerance) in FcGRIIb Deficient Lupus Mice. Cells 2019, 8, 1064. [Google Scholar] [CrossRef] [Green Version]
- Dominguez-Andres, J.; Novakovic, B.; Li, Y.; Scicluna, B.P.; Gresnigt, M.S.; Arts, R.J.W.; Oosting, M.; Moorlag, S.; Groh, L.A.; Zwaag, J.; et al. The Itaconate Pathway Is a Central Regulatory Node Linking Innate Immune Tolerance and Trained Immunity. Cell Metab 2019, 29, 211–220.e5. [Google Scholar] [CrossRef] [Green Version]
- Widdrington, J.D.; Gomez-Duran, A.; Pyle, A.; Ruchaud-Sparagano, M.H.; Scott, J.; Baudouin, S.V.; Rostron, A.J.; Lovat, P.E.; Chinnery, P.F.; Simpson, A.J. Exposure of Monocytic Cells to Lipopolysaccharide Induces Coordinated Endotoxin Tolerance, Mitochondrial Biogenesis, Mitophagy, and Antioxidant Defenses. Front. Immunol. 2018, 9, 2217. [Google Scholar] [CrossRef]
- Mosser, D.M.; Edwards, J.P. Exploring the full spectrum of macrophage activation. Nat. Rev. Immunol. 2008, 8, 958–969. [Google Scholar] [CrossRef]
- Koppenol-Raab, M.; Sjoelund, V.; Manes, N.P.; Gottschalk, R.A.; Dutta, B.; Benet, Z.L.; Fraser, I.D.; Nita-Lazar, A. Proteome and Secretome Analysis Reveals Differential Post-transcriptional Regulation of Toll-like Receptor Responses. Mol. Cell Proteom. 2017, 16, S172–S186. [Google Scholar] [CrossRef] [Green Version]
- Meissner, F.; Scheltema, R.A.; Mollenkopf, H.J.; Mann, M. Direct proteomic quantification of the secretome of activated immune cells. Science 2013, 340, 475–478. [Google Scholar] [CrossRef]
- Eichelbaum, K.; Krijgsveld, J. Rapid temporal dynamics of transcription, protein synthesis, and secretion during macrophage activation. Mol. Cell Proteom. 2014, 13, 792–810. