Different Aggregation Pathways and Structures for Aβ40 and Aβ42 Peptides
Abstract
:1. Introduction
2. Methods and Materials
2.1. Materials
2.2. Solution Preparation
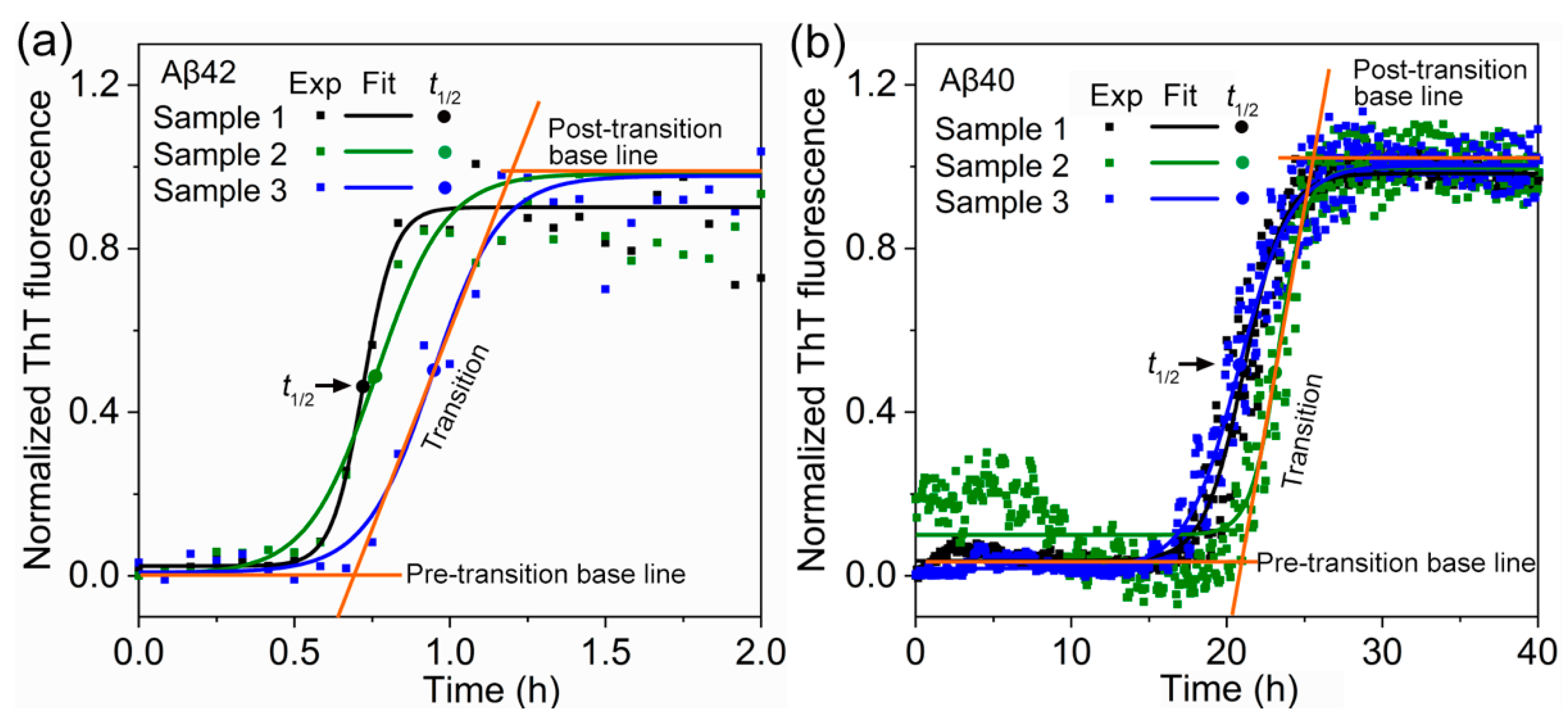
2.3. Thioflavin-T Fluorescence Assay
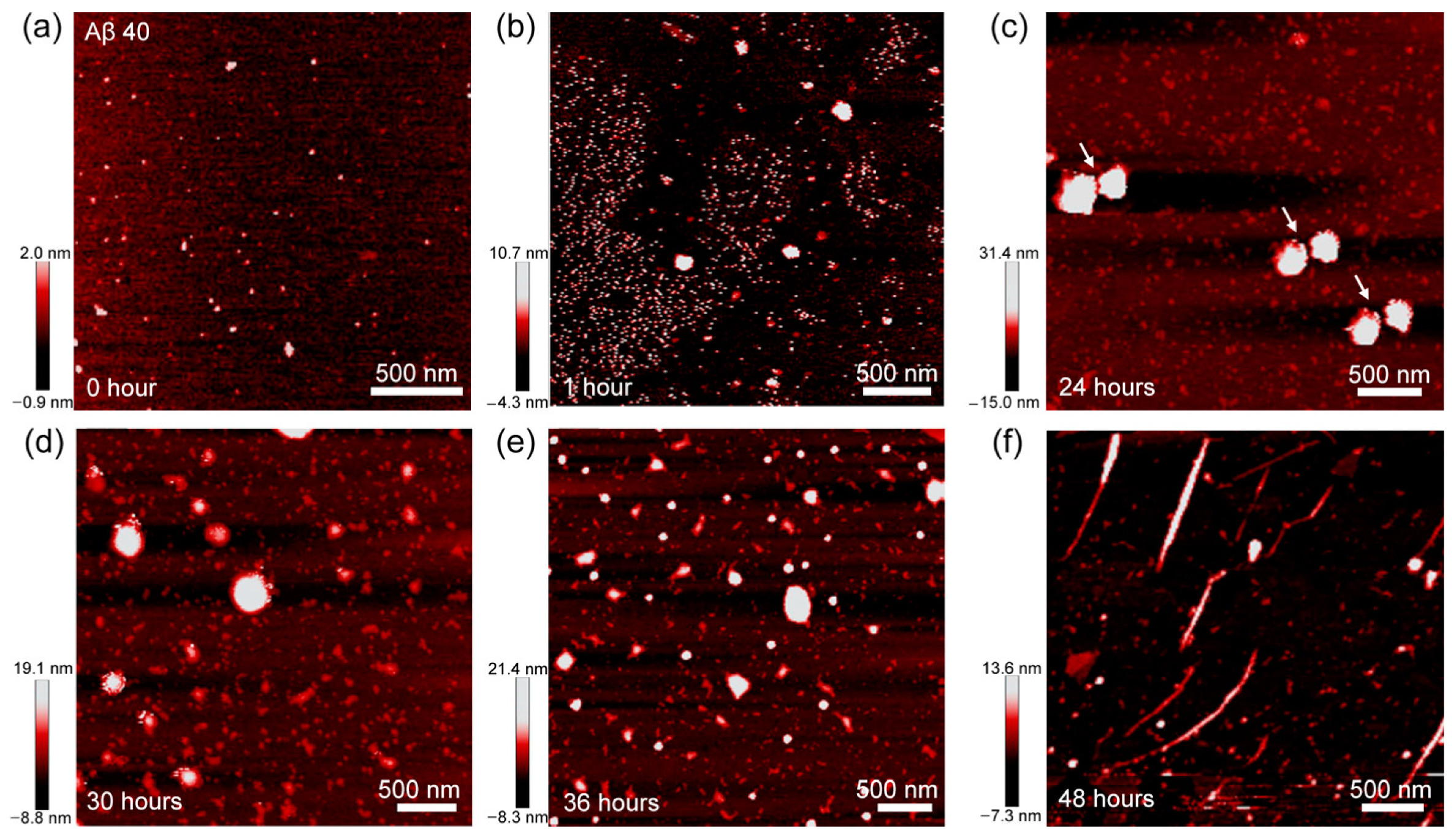
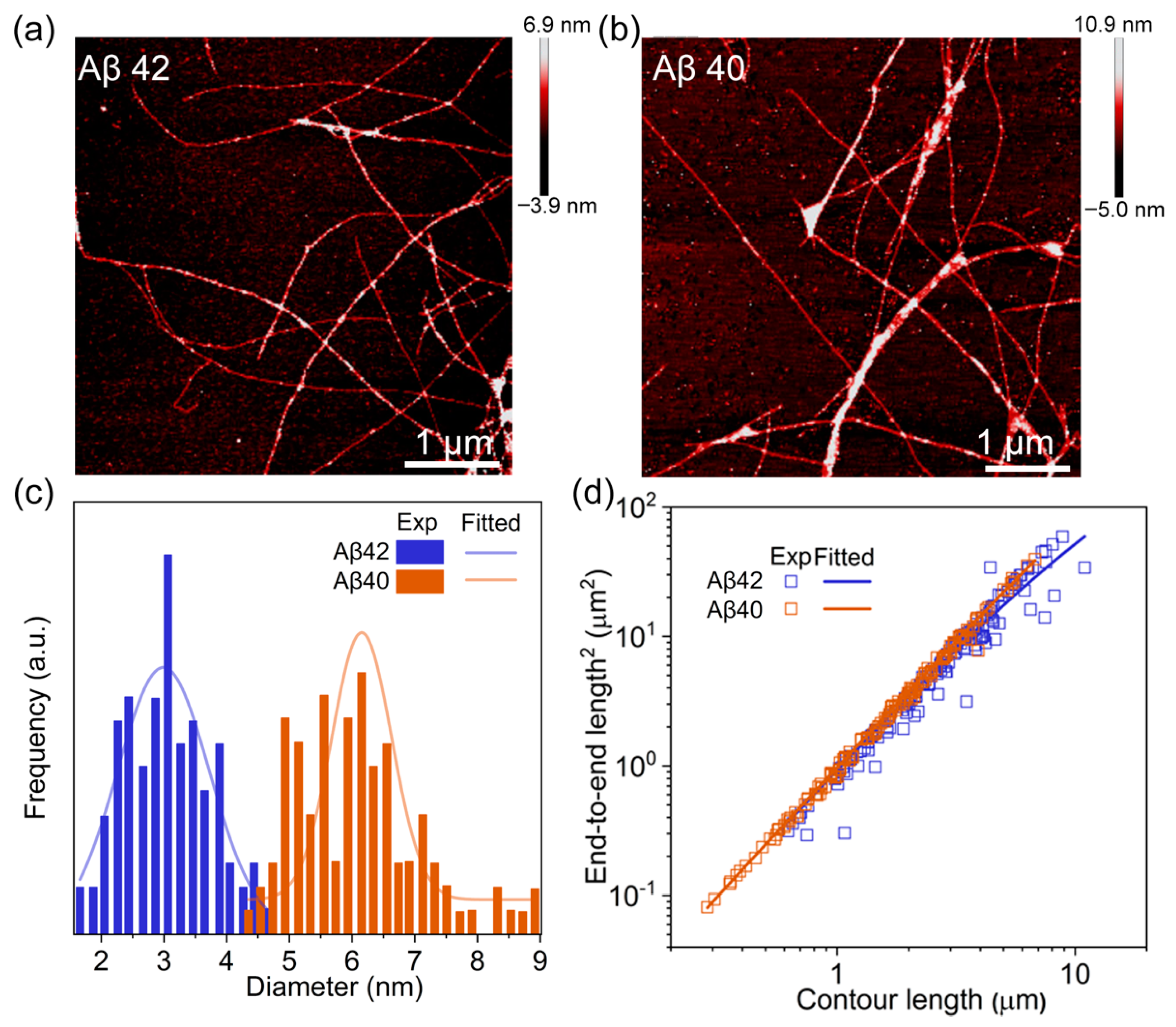
2.4. Atomic Force Microscopy Imaging
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Merlini, G.; Bellotti, V. Molecular mechanisms of amyloidosis. N. Engl. J. Med. 2003, 349, 583–596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reiss, A.B.; Arain, H.A.; Stecker, M.M.; Siegart, N.M.; Kasselman, L.J. Amyloid toxicity in Alzheimer’s disease. Rev. Neurosci. 2018, 29, 613–627. [Google Scholar] [CrossRef] [PubMed]
- Hardy, J. The amyloid hypothesis of Alzheimer’s disease: Progress and problems on the road to therapeutics. Science 2002, 297, 353–356. [Google Scholar] [CrossRef] [Green Version]
- Härd, T.; Lendel, C. Inhibition of amyloid formation. J. Mol. Biol. 2012, 421, 441–465. [Google Scholar] [CrossRef] [PubMed]
- Soto, C. Unfolding the role of protein misfolding in neurodegenerative diseases. Nat. Rev. Neurosci. 2003, 4, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Keten, S.; Xu, Z.; Ihle, B.; Buehler, M.J. Nanoconfinement controls stiffness, strength and mechanical toughness of β-sheet crystals in silk. Nat. Mater. 2010, 9, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Zhao, M.; Jiang, L.; Cheng, P.N.; Park, J.; Sawaya, M.R.; Pensalfini, A.; Gou, D.; Berk, A.J.; Glabe, C.G.; et al. Toxic fibrillar oligomers of amyloid-β have cross-β structure. Proc. Natl. Acad. Sci. USA 2012, 109, 20913–20918. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laganowsky, A.; Liu, C.; Sawaya, M.R.; Whitelegge, J.P.; Park, J.; Zhao, M.; Pensalfini, A.; Soriaga, A.B.; Landau, M.; Teng, P.K.; et al. Atomic view of a toxic amyloid small oligomer. Science 2012, 335, 1228–1231. [Google Scholar] [CrossRef] [Green Version]
- Gardner, R.C.; Yaffe, K. Dementia risk after traumatic brain injury vs nonbrain trauma: The role of age and severity. Ann. Neurol. 2014, 75, 339–341. [Google Scholar] [CrossRef] [Green Version]
- Zhu, X.C.; Tan, L.; Wang, H.F.; Jiang, T.; Cao, L.; Wang, C.; Wang, J.; Tan, C.C.; Meng, X.F.; Yu, J.T. Rate of early onset Alzheimer’s disease: A systematic review and meta-analysis. Ann. Transl. Med. 2015, 3, 38. [Google Scholar]
- Mori, H.; Takio, K.; Ogawara, M.; Selkoe, D.J. Mass spectrometry of purified amyloid beta protein in Alzheimer’s disease. J. Biol. Chem. 1992, 267, 17082–17086. [Google Scholar] [CrossRef]
- Bernstein, S.L.; Dupuis, N.F.; Lazo, N.D.; Wyttenbach, T.; Condron, M.M.; Bitan, G.; Teplow, D.B.; Shea, J.E.; Ruotolo, B.T.; Robinson, C.V.; et al. Amyloid-β protein oligomerization and the importance of tetramers and dodecamers in the aetiology of Alzheimer’s disease. Nat. Chem. 2009, 1, 326–331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sánchez, L.; Madurga, S.; Pukala, T.; Vilaseca, M.; López-Iglesias, C.; Robinson, C.V.; Giralt, E.; Carulla, N. Aβ40 and Aβ42 amyloid fibrils exhibit distinct molecular recycling properties. J. Am. Chem. Soc. 2011, 133, 6505–6508. [Google Scholar] [CrossRef] [PubMed]