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shevchenko, A.; Tomas, H.; Havlis, J.; Olsen, J.V.; Mann, M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 2006, 1, 2856–2860. [Google Scholar] [CrossRef] [PubMed]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Neuhauser, N.; Michalski, A.; Scheltema, R.A.; Olsen, J.V.; Mann, M. Andromeda: A peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 2011, 10, 1794–1805. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016, 11, 2301–2319. [Google Scholar] [CrossRef]
- Polpitiya, A.D.; Qian, W.J.; Jaitly, N.; Petyuk, V.A.; Adkins, J.N.; Camp, D.G., 2nd; Anderson, G.A.; Smith, R.D. DAnTE: A statistical tool for quantitative analysis of -omics data. Bioinformatics 2008, 24, 1556–1558. [Google Scholar] [CrossRef]
- Motani, K.; Kosako, H. Activation of stimulator of interferon genes (STING) induces ADAM17-mediated shedding of the immune semaphorin SEMA4D. J. Biol. Chem. 2018, 293, 7717–7726. [Google Scholar] [CrossRef] [Green Version]
- Hernandez, M.X.; Jiang, S.; Cole, T.A.; Chu, S.H.; Fonseca, M.I.; Fang, M.J.; Hohsfield, L.A.; Torres, M.D.; Green, K.N.; Wetsel, R.A.; et al. Prevention of C5aR1 signaling delays microglial inflammatory polarization, favors clearance pathways and suppresses cognitive loss. Mol. Neurodegener. 2017, 12, 66. [Google Scholar] [CrossRef]
- Heissig, B.; Salama, Y.; Takahashi, S.; Osada, T.; Hattori, K. The multifaceted role of plasminogen in inflammation. Cell Signal. 2020, 75, 109761. [Google Scholar] [CrossRef]
- Scheurmann, J.; Treiber, N.; Weber, C.; Renkl, A.C.; Frenzel, D.; Trenz-Buback, F.; Ruess, A.; Schulz, G.; Scharffetter-Kochanek, K.; Weiss, J.M. Mice with heterozygous deficiency of manganese superoxide dismutase (SOD2) have a skin immune system with features of “inflamm-aging”. Arch. Dermatol. Res. 2014, 306, 143–155. [Google Scholar] [CrossRef] [PubMed]