- Meisl, G.; Yang, X.; Hellstrand, E.; Frohm, B.; Kirkegaard, J.B.; Cohen, S.I.A.; Dobson, C.M.; Linse, S.; Knowles, T.P.J. Differences in nucleation behavior underlie the contrasting aggregation kinetics of the Aβ40 and Aβ42 peptides. Proc. Natl. Acad. Sci. USA 2014, 111, 9384–9389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nirmalraj, P.N.; List, J.; Battacharya, S.; Howe, G.; Xu, L.; Thompson, D.; Mayer, M. Complete aggregation pathway of amyloid β (1-40) and (1-42) resolved on an atomically clean interface. Sci. Adv. 2020, 6, eaaz6014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harper, J.D.; Wong, S.S.; Lieber, C.M.; Lansbury, P.T. Observation of metastable Aβ amyloid protofibrils by atomic force microscopy. Chem. Biol. 1997, 4, 119–125. [Google Scholar] [CrossRef] [Green Version]
- Lee, G.; Lee, W.; Lee, H.; Lee, S.W.; Yoon, D.S.; Eom, K.; Kwon, T. Mapping the surface charge distribution of amyloid fibril. Appl. Phys. Lett. 2012, 101, 043703. [Google Scholar] [CrossRef]
- Lee, G.; Lee, W.; Lee, H.; Lee, C.Y.; Eom, K.; Kwon, T. Self-assembled amyloid fibrils with controllable conformational heterogeneity. Sci. Rep. 2015, 5, 16220. [Google Scholar] [CrossRef] [Green Version]
- Lee, G.; Lee, W.; Baik, S.; Kim, Y.H.; Eom, K.; Kwon, T. Correlation between hierarchical structures and nanomechanical properties of amyloid fibrils. Nanotechnology 2018, 29, 295701. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH image to ImageJ: 25 years of image analysis. Nat. Meth. 