- Regdon, Z.; Robaszkiewicz, A.; Kovacs, K.; Rygielska, Z.; Hegedus, C.; Bodoor, K.; Szabo, E.; Virag, L. LPS protects macrophages from AIF-independent parthanatos by downregulation of PARP1 expression, induction of SOD2 expression, and a metabolic shift to aerobic glycolysis. Free Radic. Biol. Med. 2019, 131, 184–196. [Google Scholar] [CrossRef] [PubMed]
- Daniels, C.M.; Kaplan, P.R.; Bishof, I.; Bradfield, C.; Tucholski, T.; Nuccio, A.G.; Manes, N.P.; Katz, S.; Fraser, I.D.C.; Nita-Lazar, A. Dynamic ADP-Ribosylome, Phosphoproteome, and Interactome in LPS-Activated Macrophages. J. Proteome Res. 2020, 19, 3716–3731. [Google Scholar] [CrossRef] [PubMed]
- Lamark, T.; Svenning, S.; Johansen, T. Regulation of selective autophagy: The p62/SQSTM1 paradigm. Essays BioChem. 2017, 61, 609–624. [Google Scholar] [CrossRef]
- Irie, T.; Muta, T.; Takeshige, K. TAK1 mediates an activation signal from toll-like receptor(s) to nuclear factor-kappaB in lipopolysaccharide-stimulated macrophages. FEBS Lett. 2000, 467, 160–164. [Google Scholar] [CrossRef] [Green Version]
- Kehl, S.R.; Soos, B.A.; Saha, B.; Choi, S.W.; Herren, A.W.; Johansen, T.; Mandell, M.A. TAK1 converts Sequestosome 1/p62 from an autophagy receptor to a signaling platform. EMBO Rep. 2019, 20, e46238. [Google Scholar] [CrossRef] [Green Version]
- Giachelli, C.; Bae, N.; Lombardi, D.; Majesky, M.; Schwartz, S. Molecular cloning and characterization of 2B7, a rat mRNA which distinguishes smooth muscle cell phenotypes in vitro and is identical to osteopontin (secreted phosphoprotein I, 2aR). BioChem. Biophys. Res. Commun. 1991, 177, 867–873. [Google Scholar] [CrossRef]
- Miyauchi, A.; Alvarez, J.; Greenfield, E.M.; Teti, A.; Grano, M.; Colucci, S.; Zambonin-Zallone, A.; Ross, F.P.; Teitelbaum, S.L.; Cheresh, D.; et al. Recognition of osteopontin and related peptides by an alpha v beta 3 integrin stimulates immediate cell signals in osteoclasts. J. Biol. Chem. 1991, 266, 20369–20374. [Google Scholar] [CrossRef]
- Murry, C.E.; Giachelli, C.M.; Schwartz, S.M.; Vracko, R. Macrophages express osteopontin during repair of myocardial necrosis. Am. J. Pathol. 1994, 145, 1450–1462. [Google Scholar]
- Henricson, B.E.; Neta, R.; Vogel, S.N. An interleukin-1 receptor antagonist blocks lipopolysaccharide-induced colony-stimulating factor production and early endotoxin tolerance. Infect. Immun. 1991, 59, 1188–1191. [Google Scholar] [CrossRef] [Green Version]
- Nakano, M.; Saito, S.; Nakano, Y.; Yamasu, H.; Matsuura, M.; Shinomiya, H. Intracellular protein phosphorylation in murine peritoneal macrophages in response to bacterial lipopolysaccharide (LPS): Effects of kinase-inhibitors and LPS-induced tolerance. Immunobiology 1993, 187, 272–282. [Google Scholar] [CrossRef]
- Ondee, T.; Jaroonwitchawan, T.; Pisitkun, T.; Gillen, J.; Nita-Lazar, A.; Leelahavanichkul, A.; Somparn, P. Decreased Protein Kinase C-beta Type II Associated with the Prominent Endotoxin Exhaustion in the Macrophage of FcGRIIb-/- Lupus Prone Mice is Revealed by Phosphoproteomic Analysis. Int. J. Mol. Sci 2019, 20, 1354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karimi, Y.; Poznanski, S.M.; Vahedi, F.; Chen, B.; Chew, M.V.; Lee, A.J.; Ashkar, A.A. Type I interferon signalling is not required for the induction of endotoxin tolerance. Cytokine 2017, 95, 7–11. [Google Scholar] [CrossRef] [PubMed]