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Rivetti, C.; Guthold, M.; Bustamante, C. Scanning force microscopy of DNA deposited onto mica: EquilibrationversusKinetic trapping studied by statistical polymer chain analysis. J. Mol. Biol. 1996, 264, 919–932. [Google Scholar] [CrossRef] [PubMed]
- Arosio, P.; Knowles, T.P.J.; Linse, S. On the lag phase in amyloid fibril formation. Phys. Chem. Chem. Phys. 2015, 17, 7606–7618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Latshaw, D.C.; Cheon, M.; Hall, C.K. Effects of macromolecular crowding on amyloid beta (16–22) aggregation using coarse-grained simulations. J. Phys. Chem. B 2014, 118, 13513–13526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Economou, N.J.; Giammona, M.J.; Do, T.D.; Zheng, X.; Teplow, D.B.; Buratto, S.K.; Bowers, M.T. Amyloid β-protein assembly and Alzheimer’s disease: Dodecamers of Aβ42, but not of Aβ40, seed fibril formation. J. Am. Chem. Soc. 2016, 138, 1772–1775. [Google Scholar] [CrossRef] [PubMed] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.; Eom, K.; Kwon, T. Different Aggregation Pathways and Structures for Aβ40 and Aβ42 Peptides. Biomolecules 2021, 11, 198. https://doi.org/10.3390/biom11020198
Wang L, Eom K, Kwon T. Different Aggregation Pathways and Structures for Aβ40 and Aβ42 Peptides. Biomolecules. 2021; 11(2):198. https://doi.org/10.3390/biom11020198
Chicago/Turabian StyleWang, Li, Kilho Eom, and Taeyun Kwon. 2021. "Different Aggregation Pathways and Structures for Aβ40 and Aβ42 Peptides" Biomolecules 11, no. 2: 198. https://doi.org/10.3390/biom11020198