- Lopategi, A.; Flores-Costa, R.; Rius, B.; Lopez-Vicario, C.; Alcaraz-Quiles, J.; Titos, E.; Claria, J. Frontline Science: Specialized proresolving lipid mediators inhibit the priming and activation of the macrophage NLRP3 inflammasome. J. Leukoc. Biol. 2019, 105, 25–36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saha, S.; Shalova, I.N.; Biswas, S.K. Metabolic regulation of macrophage phenotype and function. Immunol. Rev. 2017, 280, 102–111. [Google Scholar] [CrossRef] [PubMed]
- Cavaillon, J.M.; Adib-Conquy, M. Bench-to-bedside review: Endotoxin tolerance as a model of leukocyte reprogramming in sepsis. Crit. Care 2006, 10, 233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ondee, T.; Surawut, S.; Taratummarat, S.; Hirankarn, N.; Palaga, T.; Pisitkun, P.; Pisitkun, T.; Leelahavanichkul, A. Fc Gamma Receptor IIB Deficient Mice: A Lupus Model with Increased Endotoxin Tolerance-Related Sepsis Susceptibility. Shock 2017, 47, 743–752. [Google Scholar] [CrossRef] [PubMed]

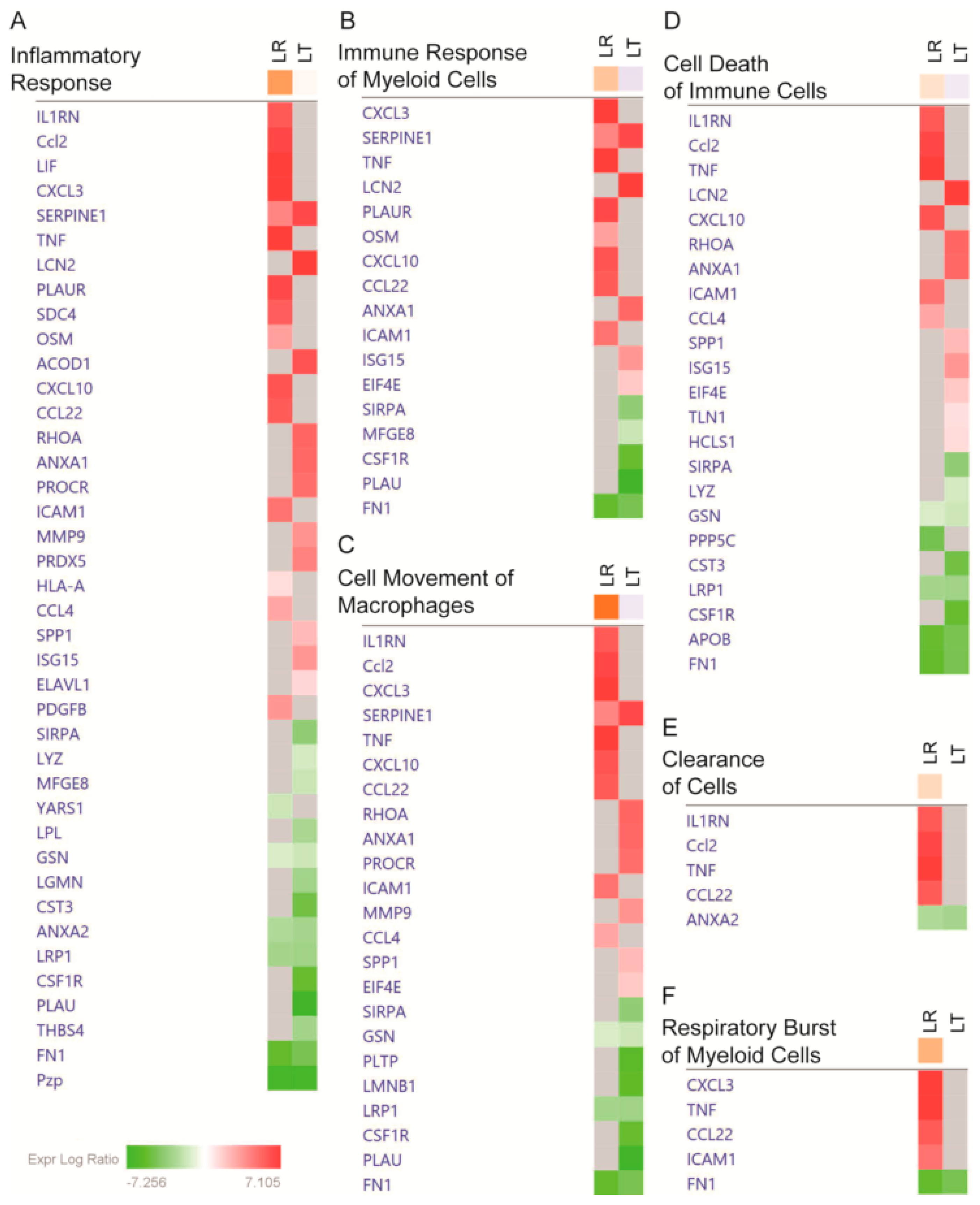
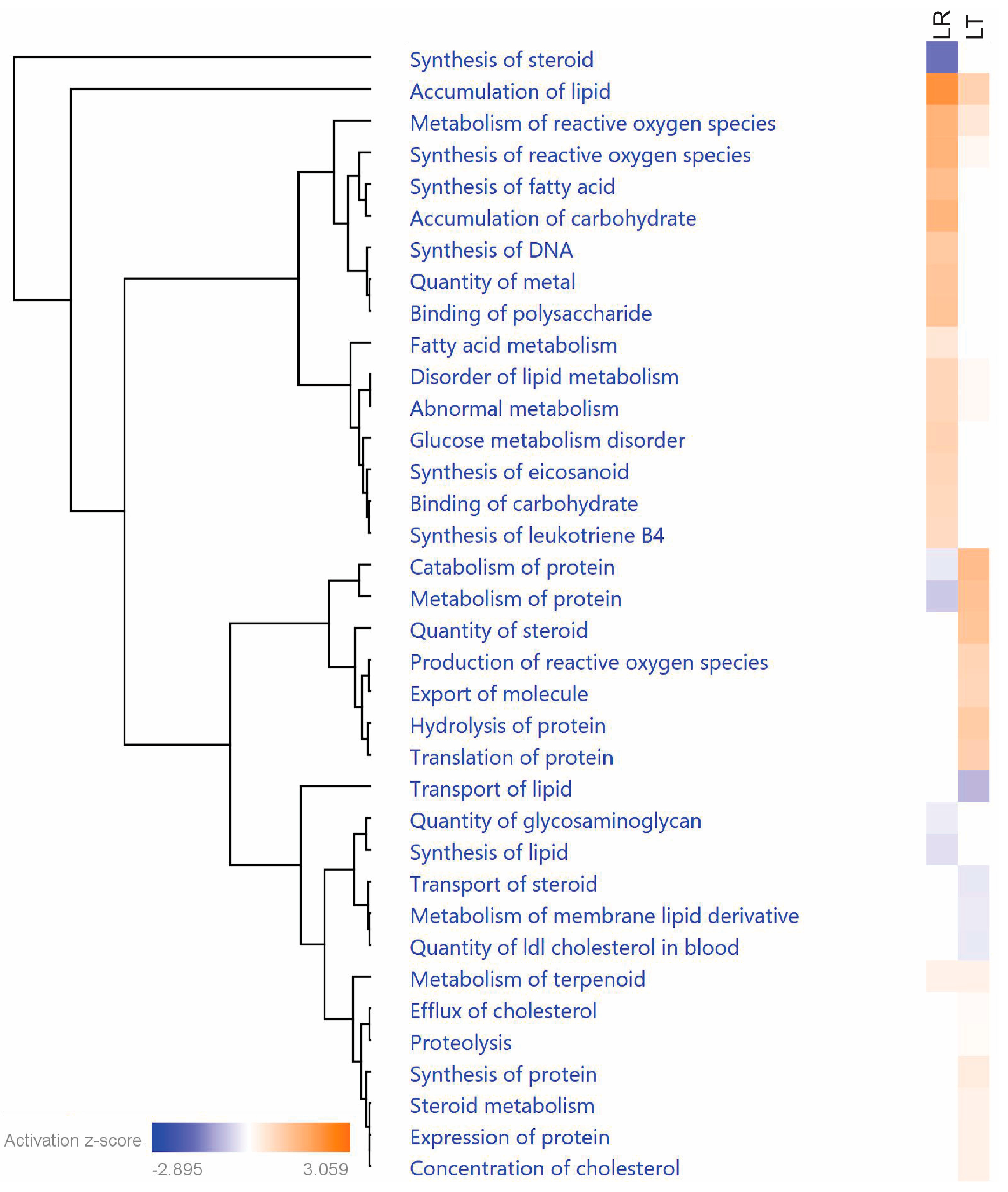
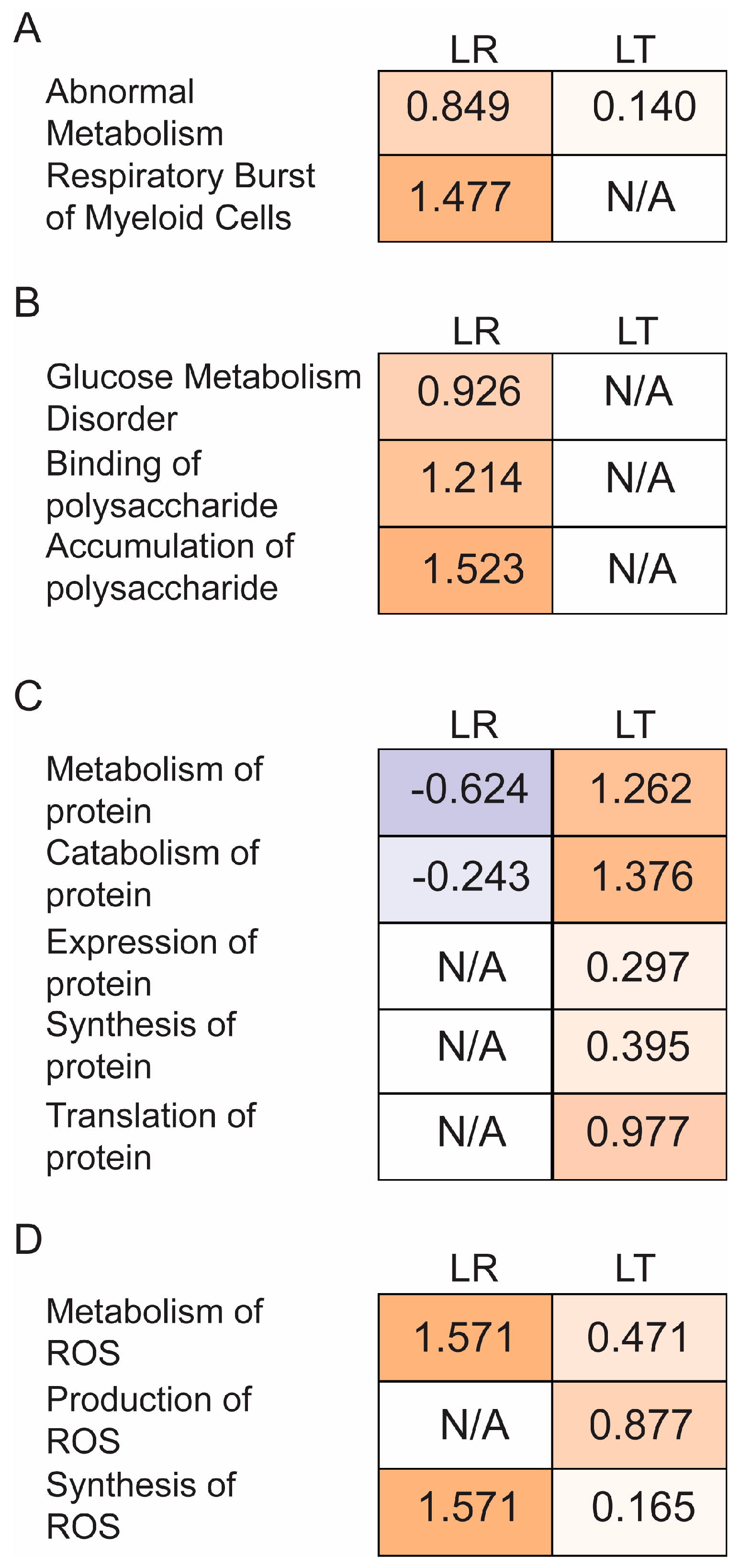
| Uniprot | Protein Name | Entrez Gene Name | LT Enrichment/LR Enrichment | Secreted | Cell Survival | Protein Metabolism | Redox |
|---|---|---|---|---|---|---|---|
| P09671 | SOD2 | superoxide dismutase 2 | 3.7913 | Yes (exosome) | Yes | Yes | Yes |
| Q64337 | SQSTM1 | sequestosome 1 | 4.6730 | Yes (exosome) | Yes | Yes | Yes |
| P10923 | SPP1 | secreted phosphoprotein 1 | 2.3679 | Yes | Yes | Yes | Yes |
| Q62351 | TFRC | transferrin receptor | 4.7225 | Yes | Yes | No | Yes |
| P60710 | ACTB | actin beta | 3.6136 | Yes (related) | Yes | No | Yes |
| P99029 | PRDX5 | peroxiredoxin 5 | 5.3956 | Yes (related) | Yes | No | Yes |
| Q3TCH7 | CUL4A | cullin 4A | 1.9345 | Yes (exosome) | Yes | Yes | No |
| Q9CXW4 | RPL11 | ribosomal protein L7 | 1.1280 | Yes (exosome) | Yes | Yes | No |
| P46471 | PSMC2 | proteasome 26S subunit, ATPase 2 | 1.6182 | Yes (related) | Yes | Yes | No |
| P11438 | LAMP1 | lysosomal associated membrane protein 1 | 2.8676 | Yes (exosome) | Yes | Yes | No |
| P22777 | SERPINE1 | serpin family E member 1 | 2.2662 | Yes | Yes | Yes | No |
| P25085 | IL1RN | interleukin 1 receptor antagonist | 1.0980 | Yes | Yes | Yes | No |
| P41245 | MMP9 | matrix metallopeptidase 9 | 3.7096 | Yes | Yes | Yes | No |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gillen, J.; Ondee, T.; Gurusamy, D.; Issara-Amphorn, J.; Manes, N.P.; Yoon, S.H.; Leelahavanichkul, A.; Nita-Lazar, A. LPS Tolerance Inhibits Cellular Respiration and Induces Global Changes in the Macrophage Secretome. Biomolecules 2021, 11, 164. https://doi.org/10.3390/biom11020164
Gillen J, Ondee T, Gurusamy D, Issara-Amphorn J, Manes NP, Yoon SH, Leelahavanichkul A, Nita-Lazar A. LPS Tolerance Inhibits Cellular Respiration and Induces Global Changes in the Macrophage Secretome. Biomolecules. 2021; 11(2):164. https://doi.org/10.3390/biom11020164
Chicago/Turabian StyleGillen, Joseph, Thunnicha Ondee, Devikala Gurusamy, Jiraphorn Issara-Amphorn, Nathan P. Manes, Sung Hwan Yoon, Asada Leelahavanichkul, and Aleksandra Nita-Lazar. 2021. "LPS Tolerance Inhibits Cellular Respiration and Induces Global Changes in the Macrophage Secretome" Biomolecules 11, no. 2: 164. https://doi.org/10.3390/biom11020164
APA StyleGillen, J., Ondee, T., Gurusamy, D., Issara-Amphorn, J., Manes, N. P., Yoon, S. H., Leelahavanichkul, A., & Nita-Lazar, A. (2021). LPS Tolerance Inhibits Cellular Respiration and Induces Global Changes in the Macrophage Secretome. Biomolecules, 11(2), 164. https://doi.org/10.3390/biom11020164







